Cfdp1 Is Essential for Cardiac Development and Function
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish Housing and Husbandry
2.2. gRNA and Cas9 mRNA Synthesis
2.3. Microinjection in Zebrafish Embryos
2.4. Tail Amputations and Genotyping
2.5. RT-PCR
2.6. RNA Extraction and Quantitative Real-Time PCR
2.7. Whole-Mount In Situ Hybridization and Immunohistochemistry
2.8. Imaging
2.9. Adult Zebrafish Heart Isolation and Histology
2.10. Estimation of Cardiac Function
2.11. Statistical Analysis
3. Results
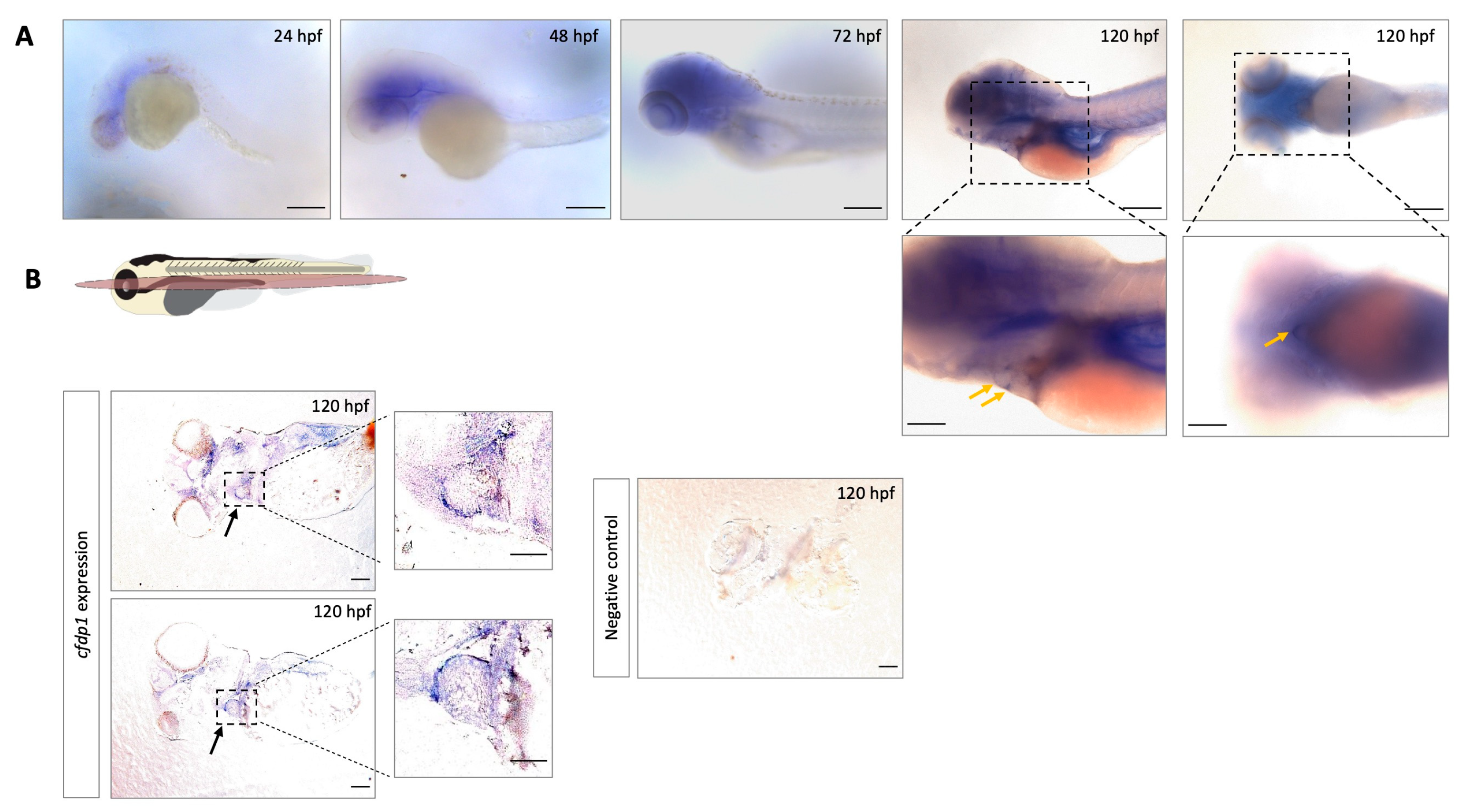
3.1. Expression Profile of Zebrafish cfdp1 Gene during Embryonic Development
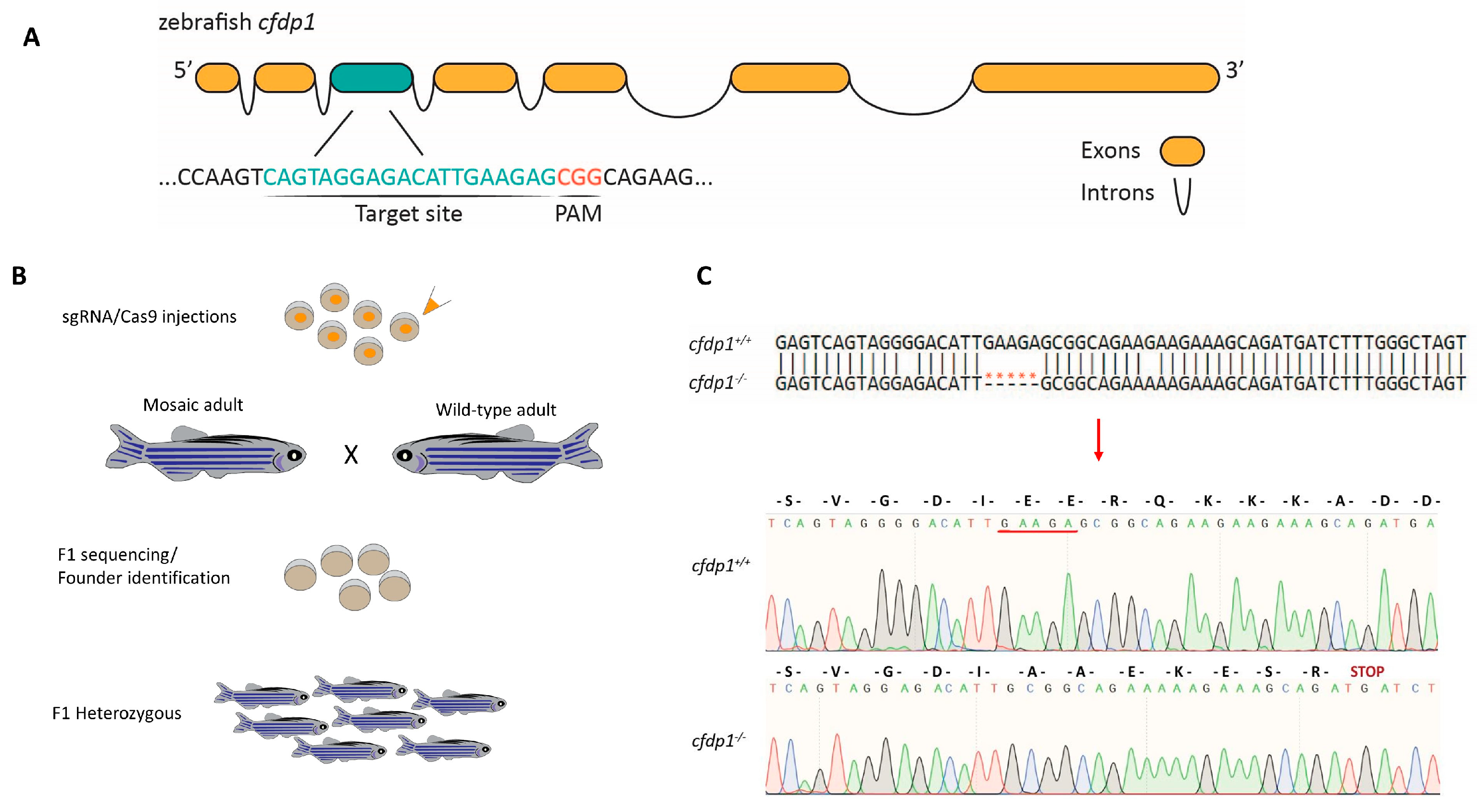
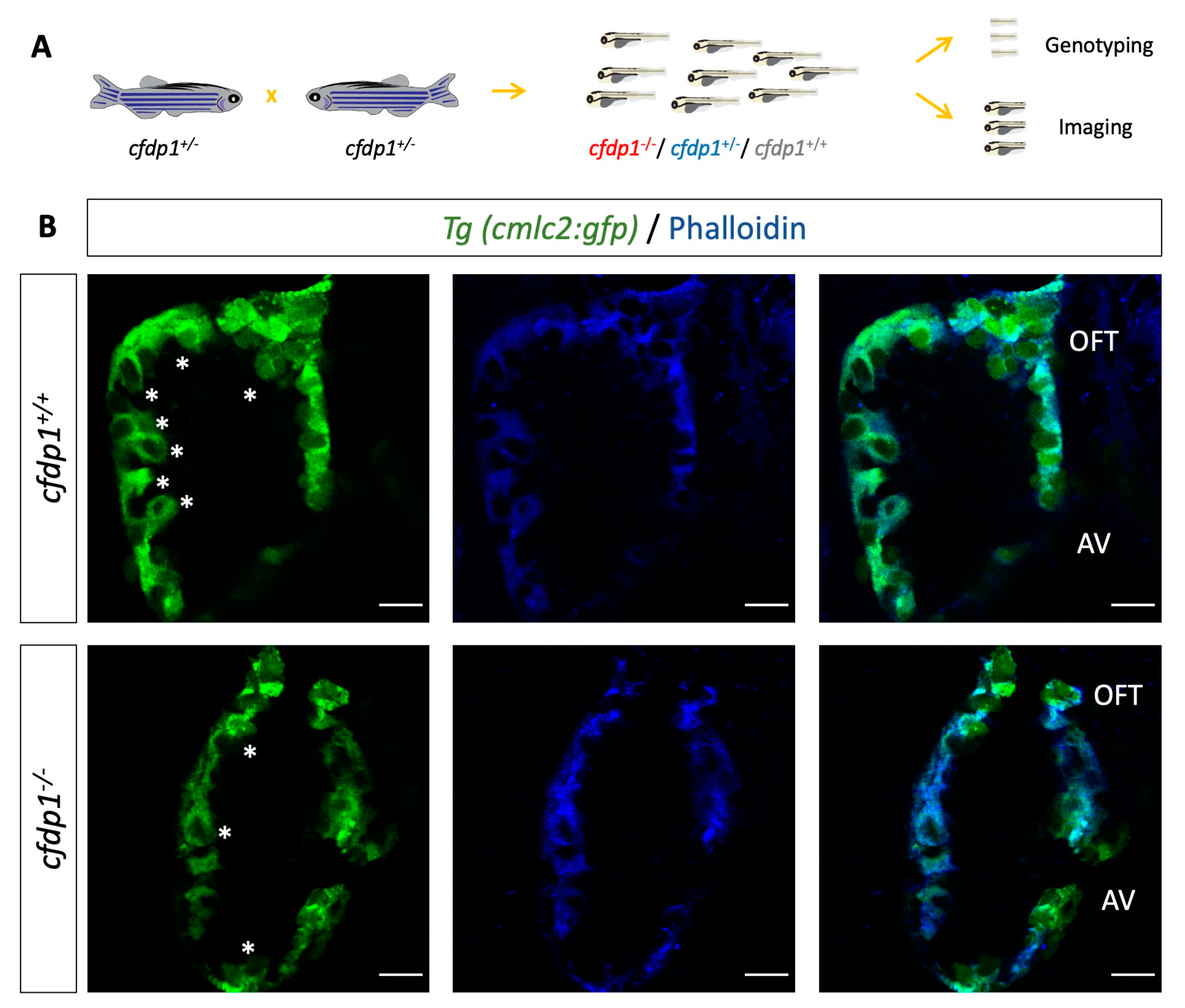
3.2. Generation of Zebrafish cfdp1 Mutant Line
3.3. Zebrafish cfdp1 Mutants Show Impaired Cardiac Performance
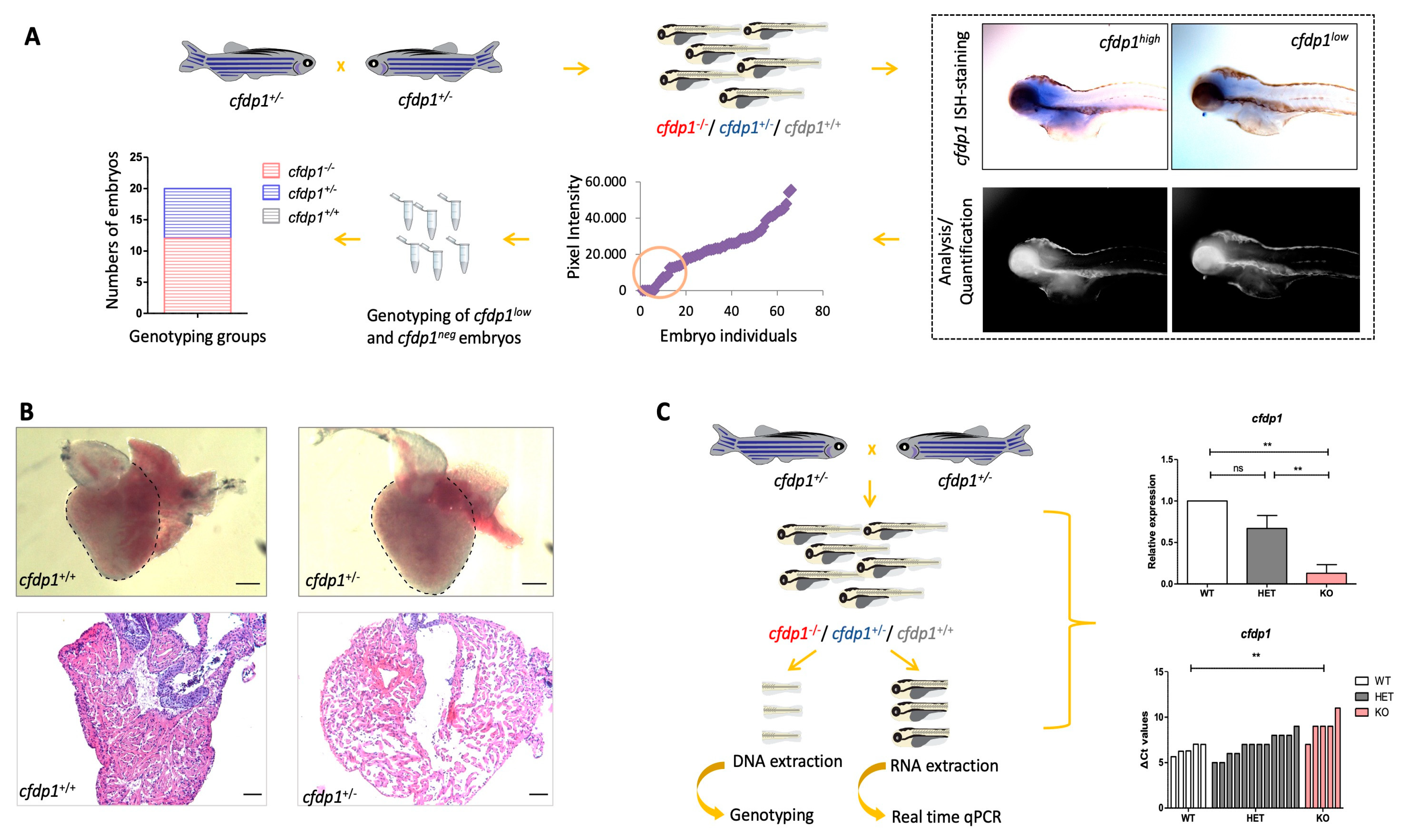
3.4. Zebrafish cfdp1 Heterozygous Develop Variation in Phenotype at Embryonic Stage
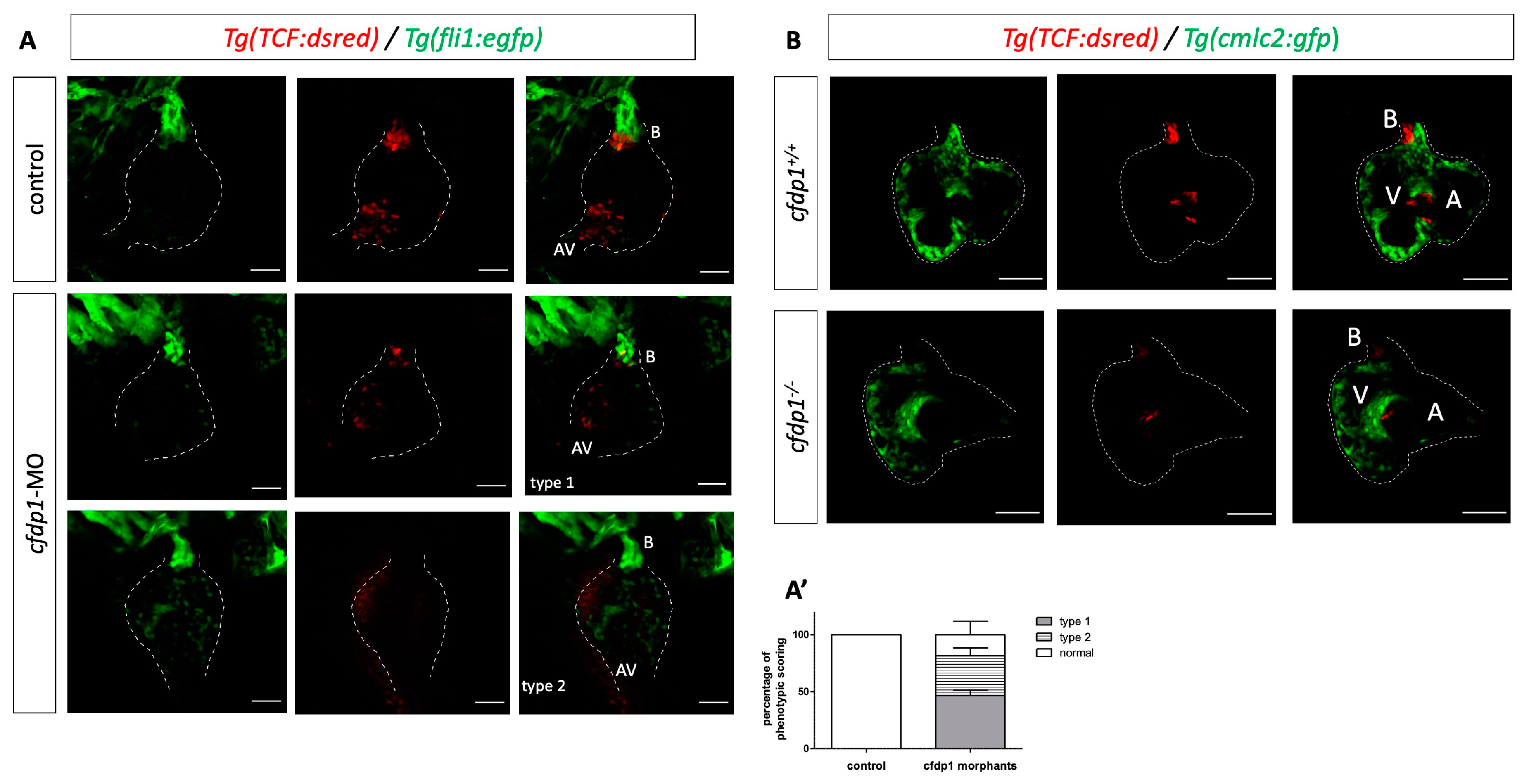
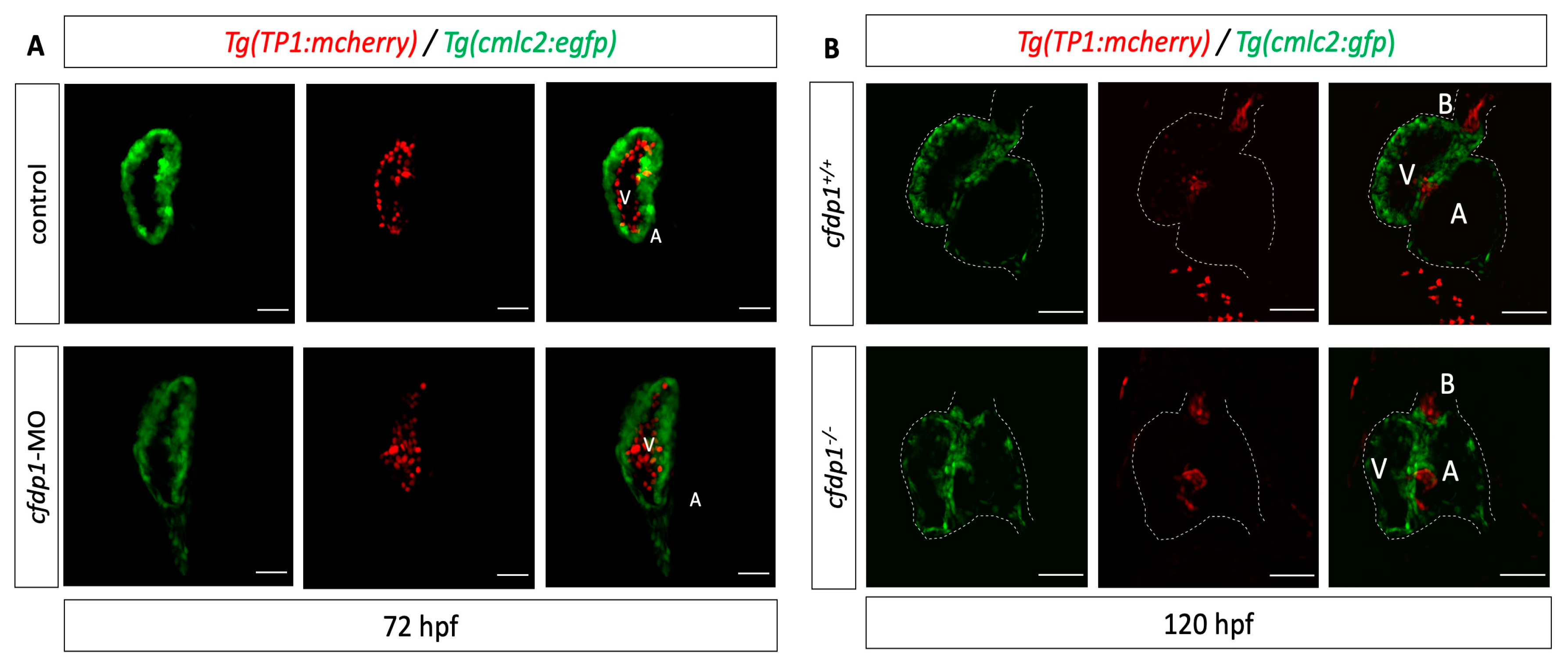
3.5. Abrogation of cfdp1 Expression Reduces Activation of Wnt Pathway in the Embryonic Heart but Notch Signaling Remains Unaffected
3.6. Cardiac Trabeculation in Developing Zebrafish Ventricle Is Defective in cfdp1 Mutants
4. Discussion
4.1. A Model for the Phenotypic and Functional Characterization of cfdp1
4.2. cfdp1 Knockdown and Knockout Zebrafish Demonstrated Similar but Not Identical Phenotypes
4.3. The Role of cfdp1 in Ventricular Trabeculation and Cardiac Function
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deloukas, P.; Kanoni, S.; Willenborg, C.; Farrall, M.; Assimes, T.L.; Thompson, J.R.; Ingelsson, E.; Saleheen, D.; Erdmann, J.; Goldstein, B.A.; et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013, 45, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Webb, T.R.; Erdmann, J.; Stirrups, K.E.; Stitziel, N.O.; Masca, N.G.D.; Jansen, H.; Kanoni, S.; Nelson, C.P.; Ferrario, P.G.; König, I.R.; et al. Systematic Evaluation of Pleiotropy Identifies 6 Further Loci Associated with Coronary Artery Disease. J. Am. Coll. Cardiol. 2017, 69, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Ntalla, I.; Kanoni, S.; Zeng, L.; Giannakopoulou, O.; Danesh, J.; Watkins, H.; Samani, N.J.; Deloukas, P.; Schunkert, H. Genetic Risk Score for Coronary Disease Identifies Predispositions to Cardiovascular and Noncardiovascular Diseases. J. Am. Coll. Cardiol. 2019, 73, 2932–2942. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.P.; Goel, A.; Butterworth, A.S.; Kanoni, S.; Webb, T.R.; Marouli, E.; Zeng, L.; Ntalla, I.; Lai, F.Y.; Hopewell, J.C.; et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017, 49, 1385–1391. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef] [Green Version]
- Sabater-Lleal, M.; Mälarstig, A.; Folkersen, L.; Artigas, M.S.; Baldassarre, D.; Kavousi, M.; Almgren, P.; Veglia, F.; Brusselle, G.; Hofman, A.; et al. Common genetic determinants of lung function, subclinical atherosclerosis and risk of coronary artery disease. PLoS ONE 2014, 9, e104082. [Google Scholar] [CrossRef] [Green Version]
- Sung, Y.J.; Winkler, T.W.; Fuentes, L.d.L.; Bentley, A.R.; Brown, M.R.; Kraja, A.T.; Schwander, K.; Ntalla, I.; Guo, X.; Franceschini, N.; et al. A Large-Scale Multi-Ancestry Genome-wide Study Accounting for Smoking Behavior Identifies Multiple Significant Loci for Blood Pressure. Am. J. Hum. Genet. 2018, 102, 375–400. [Google Scholar] [CrossRef] [Green Version]
- Boardman-Pretty, F.; Smith, A.J.; Cooper, J.; Palmen, J.; Folkersen, L.; Hamsten, A.; Catapano, A.L.; Melander, O.; Price, J.F.; Kumari, M.; et al. Functional Analysis of a Carotid Intima-Media Thickness Locus Implicates BCAR1 and Suggests a Causal Variant. Circ. Cardiovasc. Genet. 2015, 8, 696–706. [Google Scholar] [CrossRef] [Green Version]
- Gertow, K.; Sennblad, B.; Strawbridge, R.J.; Öhrvik, J.; Zabaneh, D.; Shah, S.; Veglia, F.; Fava, C.; Kavousi, M.; McLachlan, S.; et al. Identification of the BCAR1-CFDP1-TMEM170A locus as a determinant of carotid intima-media thickness and coronary artery disease risk. Circ. Cardiovasc. Genet. 2012, 5, 656–665. [Google Scholar] [CrossRef] [Green Version]
- Formicola, D.; Lasorsa, V.A.; Cantalupo, S.; Testori, A.; Cardinale, A.; Avitabile, M.; Diskin, S.; Iolascon, A.; Capasso, M. CFDP1 is a neuroblastoma susceptibility gene that regulates transcription factors of the noradrenergic cell identity. HGG Adv. 2022, 4, 100158. [Google Scholar] [CrossRef]
- Messina, G.; Celauro, E.; Atterrato, M.T.; Giordano, E.; Iwashita, S.; Dimitri, P. The Bucentaur (BCNT) protein family: A long-neglected class of essential proteins required for chromatin/chromosome organization and function. Chromosoma 2015, 124, 153–162. [Google Scholar] [CrossRef]
- Nobukuni, T.; Kobayashi, M.; Omori, A.; Ichinose, S.; Iwanaga, T.; Takahashi, I.; Hashimoto, K.; Hattori, S.; Kaibuchi, K.; Miyata, Y.; et al. An Alu-linked repetitive sequence corresponding to 280 amino acids is expressed in a novel bovine protein, but not in its human homologue. J. Biol. Chem. 1997, 272, 2801–2807. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Luk, E. Dual function of Swc5 in SWR remodeling ATPase activation and histone H2A eviction. Nucleic Acids Res. 2017, 45, 9931–9946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.-H.; Wu, C.-H.; Ladurner, A.; Mizuguchi, G.; Wei, D.; Xiao, H.; Luk, E.; Ranjan, A.; Wu, C. N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J. Biol. Chem. 2009, 284, 6200–6207. [Google Scholar] [CrossRef] [Green Version]
- Chu, W.T.; Wang, J. Influence of sequence length and charged residues on Swc5 binding with histone H2A-H2B. Proteins Struct. Funct. Bioinform. 2021, 89, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Messina, G.; Damia, E.; Fanti, L.; Atterrato, M.T.; Celauro, E.; Mariotti, F.R.; Accardo, M.C.; Walther, M.; Vernì, F.; Picchioni, D.; et al. Yeti, an essential Drosophila melanogaster gene, encodes a protein required for chromatin organization. J. Cell Sci. 2014, 127, 2577–2588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, T.; Inoue, S.; Sun, X.; Kusuda, R.; Hibi, M.; Shimizu, T. Cfdp1 controls the cell cycle and neural differentiation in the zebrafish cerebellum and retina. Dev. Dyn. 2021, 250, 1618–1633. [Google Scholar] [CrossRef] [PubMed]
- Celauro, E.; Carra, S.; Rodriguez, A.; Cotelli, F.; Dimitri, P. Functional analysis of the cfdp1 gene in zebrafish provides evidence for its crucial role in craniofacial development and osteogenesis. Exp. Cell Res. 2017, 361, 236–245. [Google Scholar] [CrossRef]
- Iwashita, S.; Suzuki, T.; Yasuda, T.; Nakashima, K.; Sakamoto, T.; Kohno, T.; Takahashi, I.; Kobayashi, T.; Ohno-Iwashita, Y.; Imajoh-Ohmi, S.; et al. Mammalian Bcnt/Cfdp1, a potential epigenetic factor characterized by an acidic stretch in the disordered N-terminal and Ser250 phosphorylation in the conserved C-terminal regions. Biosci. Rep. 2015, 35, e00228. [Google Scholar] [CrossRef] [Green Version]
- Diekwisch, T.G.; Marches, F.; Williams, A.; Luan, X. Cloning, gene expression, and characterization of CP27, a novel gene in mouse embryogenesis. Gene 1999, 235, 19–30. [Google Scholar] [CrossRef]
- Diekwisch, T.G.; Luan, X.; McIntosh, J.E. CP27 localization in the dental lamina basement membrane and in the stellate reticulum of developing teeth. J. Histochem. Cytochem. 2002, 50, 583–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, X.; Diekwisch, T.G.H. CP27 affects viability, proliferation, attachment and gene expression in embryonic fibroblasts. Cell Prolif. 2002, 35, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Iwashita, S.; Suzuki, T.; Kiriyama, Y.; Dohmae, N.; Ohoka, Y.; Song, S.-Y.; Nakashima, K. Overcoming off-targets: Assessing Western blot signals for Bcnt/Cfdp1, a tentative component of the chromatin remodeling complex. Biosci. Rep. 2020, 40, BSR20194012. [Google Scholar] [CrossRef] [PubMed]
- Messina, G.; Atterrato, M.T.; Prozzillo, Y.; Piacentini, L.; Losada, A.; Dimitri, P. The human Cranio Facial Development Protein 1 (Cfdp1) gene encodes a protein required for the maintenance of higher-order chromatin organization. Sci. Rep. 2017, 7, 45022. [Google Scholar] [CrossRef] [Green Version]
- Aragam, K.G.; Jiang, T.; Goel, A.; Kanoni, S.; Wolford, B.N.; Atri, D.S.; Weeks, E.M.; Wang, M.; Hindy, G.; Zhou, W.; et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat. Genet. 2022, 54, 1803–1815. [Google Scholar] [CrossRef]
- Giardoglou, P.; Beis, D. On zebrafish disease models and matters of the heart. Biomedicines 2019, 7, 15. [Google Scholar] [CrossRef] [Green Version]
- Bakkers, J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc. Res. 2011, 91, 279–288. [Google Scholar] [CrossRef] [Green Version]
- Bournele, D.; Beis, D. Zebrafish models of cardiovascular disease. Heart Fail. Rev. 2016, 21, 803–813. [Google Scholar] [CrossRef]
- Sehnert, A.J.; Huq, A.; Weinstein, B.M.; Walker, C.; Fishman, M.; Stainier, D.Y.R. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 2002, 31, 106–110. [Google Scholar] [CrossRef]
- Aleström, P.; D’Angelo, L.; Midtlyng, P.J.; Schorderet, D.F.; Schulte-Merker, S.; Sohm, F.; Warner, S. Zebrafish: Housing and husbandry recommendations. Lab. Anim. 2020, 54, 213–224. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.-J.; Tu, C.-T.; Hsiao, C.-D.; Hsieh, F.-J.; Tsai, H.-J. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 2003, 228, 30–40. [Google Scholar] [CrossRef]
- Lawson, N.D.; Weinstein, B.M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002, 248, 307–318. [Google Scholar] [CrossRef] [Green Version]
- Ninov, N.; Borius, M.; Stainier, D.Y.R. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development 2012, 139, 1557–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moro, E.; Ozhan-Kizil, G.; Mongera, A.; Beis, D.; Wierzbicki, C.; Young, R.M.; Bournele, D.; Domenichini, A.; Valdivia, L.E.; Lum, L.; et al. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev. Biol. 2012, 366, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Jao, L.-E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Hoage, T.; Ding, Y.; Xu, X. Cardiovascular development: Structure and molecular mechanism. Anat. Sci. Int. 2012, 84, 65–66. [Google Scholar] [CrossRef]
- Litviňuková, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Worth, C.L.; Lindberg, E.L.; Kanda, M.; Polanski, K.; Heinig, M.; Lee, M.; et al. Cells of the adult human heart. Nature 2020, 588, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Kontarakis, Z.; Gerri, C.; Nolte, H.; Hölper, S.; Krüger, M.; Stainier, D.Y.R. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 2015, 524, 230–233. [Google Scholar] [CrossRef]
- Dobrzycki, T.; Krecsmarik, M.; Monteiro, R. Genotyping and quantification of in situ hybridization staining in zebrafish. J. Vis. Exp. 2020, 155, e59956. [Google Scholar] [CrossRef] [Green Version]
- Grego-Bessa, J.; Luna-Zurita, L.; del Monte, G.; Bolós, V.; Melgar, P.; Arandilla, A.; Garratt, A.N.; Zang, H.; Mukouyama, Y.-S.; Chen, H.; et al. Notch Signaling Is Essential for Ventricular Chamber Development. Dev. Cell. 2007, 12, 415–429. [Google Scholar] [CrossRef] [Green Version]
- D’amato, G.; Luxán, G.; del Monte-Nieto, G.; Martínez-Poveda, B.; Torroja, C.; Walter, W.; Bochter, M.S.; Benedito, R.; Cole, S.; Martinez, F.; et al. Sequential Notch activation regulates ventricular chamber development. Nature 2015, 18, 7–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beis, D.; Bartman, T.; Jin, S.-W.; Scott, I.C.; D’Amico, L.A.; Ober, E.A.; Verkade, H.; Frantsve, J.; Field, H.A.; Wehman, A.; et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development 2005, 132, 4193–4204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paolini, A.; Fontana, F.; Pham, V.-C.; Rödel, C.J.; Abdelilah-Seyfried, S. Mechanosensitive Notch-Dll4 and Klf2-Wnt9 Signaling Pathways Intersect in Guiding Valvulogenesis in Zebrafish. Cell Rep. 2021, 37, 109782. [Google Scholar] [CrossRef]
- Hurlstone, A.F.L.; Haramis, A.G.; Wienholds, E. The Wnt / b -catenin pathway regulates cardiac valve formation. Nature 2003, 425, 633–637. [Google Scholar] [CrossRef]
- Walsh, E.C.; Stainier, D.Y.R. UDP-Glucose Dehydrogenase Required for Cardiac Valve Formation in Zebrafish. Science 2001, 293, 1670–1673. [Google Scholar] [CrossRef]
- Pestel, J.; Ramadass, R.; Gauvrit, S.; Helker, C.; Herzog, W.; Stainier, D.Y.R. Real-time 3D visualization of cellular rearrangements during cardiac valve formation. Development 2016, 143, 2217–2227. [Google Scholar] [CrossRef] [Green Version]
- Rasouli, S.J.; Stainier, D.Y.R. Regulation of cardiomyocyte behavior in zebrafish trabeculation by Neuregulin 2a signaling. Nat. Commun. 2017, 8, 15281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Bressan, M.; Hassel, D.; Huisken, J.; Staudt, D.; Kikuchi, K.; Poss, K.D.; Mikawa, T.; Stainier, D.Y. A dual role for ErbB2 signaling in cardiac trabeculation. Development 2010, 137, 3867–3875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peshkovsky, C.; Totong, R.; Yelon, D. Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev. Dyn. 2011, 240, 446–456. [Google Scholar] [CrossRef]
- Nurnberg, S.T.; Guerraty, M.A.; Wirka, R.C.; Rao, H.S.; Pjanic, M.; Norton, S.; Serrano, F.; Perisic, L.; Elwyn, S.; Pluta, J.; et al. Genomic profiling of human vascular cells identifies TWIST1 as a causal gene for common vascular diseases. PLoS Genet. 2020, 16, e1008538. [Google Scholar] [CrossRef]
- Klarin, D.; Zhu, Q.M.; A Emdin, C.; Chaffin, M.; Horner, S.; McMillan, B.J.; Leed, A.; E Weale, M.; A Spencer, C.C.; Aguet, F.; et al. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat. Genet. 2017, 49, 1392–1397. [Google Scholar] [CrossRef]
- Wild, P.S.; Felix, J.F.; Schillert, A.; Teumer, A.; Chen, M.-H.; Leening, M.J.; Völker, U.; Großmann, V.; Brody, J.A.; Irvin, M.R.; et al. Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J. Clin. Investig. 2017, 127, 1798–1812. [Google Scholar] [CrossRef] [Green Version]
- Yue, M.S.; Peterson, R.E.; Heideman, W. Dioxin inhibition of swim bladder development in zebrafish: Is it secondary to heart failure? Aquat. Toxicol. 2015, 162, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Kurosaki, T.; Maquat, L.E. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 2016, 129, 461–467. [Google Scholar] [CrossRef] [Green Version]
- Khajavi, M.; Inoue, K.; Lupski, J.R. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur. J. Hum. Genet. 2006, 14, 1074–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.; Pearce, D.A. Nonsense-mediated decay in genetic disease: Friend or foe? Mutat. Res. Rev. Mutat. Res. 2014, 762, 52–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.S.; Wilkinson, M.F.; Gecz, J. Nonsense-mediated mRNA decay: Inter-individual variability and human disease. Neurosci. Biobehav. Rev. 2014, 46, 175–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalogirou, S.; Malissovas, N.; Moro, E.; Argenton, F.; Stainier, D.Y.; Beis, D. Intracardiac flow dynamics regulate atrioventricular valve morphogenesis. Cardiovasc. Res. 2014, 104, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Sidhwani, P.; Yelon, D. Fluid Forces Shape the Embryonic Heart: Insights from Zebrafish, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2019; Volume 132. [Google Scholar] [CrossRef]
- Chi, N.C.; Shaw, R.M.; Jungblut, B.; Huisken, J.; Ferrer, T.; Arnaout, R.; Scott, I.; Beis, D.; Xiao, T.; Baier, H.; et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008, 6, 1006–1019. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.-C.; Kruse, F.; Vasudevarao, M.D.; Junker, J.P.; Zebrowski, D.C.; Fischer, K.; Noël, E.S.; Grün, D.; Berezikov, E.; Engel, F.B.; et al. Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration. Dev. Cell. 2016, 36, 36–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staudt, D.W.; Liu, J.; Thorn, K.S.; Stuurman, N.; Liebling, M.; Stainier, D.Y.R. High-resolution imaging of cardiomyocyte behavior reveals two distinct steps in ventricular trabeculation. Development 2014, 141, 585–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.; Han, P.; Ma, X.; Farah, E.N.; Bloomekatz, J.; Zeng, X.-X.I.; Zhang, R.; Swim, M.M.; Witty, A.D.; Knight, H.G.; et al. Canonical Wnt5b Signaling Directs Outlying Nkx2.5+ Mesoderm into Pacemaker Cardiomyocytes. Dev. Cell. 2019, 50, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Han, P.; Kim, E.H.; Mak, J.; Zhang, R.; Torrente, A.G.; Goldhaber, J.I.; Marbán, E.; Cho, H.C. Canonical Wnt signaling promotes pacemaker cell specification of cardiac mesodermal cells derived from mouse and human embryonic stem cells. Stem. Cells 2020, 38, 352–368. [Google Scholar] [CrossRef] [PubMed]
- de Pater, E.; Clijsters, L.; Marques, S.R.; Lin, Y.-F.; Garavito-Aguilar, Z.V.; Yelon, D.; Bakkers, J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 2009, 136, 1633–1641. [Google Scholar] [CrossRef] [Green Version]
- Burkhard, S.B.; Bakkers, J. Spatially resolved RNA-sequencing of the embryonic heart identifies a role for Wnt/β-catenin signaling in autonomic control of heart rate. eLife 2018, 7, e31515. [Google Scholar] [CrossRef]
- Gillers, B.S.; Chiplunkar, A.; Aly, H.; Valenta, T.; Basler, K.; Christoffels, V.M.; Efimov, I.R.; Boukens, B.J.; Rentschler, S. Canonical Wnt signaling regulates atrioventricular junction programming and electrophysiological properties. Circ. Res. 2014, 116, 398–406. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giardoglou, P.; Deloukas, P.; Dedoussis, G.; Beis, D. Cfdp1 Is Essential for Cardiac Development and Function. Cells 2023, 12, 1994. https://doi.org/10.3390/cells12151994
Giardoglou P, Deloukas P, Dedoussis G, Beis D. Cfdp1 Is Essential for Cardiac Development and Function. Cells. 2023; 12(15):1994. https://doi.org/10.3390/cells12151994
Chicago/Turabian StyleGiardoglou, Panagiota, Panos Deloukas, George Dedoussis, and Dimitris Beis. 2023. "Cfdp1 Is Essential for Cardiac Development and Function" Cells 12, no. 15: 1994. https://doi.org/10.3390/cells12151994
APA StyleGiardoglou, P., Deloukas, P., Dedoussis, G., & Beis, D. (2023). Cfdp1 Is Essential for Cardiac Development and Function. Cells, 12(15), 1994. https://doi.org/10.3390/cells12151994






