N-Acetylaspartate Drives Oligodendroglial Differentiation via Histone Deacetylase Activation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Media
2.2. Compound Treatments
2.3. Quantitative Immunofluorescence Microscopy Analysis
2.4. Evaluation of Cell Engagement in PS Microfibers
2.5. Total RNA Extraction and qPCR
2.6. Crude Extract Preparation and Immunoblot Analysis
2.7. Statistical Methods
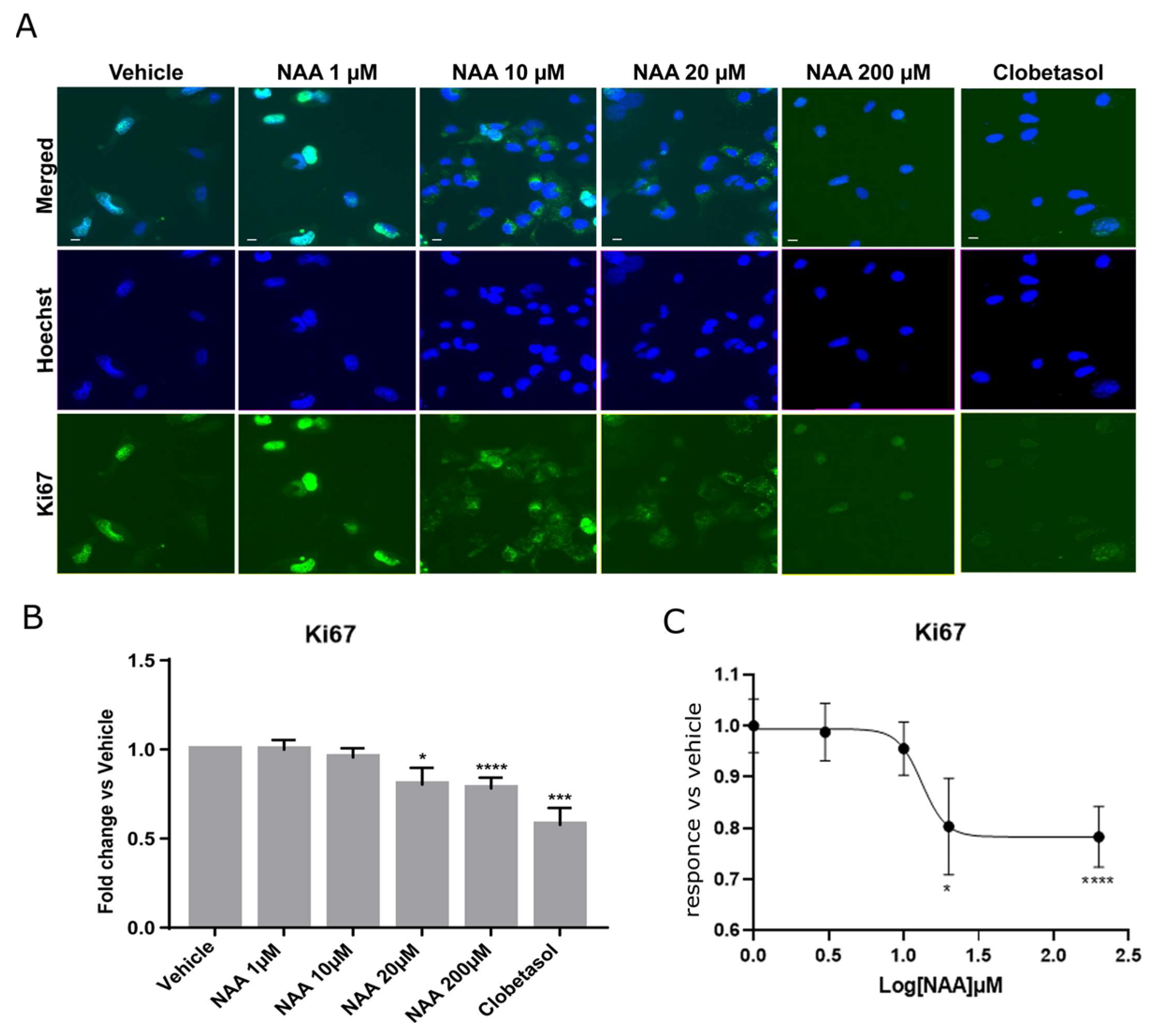
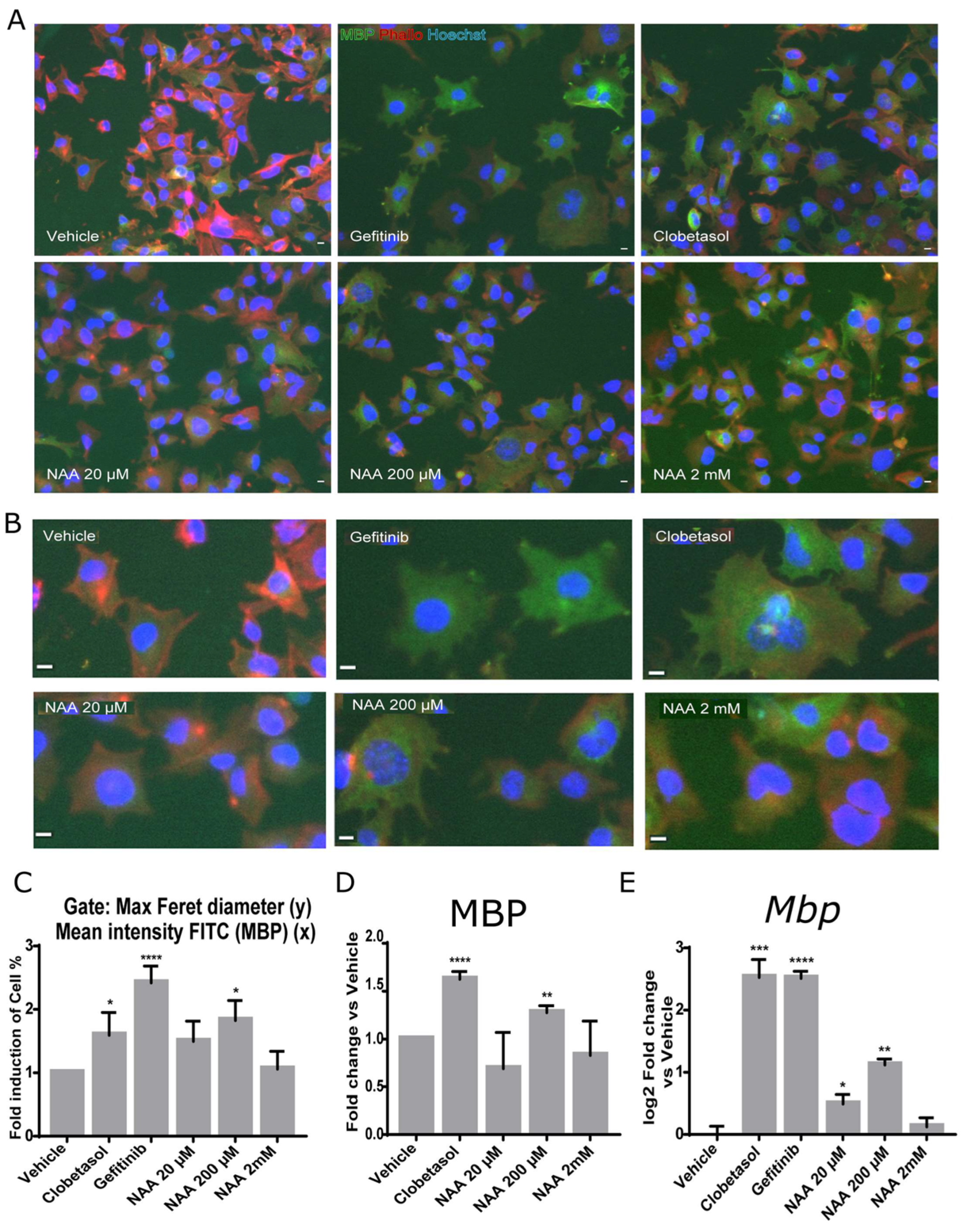
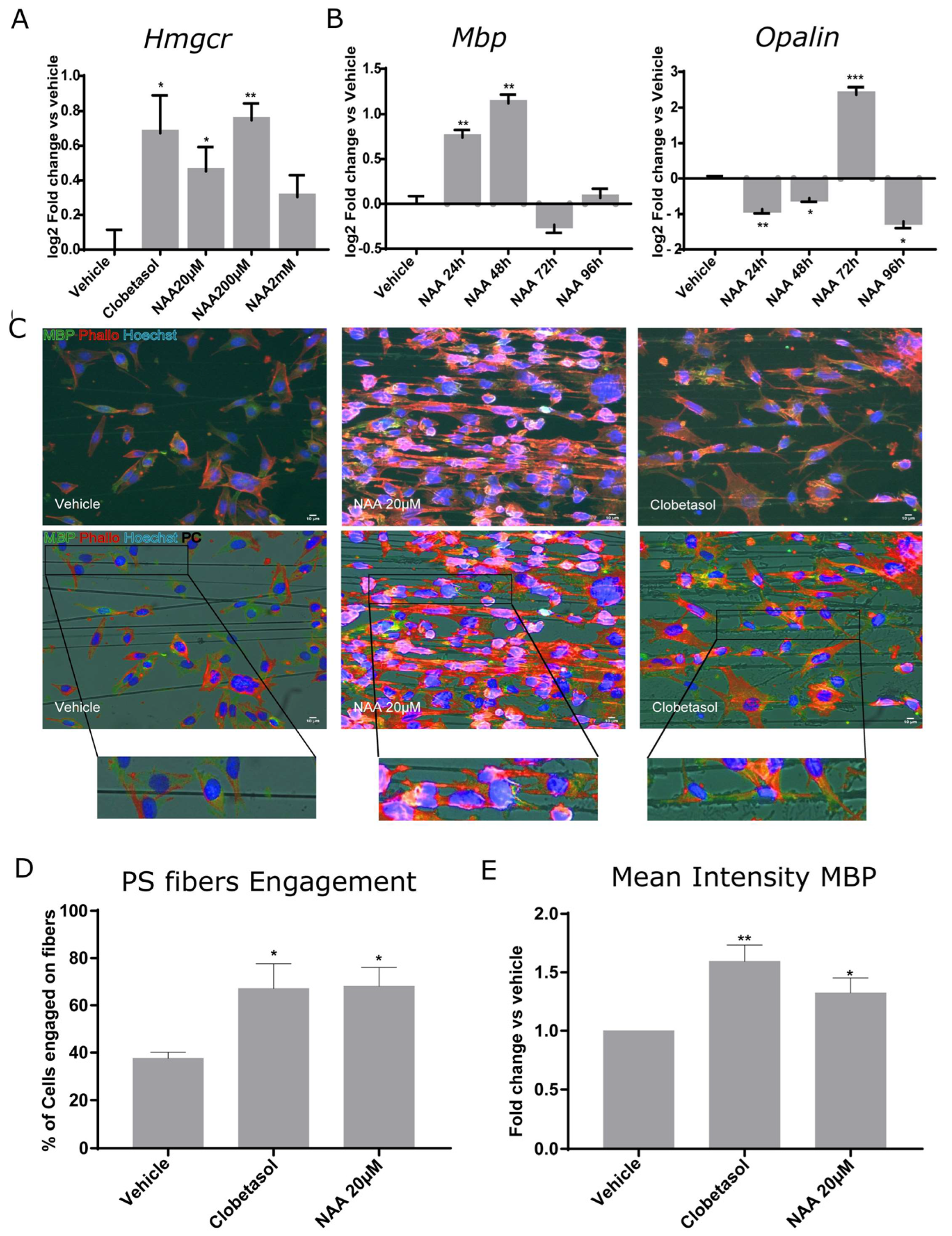
3. Results
3.1. Oligodendroglia Differentiation and MBP Expression Are Stimulated by 200 mM NAA Treatment
3.2. NAA Treatment Promotes 3-Hydroxy-3-Methylglutaryl-CoA Reductase Increase, Temporally-Regulated Myelin Gene Expression, and Synthetic Axon Myelination
3.3. Oli-neuM Deacetylation Regulates MBP Gene Expression under NAA Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amaral, A.I.; Hadera, M.G.; Kotter, M.; Sonnewald, U. Oligodendrocytes Do Not Export NAA-Derived Aspartate In Vitro. Neurochem. Res. 2017, 42, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Urenjak, J.; Williams, S.R.; Gadian, D.G.; Noble, M. Specific Expression of N-Acetylaspartate in Neurons, Oligodendrocyte-Type-2 Astrocyte Progenitors, and Immature Oligodendrocytes In Vitro. J. Neurochem. 1992, 59, 55–61. [Google Scholar] [CrossRef]
- Urenjak, J.; Williams, S.R.; Gadian, D.G.; Noble, M. Proton Nuclear Magnetic Resonance Spectroscopy Unambiguously Identifies Different Neural Cell Types. J. Neurosci. 1993, 13, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, G.; Mekala, P.; Yahya, D.; Wu, G.; Ledeen, R.W. Intraneuronal N-Acetylaspartate Supplies Acetyl Groups for Myelin Lipid Synthesis: Evidence for Myelin-Associated Aspartoacylase. J. Neurochem. 2001, 78, 736–745. [Google Scholar] [CrossRef]
- Moffett, J.R.; Arun, P.; Ariyannur, P.S.; Namboodiri, A.M.A. N-Acetylaspartate Reductions in Brain Injury: Impact on Post-Injury Neuroenergetics, Lipid Synthesis, and Protein Acetylation. Front. Neuroenergetics 2013, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Rigotti, D.J.; Kirov, I.I.; Djavadi, B.; Perry, N.; Babb, J.S.; Gonen, O. Longitudinal Whole-Brain N -Acetylaspartate Concentration in Healthy Adults. AJNR Am. J. Neuroradiol. 2011, 32, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- Shannon, R.J.; van der Heide, S.; Carter, E.L.; Jalloh, I.; Menon, D.K.; Hutchinson, P.J.; Carpenter, K.L.H. Extracellular N -Acetylaspartate in Human Traumatic Brain Injury. J. Neurotrauma 2016, 33, 319–329. [Google Scholar] [CrossRef]
- Aboul-Enein, F.; Krššák, M.; Höftberger, R.; Prayer, D.; Kristoferitsch, W. Reduced NAA-Levels in the NAWM of Patients with MS Is a Feature of Progression. A Study with Quantitative Magnetic Resonance Spectroscopy at 3 Tesla. PLoS ONE 2010, 5, e11625. [Google Scholar] [CrossRef]
- Traiffort, E.; Angot, E.; Ruat, M. Sonic Hedgehog Signaling in the Mammalian Brain. J. Neurochem. 2010, 113, 576–590. [Google Scholar] [CrossRef]
- Allanach, J.R.; Farrell, J.W.; Mésidor, M.; Karimi-Abdolrezaee, S. Current Status of Neuroprotective and Neuroregenerative Strategies in Multiple Sclerosis: A Systematic Review. Mult. Scler. 2022, 28, 29–48. [Google Scholar] [CrossRef]
- Watanabe, M.; Toyama, Y.; Nishiyama, A. Differentiation of Proliferated NG2-Positive Glial Progenitor Cells in a Remyelinating Lesion. J. Neurosci. Res. 2002, 69, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Duncan, I.D.; Marik, R.L.; Broman, A.T.; Heidari, M. Thin Myelin Sheaths as the Hallmark of Remyelination Persist over Time and Preserve Axon Function. Proc. Natl. Acad. Sci. USA 2017, 114, E9685–E9691. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.L.; Röth, P.T.; Stratton, J.A.S.; Chuang, B.H.A.; Danne, J.; Ellis, S.L.; Ng, S.W.; Kilpatrick, T.J.; Merson, T.D. Adult Neural Precursor Cells from the Subventricular Zone Contribute Significantly to Oligodendrocyte Regeneration and Remyelination. J. Neurosci. 2014, 34, 14128–14146. [Google Scholar] [CrossRef]
- Duncan, I.D.; Radcliff, A.B.; Heidari, M.; Kidd, G.; August, B.K.; Wierenga, L.A. The Adult Oligodendrocyte Can Participate in Remyelination. Proc. Natl. Acad. Sci. USA 2018, 115, E11807–E11816. [Google Scholar] [CrossRef]
- Butt, A.M.; Rivera, A.D.; Fulton, D.; Azim, K. Targeting the Subventricular Zone to Promote Myelin Repair in the Aging Brain. Cells 2022, 11, 1809. [Google Scholar] [CrossRef]
- Hughes, J.; Rees, S.; Kalindjian, S.; Philpott, K. Principles of Early Drug Discovery: Principles of Early Drug Discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Serwanski, D.R.; Rasmussen, A.L.; Brunquell, C.B.; Perkins, S.S.; Nishiyama, A. Sequential Contribution of Parenchymal and Neural Stem Cell-Derived Oligodendrocyte Precursor Cells toward Remyelination. Neuroglia 2018, 1, 91–105. [Google Scholar] [CrossRef]
- Moyon, S.; Holloman, M.; Salzer, J.L. Neural Stem Cells and Oligodendrocyte Progenitor Cells Compete for Remyelination in the Corpus Callosum. Front. Cell. Neurosci. 2023, 17, 1114781. [Google Scholar] [CrossRef]
- Remaud, S.; Ortiz, F.C.; Perret-Jeanneret, M.; Aigrot, M.-S.; Gothié, J.-D.; Fekete, C.; Kvárta-Papp, Z.; Gereben, B.; Langui, D.; Lubetzki, C.; et al. Transient Hypothyroidism Favors Oligodendrocyte Generation Providing Functional Remyelination in the Adult Mouse Brain. eLife 2017, 6, e29996. [Google Scholar] [CrossRef]
- Balestri, S.; Del Giovane, A.; Sposato, C.; Ferrarelli, M.; Ragnini-Wilson, A. The Current Challenges for Drug Discovery in CNS Remyelination. Int. J. Mol. Sci. 2021, 22, 2891. [Google Scholar] [CrossRef]
- Bebo, B.F.; Allegretta, M.; Landsman, D.; Zackowski, K.M.; Brabazon, F.; Kostich, W.A.; Coetzee, T.; Ng, A.V.; Marrie, R.A.; Monk, K.R.; et al. Pathways to Cures for Multiple Sclerosis: A Research Roadmap. Mult. Scler. 2022, 28, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Cunniffe, N.; Coles, A. Promoting Remyelination in Multiple Sclerosis. J. Neurol. 2021, 268, 30. [Google Scholar] [CrossRef]
- Beyer, B.A.; Fang, M.; Sadrian, B.; Montenegro-Burke, J.R.; Plaisted, W.C.; Kok, B.P.C.; Saez, E.; Kondo, T.; Siuzdak, G.; Lairson, L.L. Metabolomics-Based Discovery of a Metabolite That Enhances Oligodendrocyte Maturation. Nat. Chem. Biol. 2018, 14, 22–28. [Google Scholar] [CrossRef]
- Singhal, N.K.; Huang, H.; Li, S.; Clements, R.; Gadd, J.; Daniels, A.; Kooijman, E.E.; Bannerman, P.; Burns, T.; Guo, F.; et al. The Neuronal Metabolite NAA Regulates Histone H3 Methylation in Oligodendrocytes and Myelin Lipid Composition. Exp. Brain Res. 2017, 235, 279–292. [Google Scholar] [CrossRef]
- Wijayasinghe, Y.S.; Pavlovsky, A.G.; Viola, R.E. Aspartoacylase Catalytic Deficiency as the Cause of Canavan Disease: A Structural Perspective. Biochemistry 2014, 53, 4970–4978. [Google Scholar] [CrossRef] [PubMed]
- von Jonquieres, G.; Spencer, Z.H.T.; Rowlands, B.D.; Klugmann, C.B.; Bongers, A.; Harasta, A.E.; Parley, K.E.; Cederholm, J.; Teahan, O.; Pickford, R.; et al. Uncoupling N-Acetylaspartate from Brain Pathology: Implications for Canavan Disease Gene Therapy. Acta Neuropathol. 2018, 135, 95–113. [Google Scholar] [CrossRef] [PubMed]
- De Vries, G.H.; Boullerne, A.I. Glial Cell Lines: An Overview. Neurochem. Res. 2010, 35, 1978–2000. [Google Scholar] [CrossRef]
- Del Giovane, A.; Russo, M.; Tirou, L.; Faure, H.; Ruat, M.; Balestri, S.; Sposato, C.; Basoli, F.; Rainer, A.; Kassoussi, A.; et al. Smoothened/AMP-Activated Protein Kinase Signaling in Oligodendroglial Cell Maturation. Front. Cell. Neurosci. 2022, 15, 801704. [Google Scholar] [CrossRef]
- Nocita, E.; Del Giovane, A.; Tiberi, M.; Boccuni, L.; Fiorelli, D.; Sposato, C.; Romano, E.; Basoli, F.; Trombetta, M.; Rainer, A.; et al. EGFR/ErbB Inhibition Promotes OPC Maturation up to Axon Engagement by Co-Regulating PIP2 and MBP. Cells 2019, 8, 844. [Google Scholar] [CrossRef]
- Porcu, G.; Serone, E.; De Nardis, V.; Di Giandomenico, D.; Lucisano, G.; Scardapane, M.; Poma, A.; Ragnini-Wilson, A. Clobetasol and Halcinonide Act as Smoothened Agonists to Promote Myelin Gene Expression and RxRγ Receptor Activation. PLoS ONE 2015, 10, e0144550. [Google Scholar] [CrossRef]
- Jochems, J.; Boulden, J.; Lee, B.G.; Blendy, J.A.; Jarpe, M.; Mazitschek, R.; Van Duzer, J.H.; Jones, S.; Berton, O. Antidepressant-Like Properties of Novel HDAC6-Selective Inhibitors with Improved Brain Bioavailability. Neuropsychopharmacology 2014, 39, 389–400. [Google Scholar] [CrossRef] [PubMed]
- White, R.; Krämer-Albers, E.-M. Axon-Glia Interaction and Membrane Traffic in Myelin Formation. Front. Cell. Neurosci. 2014, 7, 284. [Google Scholar] [CrossRef] [PubMed]
- Zuchero, J.B.; Fu, M.; Sloan, S.A.; Ibrahim, A.; Olson, A.; Zaremba, A.; Dugas, J.C.; Wienbar, S.; Caprariello, A.V.; Kantor, C.; et al. CNS Myelin Wrapping Is Driven by Actin Disassembly. Dev. Cell 2015, 34, 152–167. [Google Scholar] [CrossRef]
- Bijlard, M.; Klunder, B.; de Jonge, J.C.; Nomden, A.; Tyagi, S.; de Vries, H.; Hoekstra, D.; Baron, W. Transcriptional Expression of Myelin Basic Protein in Oligodendrocytes Depends on Functional Syntaxin 4: A Potential Correlation with Autocrine Signaling. Mol. Cell. Biol. 2015, 35, 675–687. [Google Scholar] [CrossRef]
- Gonen, O.; Viswanathan, A.K.; Catalaa, I.; Babb, J.; Udupa, J.; Grossman, R.I. Total Brain N-Acetylaspartate Concentration in Normal, Age-Grouped Females: Quantitation with Non-Echo Proton NMR Spectroscopy. Magn. Reson. Med. 1998, 40, 684–689. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qi, X.; Faulkner, R.A.; Schumacher, M.M.; Donnelly, L.M.; DeBose-Boyd, R.A.; Li, X. Regulated Degradation of HMG CoA Reductase Requires Conformational Changes in Sterol-Sensing Domain. Nat. Commun. 2022, 13, 4273. [Google Scholar] [CrossRef]
- Gregath, A.; Lu, Q.R. Epigenetic Modifications—Insight into Oligodendrocyte Lineage Progression, Regeneration, and Disease. FEBS Lett. 2018, 592, 1063–1078. [Google Scholar] [CrossRef]
- Golan, N.; Adamsky, K.; Kartvelishvily, E.; Brockschnieder, D.; Möbius, W.; Spiegel, I.; Roth, A.D.; Thomson, C.E.; Rechavi, G.; Peles, E. Identification of Tmem10/Opalin as an Oligodendrocyte Enriched Gene Using Expression Profiling Combined with Genetic Cell Ablation: Tmem10/Opalin in Oligodendrocytes. Glia 2008, 56, 1176–1186. [Google Scholar] [CrossRef]
- Lee, S.; Leach, M.K.; Redmond, S.A.; Chong, S.Y.C.; Mellon, S.H.; Tuck, S.J.; Feng, Z.-Q.; Corey, J.M.; Chan, J.R. A Culture System to Study Oligodendrocyte Myelination Processes Using Engineered Nanofibers. Nat. Methods 2012, 9, 917–922. [Google Scholar] [CrossRef]
- Hubler, Z.; Allimuthu, D.; Bederman, I.; Elitt, M.S.; Madhavan, M.; Allan, K.C.; Shick, H.E.; Garrison, E.; Karl, M.T.; Factor, D.C.; et al. Accumulation of 8,9-Unsaturated Sterols Drives Oligodendrocyte Formation and Remyelination. Nature 2018, 560, 372–376. [Google Scholar] [CrossRef]
- Sax, J.L.; Hershman, S.N.; Hubler, Z.; Allimuthu, D.; Elitt, M.S.; Bederman, I.; Adams, D.J. Enhancers of Human and Rodent Oligodendrocyte Formation Predominantly Induce Cholesterol Precursor Accumulation. ACS Chem. Biol. 2022, 17, 2188–2200. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Clements, R.; Sulak, M.; Gregory, R.; Freeman, E.; McDonough, J. Decreased NAA in Gray Matter Is Correlated with Decreased Availability of Acetate in White Matter in Postmortem Multiple Sclerosis Cortex. Neurochem. Res. 2013, 38, 2385–2396. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Cueto, M.A.; Asselbergs, F.; Atadja, P. Cloning and Functional Characterization of HDAC11, a Novel Member of the Human Histone Deacetylase Family. J. Biol. Chem. 2002, 277, 25748–25755. [Google Scholar] [CrossRef] [PubMed]
- Hisahara, S.; Iwahara, N.; Matsushita, T.; Suzuki, S.; Matsumura, A.; Fujikura, M.; Yokokawa, K.; Saito, T.; Manabe, T.; Kawamata, J.; et al. SIRT1 Decelerates Morphological Processing of Oligodendrocyte Cell Lines and Regulates the Expression of Cytoskeleton-Related Oligodendrocyte Proteins. Biochem. Biophys. Res. Commun. 2021, 546, 7–14. [Google Scholar] [CrossRef]
- Jablonska, B.; Adams, K.L.; Kratimenos, P.; Li, Z.; Strickland, E.; Haydar, T.F.; Kusch, K.; Nave, K.-A.; Gallo, V. Sirt2 Promotes White Matter Oligodendrogenesis during Development and in Models of Neonatal Hypoxia. Nat. Commun. 2022, 13, 4771. [Google Scholar] [CrossRef]
- Liu, H.; Hu, Q.; D’ercole, A.J.; Ye, P. Histone Deacetylase 11 Regulates Oligodendrocyte-Specific Gene Expression and Cell Development in OL-1 Oligodendroglia Cells. Glia 2009, 57, 1–12. [Google Scholar] [CrossRef]
- LoPresti, P. The Selective HDAC6 Inhibitor ACY-738 Impacts Memory and Disease Regulation in an Animal Model of Multiple Sclerosis. Front. Neurol. 2019, 10, 519. [Google Scholar] [CrossRef]
- Hughes, E.G.; Stockton, M.E. Premyelinating Oligodendrocytes: Mechanisms Underlying Cell Survival and Integration. Front. Cell Dev. Biol. 2021, 9, 714169. [Google Scholar] [CrossRef]
- Zveik, O.; Fainstein, N.; Rechtman, A.; Haham, N.; Ganz, T.; Lavon, I.; Brill, L.; Vaknin-Dembinsky, A. Cerebrospinal Fluid of Progressive Multiple Sclerosis Patients Reduces Differentiation and Immune Functions of Oligodendrocyte Progenitor Cells. Glia 2022, 70, 1191–1209. [Google Scholar] [CrossRef]
- Shen, S.; Li, J.; Casaccia-Bonnefil, P. Histone Modifications Affect Timing of Oligodendrocyte Progenitor Differentiation in the Developing Rat Brain. J. Cell Biol. 2005, 169, 577–589. [Google Scholar] [CrossRef]
- Canettieri, G.; Di Marcotullio, L.; Greco, A.; Coni, S.; Antonucci, L.; Infante, P.; Pietrosanti, L.; De Smaele, E.; Ferretti, E.; Miele, E.; et al. Histone Deacetylase and Cullin3–RENKCTD11 Ubiquitin Ligase Interplay Regulates Hedgehog Signalling through Gli Acetylation. Nat. Cell Biol. 2010, 12, 132–142. [Google Scholar] [CrossRef]
- Gupta, R.; Ambasta, R.K.; Kumar, P. Chapter Four—Histone Deacetylase in Neuropathology. In Advances in Clinical Chemistry; Makowski, G.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2021; Volume 104, pp. 151–231. [Google Scholar]
- Del Giovane, A.; Ragnini-Wilson, A. Targeting Smoothened as a New Frontier in the Functional Recovery of Central Nervous System Demyelinating Pathologies. Int. J. Mol. Sci. 2018, 19, 3677. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Tang, T.; Qiu, M.; Xu, X. Hedgehog Signaling in CNS Remyelination. Cells 2022, 11, 2260. [Google Scholar] [CrossRef] [PubMed]
- Jagielska, A.; Lowe, A.L.; Makhija, E.; Wroblewska, L.; Guck, J.; Franklin, R.J.M.; Shivashankar, G.V.; Van Vliet, K.J. Mechanical Strain Promotes Oligodendrocyte Differentiation by Global Changes of Gene Expression. Front. Cell. Neurosci. 2017, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Pedre, X.; Mastronardi, F.; Bruck, W.; López-Rodas, G.; Kuhlmann, T.; Casaccia, P. Changed Histone Acetylation Patterns in Normal-Appearing White Matter and Early Multiple Sclerosis Lesions. J. Neurosci. 2011, 31, 3435–3445. [Google Scholar] [CrossRef]
- Ryan, D.G.; Frezza, C.; O’Neill, L.A. TCA Cycle Signalling and the Evolution of Eukaryotes. Curr. Opin. Biotechnol. 2021, 68, 72–88. [Google Scholar] [CrossRef]

| Gene Name | Forward | Reverse |
|---|---|---|
| Gapdh | 5′-CCAATGTGTCCGTCGTGGATCT-3′ | 5′-GTTGAAGTCGCAGGAGACAACC-3′ |
| Hdac1 | 5′-ACAGCAATAGGAGGCCAGTT-3′ | 5′-TCCCTCCTTGCTTTCTCAGG-3′ |
| Hdac6 | 5′-GGAGACAACCCAGTACATGAATGAA- 3′ | 5′-CGGAGGACACGGAGGACAGAGCCTGTAG-3′ |
| Hdac11 | 5′-GGGGGATCTCAGTGATGGTA-3′ | 5′-AAGAGAAGCTGCTGTCCGAT-3′ |
| Hmgcr1 | 5′-CTTGTGGAATGCCTTGTGATTG-3′ | 5′-AGCCGAAGCAGCACATGAT-3′ |
| Mbp | 5′-TACCCTGGCTAAAGCAGAGC-3′ | 5′-GAGGTGGTGTTCGAGGTGTC-3′ |
| Opalin | 5′-CAGCTGCCTCTCACTCAACATC-3′ | 5′-TCCCAAAGGCAGACTTCTCTCG-3′ |
| Sirt1 | 5′-GTAAGCGGCTTGAGGG-3′ | 5′-TTCGGGCCTCTCCGTA-3′ |
| Sirt2 | 5′-ATCCACCGGCCTCTATGACAA-3′ | 5′-CGCATGAAGTAGTGACAGATGG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dominicis, A.; Del Giovane, A.; Torreggiani, M.; Recchia, A.D.; Ciccarone, F.; Ciriolo, M.R.; Ragnini-Wilson, A. N-Acetylaspartate Drives Oligodendroglial Differentiation via Histone Deacetylase Activation. Cells 2023, 12, 1861. https://doi.org/10.3390/cells12141861
Dominicis A, Del Giovane A, Torreggiani M, Recchia AD, Ciccarone F, Ciriolo MR, Ragnini-Wilson A. N-Acetylaspartate Drives Oligodendroglial Differentiation via Histone Deacetylase Activation. Cells. 2023; 12(14):1861. https://doi.org/10.3390/cells12141861
Chicago/Turabian StyleDominicis, Alessandra, Alice Del Giovane, Matteo Torreggiani, Antonella Damiana Recchia, Fabio Ciccarone, Maria Rosa Ciriolo, and Antonella Ragnini-Wilson. 2023. "N-Acetylaspartate Drives Oligodendroglial Differentiation via Histone Deacetylase Activation" Cells 12, no. 14: 1861. https://doi.org/10.3390/cells12141861
APA StyleDominicis, A., Del Giovane, A., Torreggiani, M., Recchia, A. D., Ciccarone, F., Ciriolo, M. R., & Ragnini-Wilson, A. (2023). N-Acetylaspartate Drives Oligodendroglial Differentiation via Histone Deacetylase Activation. Cells, 12(14), 1861. https://doi.org/10.3390/cells12141861







