The Role of circTmeff-1 in Morphine Addiction Memory of Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Drugs
2.3. Behavior Test
2.3.1. Conditioned Place Preference
2.3.2. Light–Dark Box
2.3.3. Locomotion Test
2.3.4. Barnes Maze
2.4. RNA Sequencing
2.5. Quantitative Real-Time PCR
- circTmeff-1:
- upstream primer 5′-CAGAAGGCCTCTTG TAGAATTA-3′,
- downstream primer 5′-CATTTCAAACCATCTCCATCTTC-3′;
- circDcun1d4:
- upstream primer 5′-GTGGTTTTATGAATATGCAGACTT-3′,
- downstream primer 5′-ACGTCTTCTGTCAGATTCAG-3′;
- GAPDH:
- upstream primer 5′-GCTCGTCGTCGACAACGGCTG-3′,
- downstream primer 5′-CAAACATGAT CTGGGTCATCTTTC-3′.
2.6. CircRNA Target Prediction
2.7. Functional Analysis
2.8. RNA Fluorescence In Situ Hybridization
2.9. Adeno-Associated Virus Injection
2.10. Statistics
3. Results
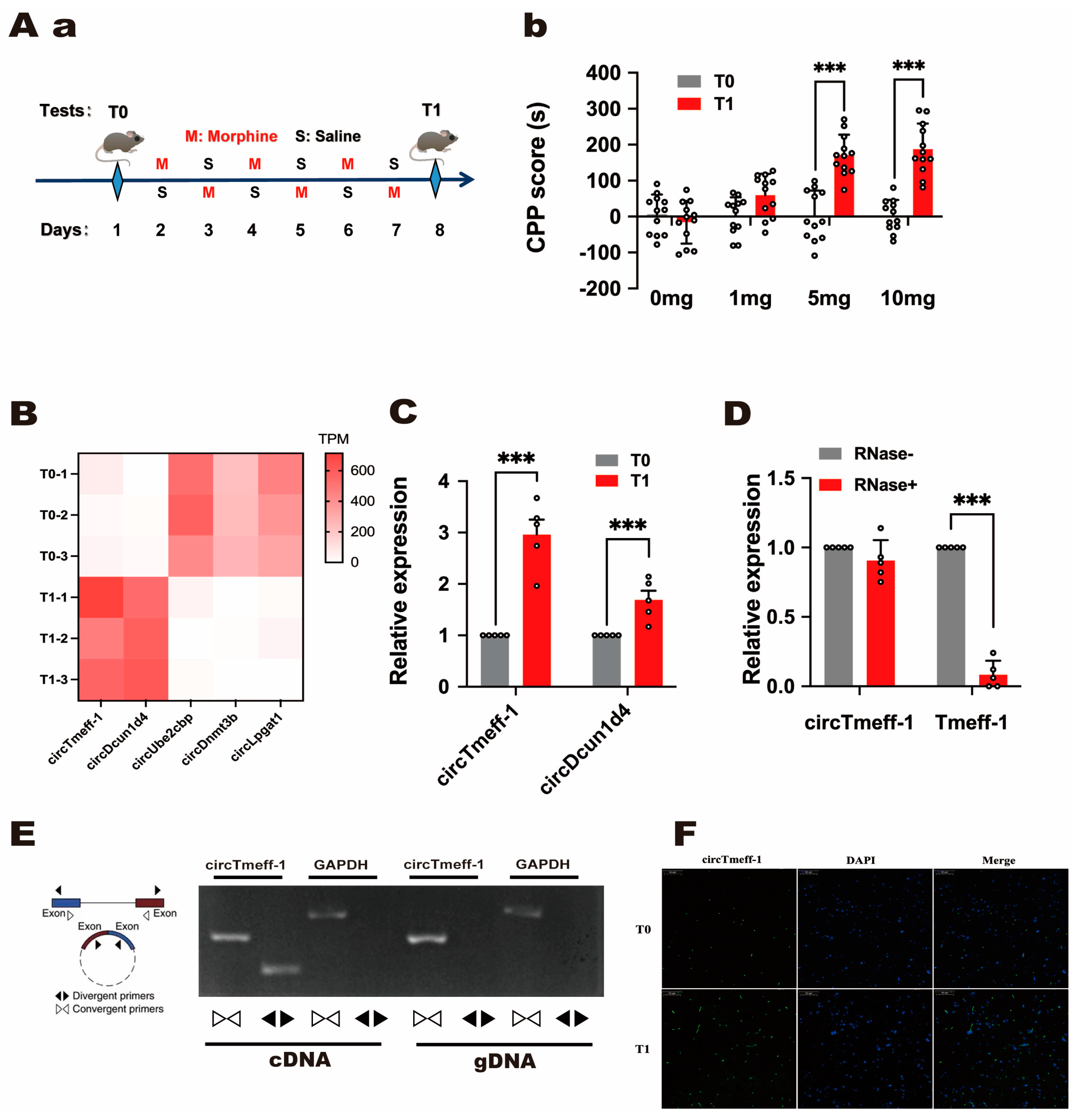
3.1. CircRNAs Expression Profile Changed in the Nucleus Accumbens of Mice with Morphine-Addicted Memory Formation
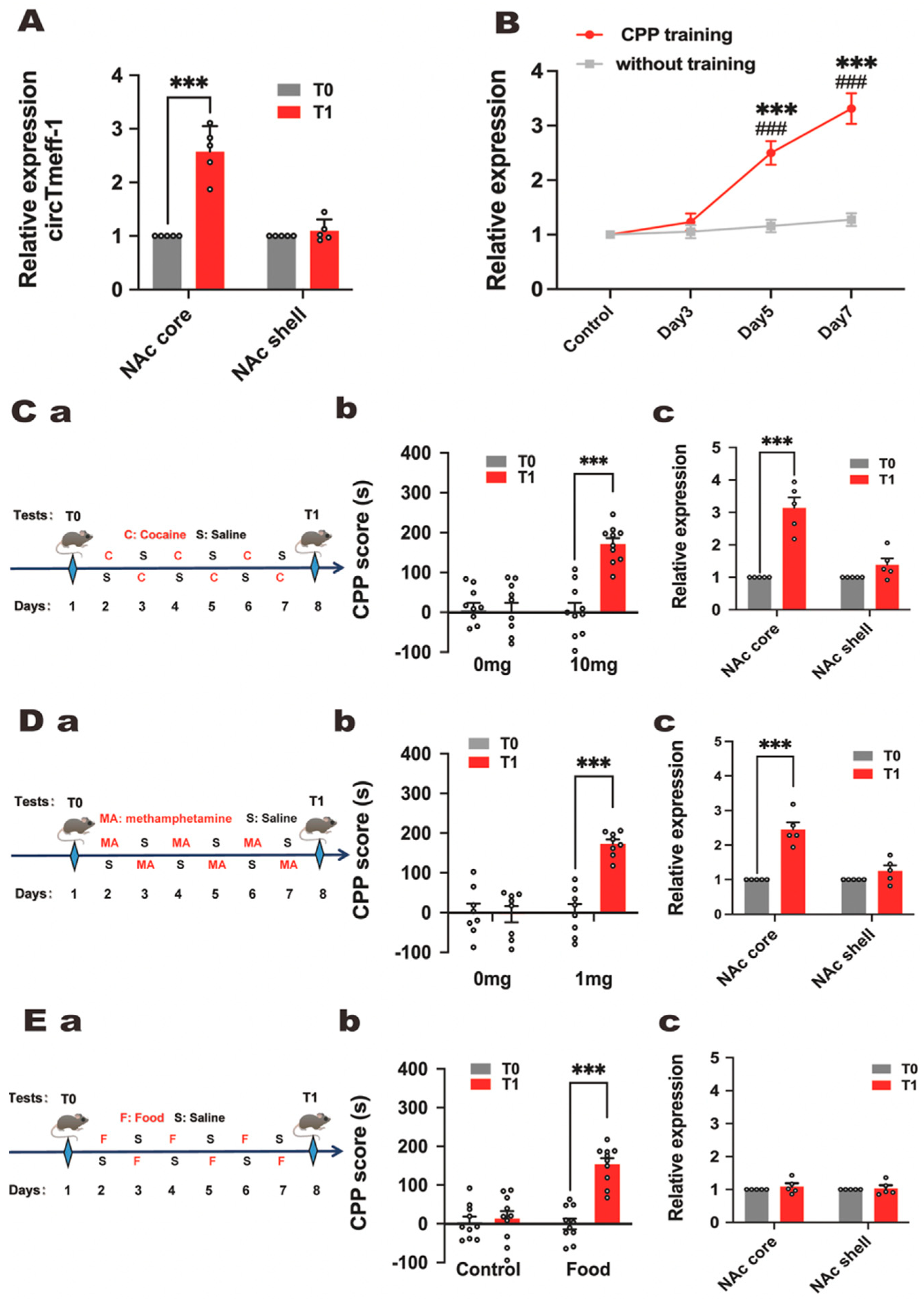
3.2. The Expression of circTmeff-1 Increased in the NAc Core in Mice with Morphine-Addicted Memory Formation
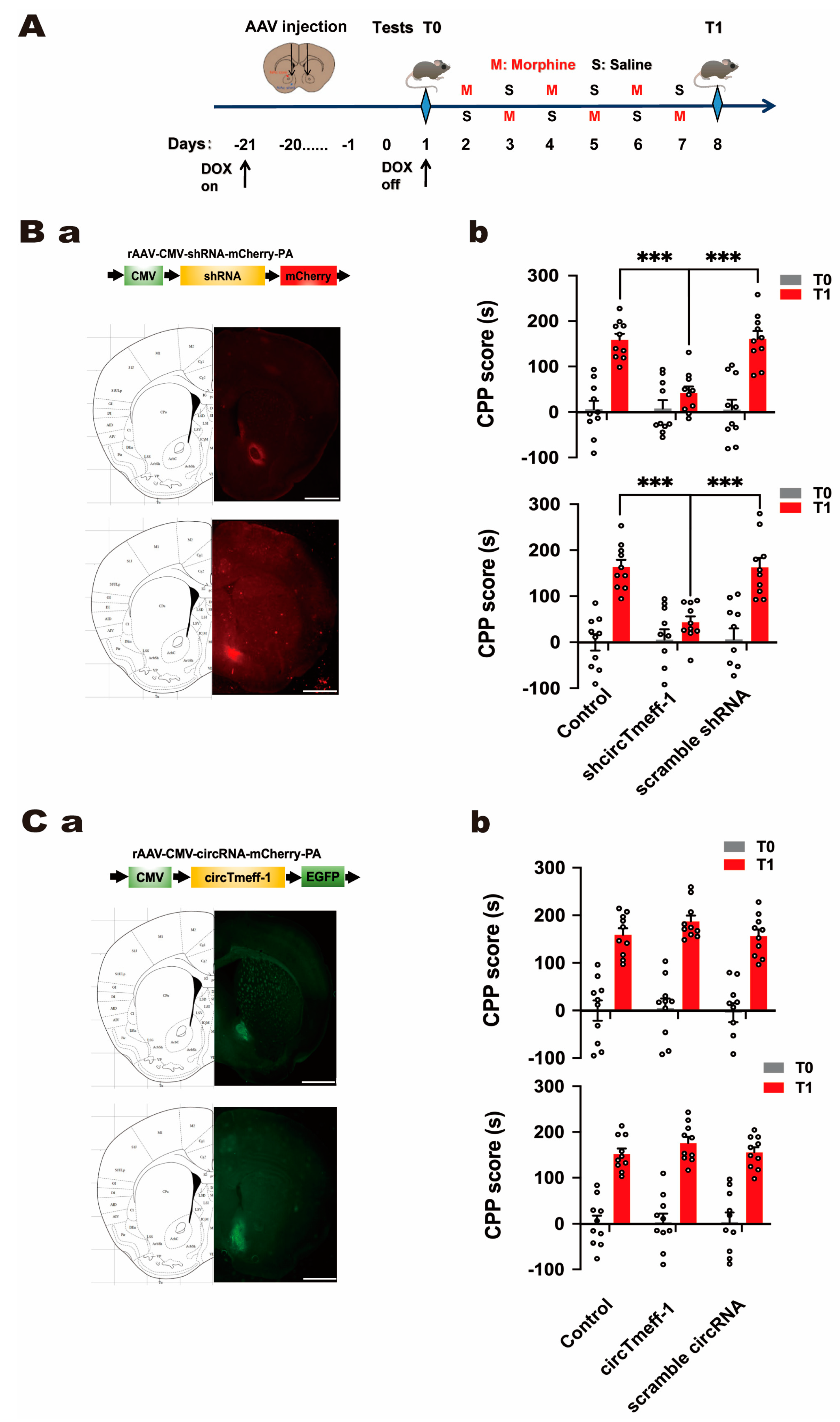
3.3. CircTmeff-1 in NAc Core and Shell Plays a Role in Morphine-Dependent Memory Formation
3.4. CircTmeff-1 in NAc Core and Shell Did Not Affect Motor Activity, Anxiety-Like Behavior, and Spatial Memory of Mice
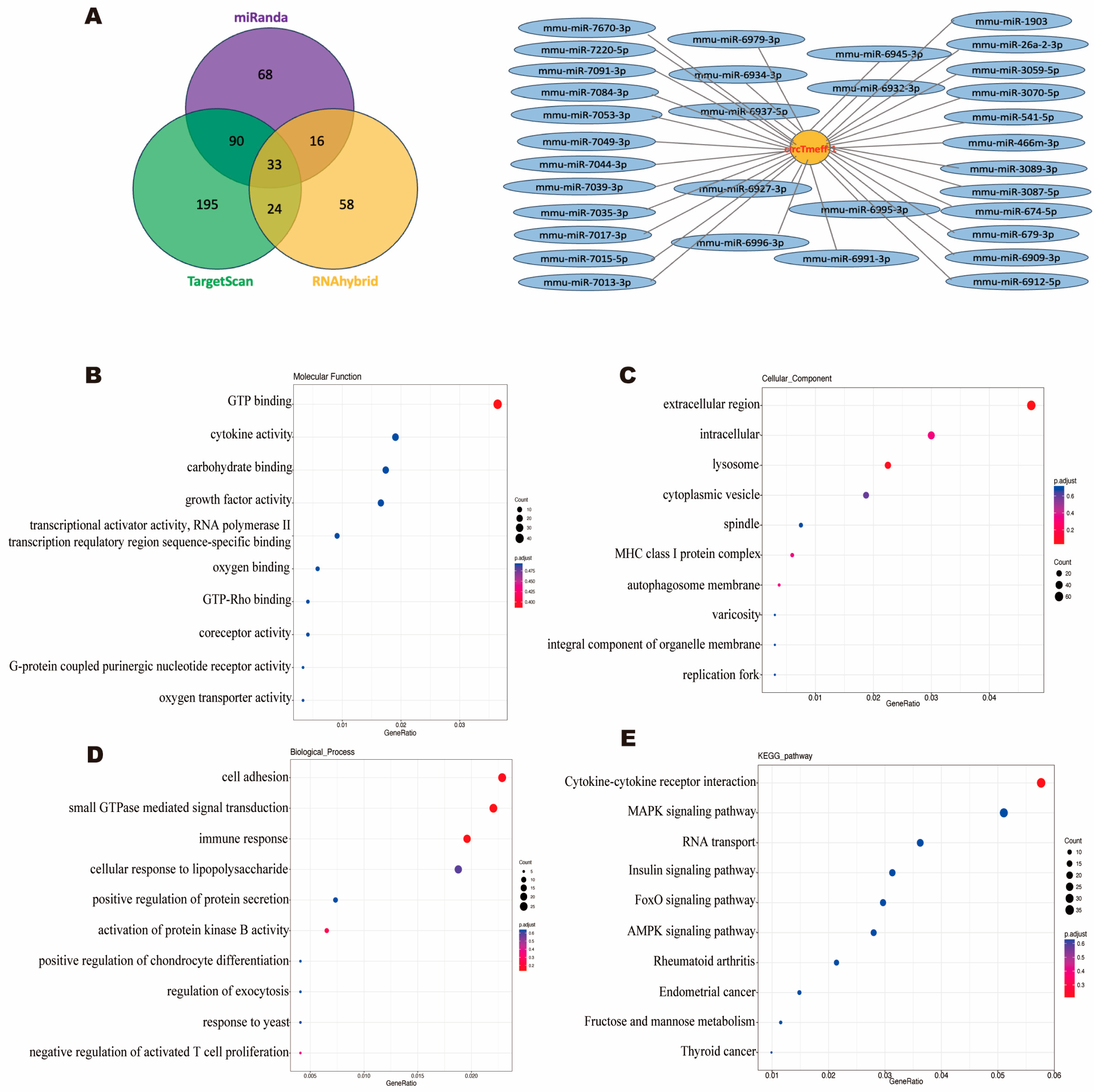
3.5. GO and KEGG Pathway Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leshner, A.I. Addiction Is a Brain Disease, and It Matters. Science 1997, 278, 45–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camí, J.; Farré, M. Drug Addiction. N. Engl. J. Med. 2003, 349, 975–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schifano, F.; Chiappini, S.; Corkery, J.M.; Guirguis, A. Assessing the 2004–2018 Fentanyl Misusing Issues Reported to an International Range of Adverse Reporting Systems. Front. Pharmacol. 2019, 10, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkow, N.D.; Jones, E.B.; Einstein, E.B.; Wargo, E.M. Prevention and Treatment of Opioid Misuse and Addiction. JAMA Psychiatry 2019, 76, 208–216. [Google Scholar] [CrossRef]
- Kohler, R.J.; Cibelli, J.; Baker, L.E. Combined effects of mephedrone and cocaine on locomotor activity and conditioned place preference in male Sprague–Dawley rats. Behav. Pharmacol. 2020, 31, 368–377. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, H.; Liu, Z.; Chen, X.; Su, X.; Ma, C.; Tian, Z.; Huang, B.; Yan, E.; Liu, X.; et al. A ventral CA1 to nucleus accumbens core engram circuit mediates conditioned place preference for cocaine. Nat. Neurosci. 2019, 22, 1986–1999. [Google Scholar] [CrossRef]
- de Jong, J.W.; Afjei, S.A.; Dorocic, I.P.; Peck, J.R.; Liu, C.; Kim, C.K.; Tian, L.; Deisseroth, K.; Lammel, S. A neural circuit mechanism for encoding aversive stimuli in the mesolimbic dopamine system. Neuron 2019, 101, 133.e7–151.e7. [Google Scholar] [CrossRef] [Green Version]
- Soares-Cunha, C.; de Vasconcelos, N.A.P.; Coimbra, B.; Domingues, A.V.; Silva, J.M.; Loureiro-Campos, E.; Gaspar, R.; Sotiropoulos, I.; Sousa, N.; Rodrigues, A.J. Nucleus accumbens medium spiny neurons subtypes signal both reward and aversion. Mol. Psychiatry 2020, 25, 3241–3255. [Google Scholar] [CrossRef] [Green Version]
- Bahi, A.; Dreyer, J.-L. Overexpression of plasminogen activators in the nucleus accumbens enhances cocaine-, amphetamine- and morphine-induced reward and behavioral sensitization. Genes Brain Behav. 2008, 7, 244–256. [Google Scholar] [CrossRef] [Green Version]
- Nakamoto, K.; Kawasaki, S.; Kobori, T.; Fujita-Hamabe, W.; Mizoguchi, H.; Yamada, K.; Nabeshima, T.; Tokuyama, S. Involvement of matrix metalloproteinase-9 in the development of morphine tolerance. Eur. J. Pharmacol. 2012, 683, 86–92. [Google Scholar] [CrossRef]
- Zaiou, M. circRNAs Signature as Potential Diagnostic and Prognostic Biomarker for Diabetes Mellitus and Related Cardiovascular Complications. Cells 2020, 9, 659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Eger, N.; Schoppe, L.; Schuster, S.; Laufs, U.; Boeckel, J.-N. Circular RNA Splicing. Circ. RNAs 2018, 1087, 41–52. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular Intronic Long Noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Circ. RNAs 2018, 1087, 67–79. [Google Scholar] [CrossRef]
- Bu, Q.; Long, H.; Shao, X.; Gu, H.; Kong, J.; Luo, L.; Liu, B.; Guo, W.; Wang, H.; Tian, J.; et al. Cocaine induces differential circular RNA expression in striatum. Transl. Psychiatry 2019, 9, 199. [Google Scholar] [CrossRef] [Green Version]
- Szabo, L.A.; Morey, R.; Palpant, N.J.; Wang, P.L.; Afari, N.; Jiang, C.; Parast, M.M.; Murry, C.E.; Laurent, L.C.; Salzman, J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015, 16, 126. [Google Scholar] [CrossRef] [Green Version]
- Veno, M.T.; Hansen, T.B.; Venø, S.T.; Clausen, B.H.; Grebing, M.; Finsen, B.; Holm, I.E.; Kjems, J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015, 16, 245. [Google Scholar] [CrossRef] [Green Version]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, present, and future of circ RNA s. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef]
- Mehta, S.L.; Dempsey, R.J.; Vemuganti, R. Role of circular RNAs in brain development and CNS diseases. Prog. Neurobiol. 2020, 186, 101746. [Google Scholar] [CrossRef]
- Xu, K.; Zhang, Y.; Li, J. Expression and function of circular RNAs in the mammalian brain. Cell. Mol. Life Sci. 2021, 78, 4189–4200. [Google Scholar] [CrossRef] [PubMed]
- Bian, A.; Wang, Y.; Liu, J.; Wang, X.; Liu, D.; Jiang, J.; Ding, L.; Hui, X. Circular RNA Complement Factor H (CFH) Promotes Glioma Progression by Sponging miR-149 and Regulating AKT1. Experiment 2018, 24, 5704–5712. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Xie, B.; Zhang, J.; Luo, Y.; Galaj, E.; Zhang, X.; Shen, Q.; Liu, Y.; Cong, B.; Wen, D.; et al. The role of circTmeff-1 in incubation of context-induced morphine craving. Pharmacol. Res. 2021, 170, 105722. [Google Scholar] [CrossRef] [PubMed]
- Eib, D.W.; Holling, T.M.; Zwijsen, A.; Dewulf, N.; de Groot, E.; Raaij, A.J.v.D.E.-V.; Huylebroeck, D.; Martens, G.J. Expression of the follistatin/EGF-containing transmembrane protein M7365 (tomoregulin-1) during mouse development. Mech. Dev. 2000, 97, 167–171. [Google Scholar] [CrossRef]
- Arano, T.; Fujisaki, S.; Ikemoto, M.J. Identification of tomoregulin-1 as a novel addicsin-associated factor. Neurochem. Int. 2014, 71, 22–35. [Google Scholar] [CrossRef]
- Tzschentke, T.M. Measuring reward with the conditioned place preference (CPP) paradigm: Update of the last decade. Addict. Biol. 2007, 12, 227–462. [Google Scholar] [CrossRef]
- Falco, A.M.; McDonald, C.G.; Smith, R.F. Anxiety status affects nicotine- and baclofen-induced locomotor activity, anxiety, and single-trial conditioned place preference in male adolescent rats. Dev. Psychobiol. 2014, 56, 1352–1364. [Google Scholar] [CrossRef]
- Asih, P.R.; Prikas, E.; Stefanoska, K.; Tan, A.R.P.; Ahel, H.I.; Ittner, A. Functions of p38 MAP Kinases in the Central Nervous System. Front. Mol. Neurosci. 2020, 13, 570586. [Google Scholar] [CrossRef]
- López-Gambero, A.J.; de Fonseca, F.R.; Suárez, J. Energy sensors in drug addiction: A potential therapeutic target. Addict. Biol. 2021, 26, e12936. [Google Scholar] [CrossRef]
- Dell’orco, M.; Oliver, R.J.; Perrone-Bizzozero, N. HuD Binds to and Regulates Circular RNAs Derived from Neuronal Development- and Synaptic Plasticity-Associated Genes. Front. Genet. 2020, 11, 790. [Google Scholar] [CrossRef]
- Larson, A.A.; Takemori, A.E. Effect of narcotics on the uptake of serotonin precursors by the rat brain. J. Pharmacol. Exp. Ther. 1977, 200, 216–223. [Google Scholar] [PubMed]
- Cheron, J.; D’exaerde, A.d.K. Drug addiction: From bench to bedside. Transl. Psychiatry 2021, 11, 424. [Google Scholar] [CrossRef] [PubMed]
- Castro, D.C.; Bruchas, M.R. A Motivational and Neuropeptidergic Hub: Anatomical and Functional Diversity within the Nucleus Accumbens Shell. Neuron 2019, 102, 529–552. [Google Scholar] [CrossRef] [PubMed]
- Scofield, M.D.; Heinsbroek, J.A.; Gipson, C.D.; Kupchik, Y.M.; Spencer, S.; Smith, A.C.W.; Roberts-Wolfe, D.; Kalivas, P.W. The Nucleus Accumbens: Mechanisms of Addiction across Drug Classes Reflect the Importance of Glutamate Homeostasis. Pharmacol. Rev. 2016, 68, 816–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkow, N.D.; Michaelides, M.; Baler, R. The Neuroscience of Drug Reward and Addiction. Physiol. Rev. 2019, 99, 2115–2140. [Google Scholar] [CrossRef]
- Brog, J.S.; Salyapongse, A.; Deutch, A.Y.; Zahm, D.S. The patterns of afferent innervation of the core and shell in the “Accumbens” part of the rat ventral striatum: Immunohistochemical detection of retrogradely transported fluoro-gold. J. Comp. Neurol. 1993, 338, 255–278. [Google Scholar] [CrossRef]
- Everitt, B.J.; Robbins, T.W. Neural systems of reinforcement for drug addiction: From actions to habits to compulsion. Nat. Neurosci. 2005, 8, 1481–1489. [Google Scholar] [CrossRef]
- Ambroggi, F.; Ghazizadeh, A.; Nicola, S.M.; Fields, H.L. Roles of Nucleus Accumbens Core and Shell in Incentive-Cue Responding and Behavioral Inhibition. J. Neurosci. 2011, 31, 6820–6830. [Google Scholar] [CrossRef] [Green Version]
- Portugal, G.S.; Al-Hasani, R.; Fakira, A.K.; -Romero, J.L.G.; Melyan, Z.; McCall, J.G.; Bruchas, M.R.; Morón, J.A. Hippocampal Long-Term Potentiation Is Disrupted during Expression and Extinction but Is Restored after Reinstatement of Morphine Place Preference. J. Neurosci. 2014, 34, 527–538. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; DeJoseph, M.; Urban, J.H.; Bahi, A.; Dreyer, J.-L.; Meredith, G.E.; Ford, K.A.; Ferrario, C.R.; Loweth, J.A.; Wolf, M.E. Different Roles of BDNF in Nucleus Accumbens Core versus Shell during the Incubation of Cue-Induced Cocaine Craving and Its Long-Term Maintenance. J. Neurosci. 2013, 33, 1130–1142. [Google Scholar] [CrossRef] [Green Version]
- Lowenstein, E.G.; Velazquez-Ulloa, N.A. A Fly’s Eye View of Natural and Drug Reward. Front. Physiol. 2018, 9, 407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oginsky, M.F.; Goforth, P.B.; Nobile, C.W.; Lopez-Santiago, L.F.; Ferrario, C.R. Eating ‘Junk-Food’ Produces Rapid and Long-Lasting Increases in NAc CP-AMPA Receptors: Implications for Enhanced Cue-Induced Motivation and Food Addiction. Neuropsychopharmacology 2016, 41, 2977–2986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epstein, D.H.; Shaham, Y. Cheesecake-eating rats and the question of food addiction. Nat. Neurosci. 2010, 13, 529–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibani, F.; Sahamsizadeh, A.; Fatemi, I.; Allahtavakoli, M.; Hasanshahi, J.; Rahmani, M.; Azin, M.; Hassanipour, M.; Mozafari, N.; Kaeidi, A. Effect of oleuropein on morphine-induced hippocampus neurotoxicity and memory impairments in rats. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2019, 392, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Kühn, S.; Schubert, F.; Gallinat, J. Structural correlates of trait anxiety: Reduced thickness in medial orbitofrontal cortex accompanied by volume increase in nucleus accumbens. J. Affect. Disord. 2011, 134, 315–319. [Google Scholar] [CrossRef]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef]
- Ebbesen, K.K.; Hansen, T.B.; Kjems, J. Insights into circular RNA biology. RNA Biol. 2017, 14, 1035–1045. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.-Y.; Cai, Z.-R.; Liu, J.; Wang, D.-S.; Ju, H.-Q.; Xu, R.-H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Wu, S.; Zhou, Z.; Ding, X.; Shi, R.; Thorne, R.F.; Zhang, X.D.; Hu, W.; Wu, M. CircACC1 Regulates Assembly and Activation of AMPK Complex under Metabolic Stress. Cell Metab. 2019, 30, 157–173.e7. [Google Scholar] [CrossRef]
- Valjent, E.; Pages, C.; Herve, D.; Girault, J.-A.; Caboche, J. Addictive and non-addictive drugs induce distinct and specific patterns of ERK activation in mouse brain. Eur. J. Neurosci. 2004, 19, 1826–1836. [Google Scholar] [CrossRef]
- Amaral, I.M.; Scheffauer, L.; Hofer, A.; El Rawas, R. Protein kinases in natural versus drug reward. Pharmacol. Biochem. Behav. 2022, 221, 173472. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.M.; Huganir, R.L. MAPK cascade signalling and synaptic plasticity. Nat. Rev. Neurosci. 2004, 5, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.M.; Messing, R.O. Protein Kinases and Addiction. Ann. N. Y. Acad. Sci. 2008, 1141, 22–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valjent, E.; Pascoli, V.; Svenningsson, P.; Paul, S.; Enslen, H.; Corvol, J.-C.; Stipanovich, A.; Caboche, J.; Lombroso, P.J.; Nairn, A.C.; et al. From The Cover: Regulation of a protein phosphatase cascade allows convergent dopamine and glutamate signals to activate ERK in the striatum. Proc. Natl. Acad. Sci. USA 2005, 102, 491–496. [Google Scholar] [CrossRef]
- Lu, L.; Dempsey, J.; Liu, S.Y.; Bossert, J.M.; Shaham, Y. A Single Infusion of Brain-Derived Neurotrophic Factor into the Ventral Tegmental Area Induces Long-Lasting Potentiation of Cocaine Seeking after Withdrawal. J. Neurosci. 2004, 24, 1604–1611. [Google Scholar] [CrossRef] [Green Version]
- Potter, W.B.; O’Riordan, K.J.; Barnett, D.; Osting, S.M.K.; Wagoner, M.; Burger, C.; Roopra, A. Metabolic Regulation of Neuronal Plasticity by the Energy Sensor AMPK. PLoS ONE 2010, 5, e8996. [Google Scholar] [CrossRef] [Green Version]
- Huhn, A.S.; Berry, M.S.; Dunn, K.E. Systematic review of sex-based differences in opioid-based effects. Int. Rev. Psychiatry 2018, 30, 107–116. [Google Scholar] [CrossRef]
- Limonta, P.; Dondi, D.; Maggi, R.; Piva, F. Testosterone and postnatal ontogenesis of hypothalamic μ ([3H]dihydromorphine) opioid receptors in the rat. Dev. Brain Res. 1991, 62, 131–136. [Google Scholar] [CrossRef]
- Cruz, W.S.; Pereira, L.A.; Cezar, L.C.; Camarini, R.; Felicio, L.F.; Bernardi, M.M.; Teodorov, E. Role of steroid hormones and morphine treatment in the modulation of opioid receptor gene expression in brain structures in the female rat. Springerplus 2015, 4, 355. [Google Scholar] [CrossRef] [Green Version]
- Knodell, R.G.; Allen, R.C.; Kyner, W.T. Effects of ethinyl estradiol on pharmacokinetics of meperidine and pentobarbital in the rat. Experiment 1982, 221, 1–6. [Google Scholar]
- Munro, C.A.; McCaul, M.E.; Wong, D.F.; Oswald, L.M.; Zhou, Y.; Brasic, J.; Kuwabara, H.; Kumar, A.; Alexander, M.; Ye, W.; et al. Sex Differences in Striatal Dopamine Release in Healthy Adults. Biol. Psychiatry 2006, 59, 966–974. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.B.; Perry, A.N.; Westenbroek, C. Sex differences in the neural mechanisms mediating addiction: A new synthesis and hypothesis. Biol. Sex Differ. 2012, 3, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cummings, J.A.; Jagannathan, L.; Jackson, L.R.; Becker, J.B. Sex differences in the effects of estradiol in the nucleus accumbens and striatum on the response to cocaine: Neurochemistry and behavior. Drug Alcohol Depend. 2013, 135, 22–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Step | Number of Cycles | Temperature | Time |
|---|---|---|---|
| Initial denaturation | 1 | 95 °C | 30 s |
| PCR reaction | 40 | 95 °C 60 °C | 5 s 30 s |
| Solution curve analysis | 1 | 60–95 °C | 2 min |
| Name | Titer |
|---|---|
| rAAV-TRE-5′-circRNA-nEfla-CMV-EGFP-PA | 5.00 × 1012 vg mL−1 |
| rAAV-TRE-5′-CMV-BSSI-nEfla-EGFP | 2.86 × 1012 vg mL−1 |
| rAAV-TRE-5′-mir30-shRNA(circRNA)-miR30-3′-CMV-mcherry-PA | 8.23 × 1012 vg mL−1 |
| rAAV-Tre3g-speI-5′-miR30a-xhoI-EcoRI-miR30a-3′-NheI-CMV-mcherry-PA | 5.20 × 1012 vg mL−1 |
| rAAV-CMV-tTA-WPRE-PA | 6.93 × 1012 vg mL−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Wen, B.; Lu, Y.; Xie, B.; Yu, F.; Zhang, M.; Ma, C.; Cong, B.; Wen, D.; Bi, H. The Role of circTmeff-1 in Morphine Addiction Memory of Mice. Cells 2023, 12, 1985. https://doi.org/10.3390/cells12151985
Yu H, Wen B, Lu Y, Xie B, Yu F, Zhang M, Ma C, Cong B, Wen D, Bi H. The Role of circTmeff-1 in Morphine Addiction Memory of Mice. Cells. 2023; 12(15):1985. https://doi.org/10.3390/cells12151985
Chicago/Turabian StyleYu, Hailei, Boyang Wen, Yun Lu, Bing Xie, Feng Yu, Minglong Zhang, Chunling Ma, Bin Cong, Di Wen, and Haitao Bi. 2023. "The Role of circTmeff-1 in Morphine Addiction Memory of Mice" Cells 12, no. 15: 1985. https://doi.org/10.3390/cells12151985
APA StyleYu, H., Wen, B., Lu, Y., Xie, B., Yu, F., Zhang, M., Ma, C., Cong, B., Wen, D., & Bi, H. (2023). The Role of circTmeff-1 in Morphine Addiction Memory of Mice. Cells, 12(15), 1985. https://doi.org/10.3390/cells12151985





