3D Computational Modeling of Defective Early Endosome Distribution in Human iPSC-Based Cardiomyopathy Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation, Culture, CRISPR/Cas9 Gene Editing, and Cardiac Differentiation of Human iPSCs
2.2. STED Imaging
2.3. Vesicular Distribution Computational Modeling
2.4. Statistical Analysis
3. Results
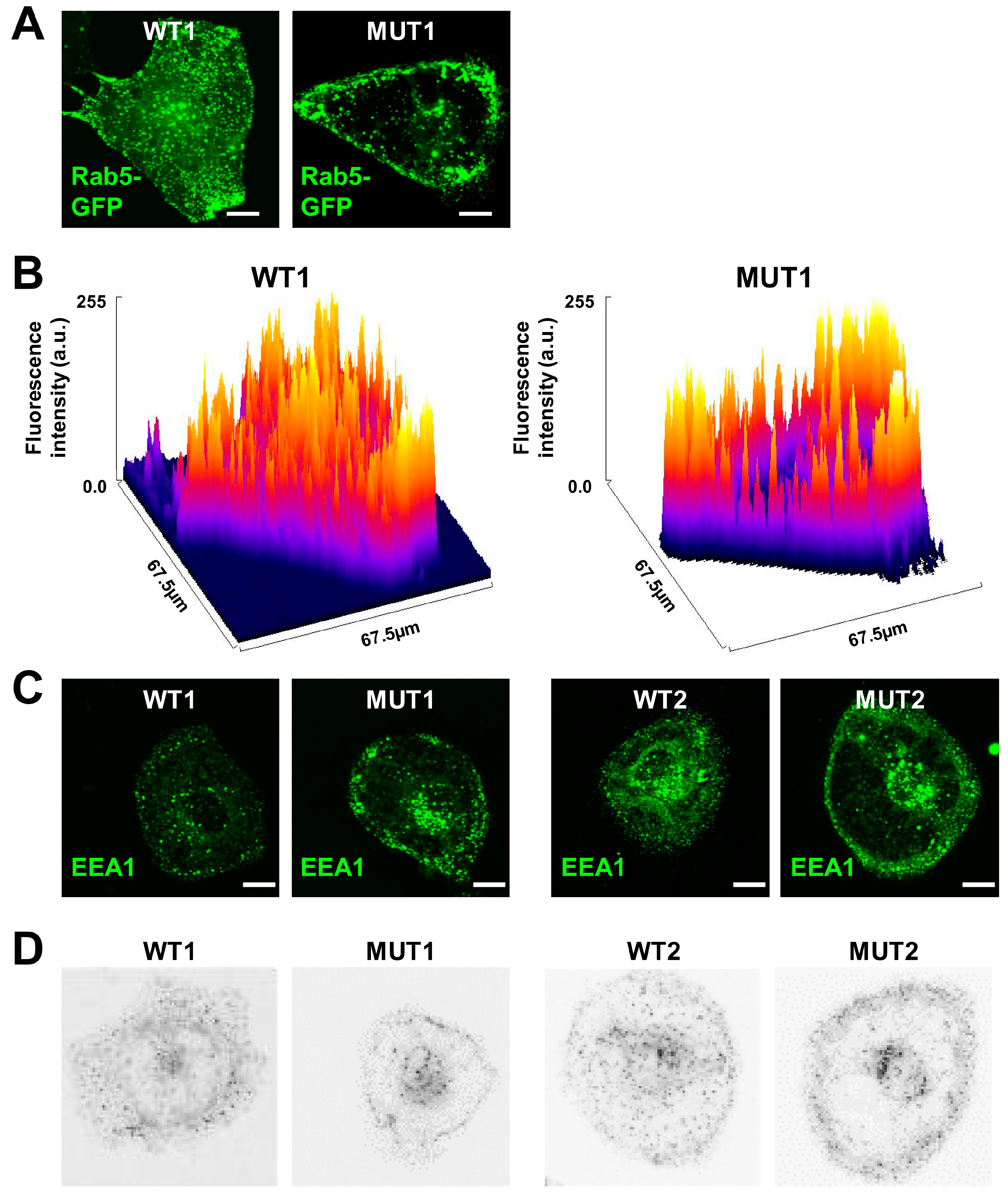
3.1. Early Endosomes Fail to Distribute in iPSC-CMs Carrying DCM Mutations
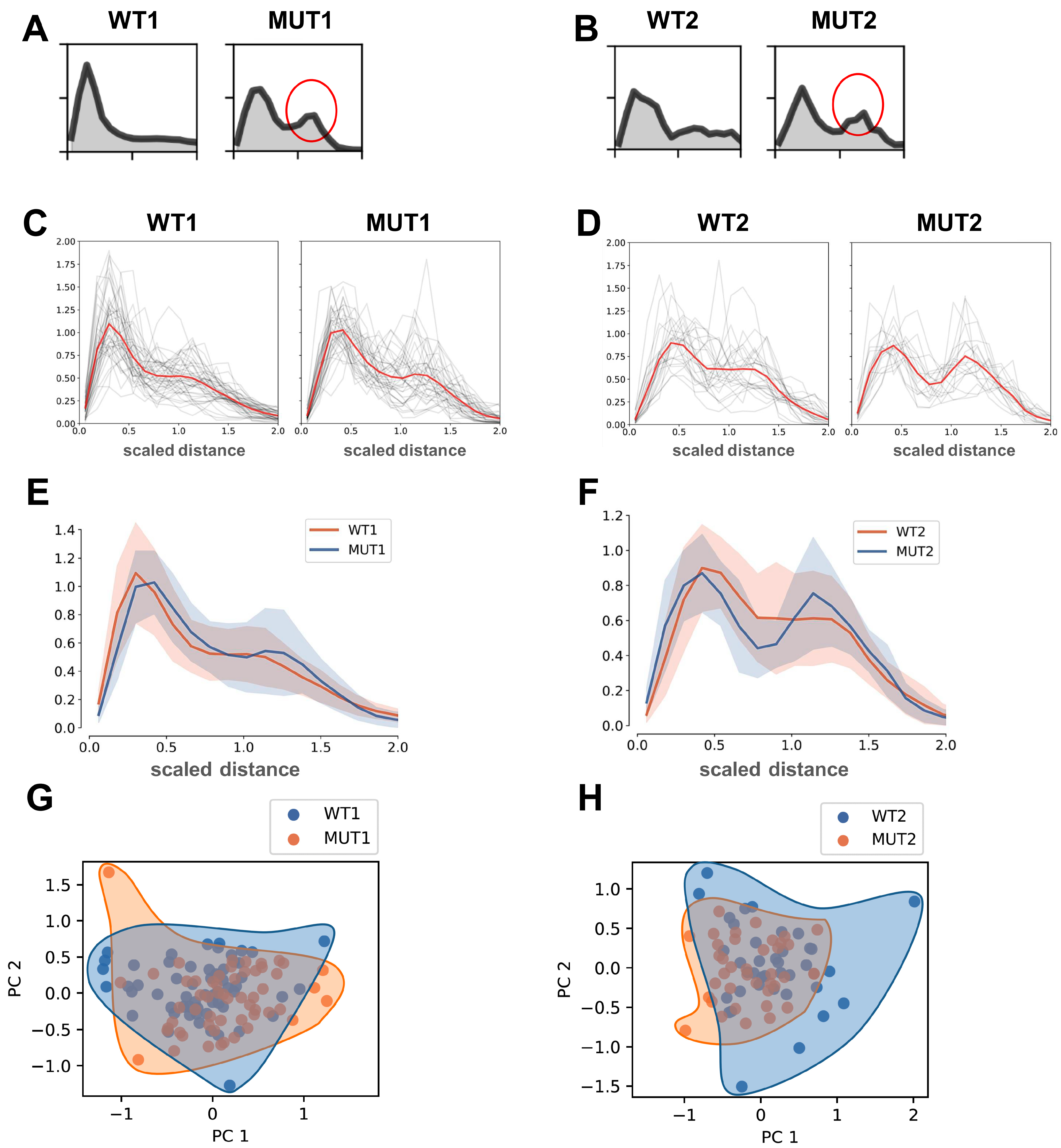
3.2. Analysis of Normalized Distance Distributions for DCM/MUT vs. WT
3.3. Spherical Modeling Recapitulates Altered Vesicular Distributions in MUT iPSC-CMs
3.4. Analysis of Distance Distributions for Vesicular Populations Using Spherical Modelling
3.5. Robustness of Distance Distributions Using 3D Spherical Modeling
3.6. Scaled Distancing of 3D Spherical Models Accurately Reflects Altered Distribution of Early Endosomes in MUT iPSC-CMs
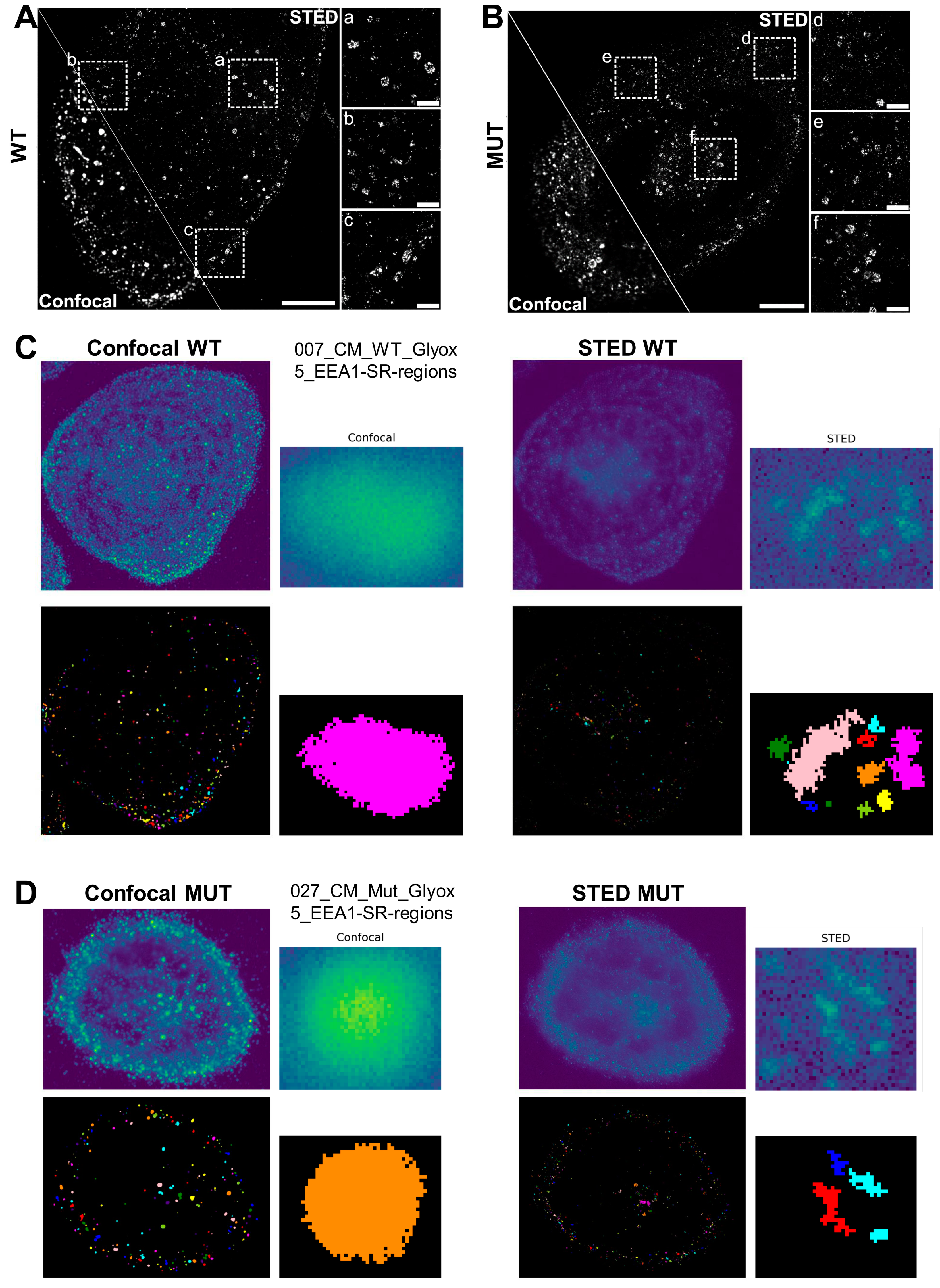
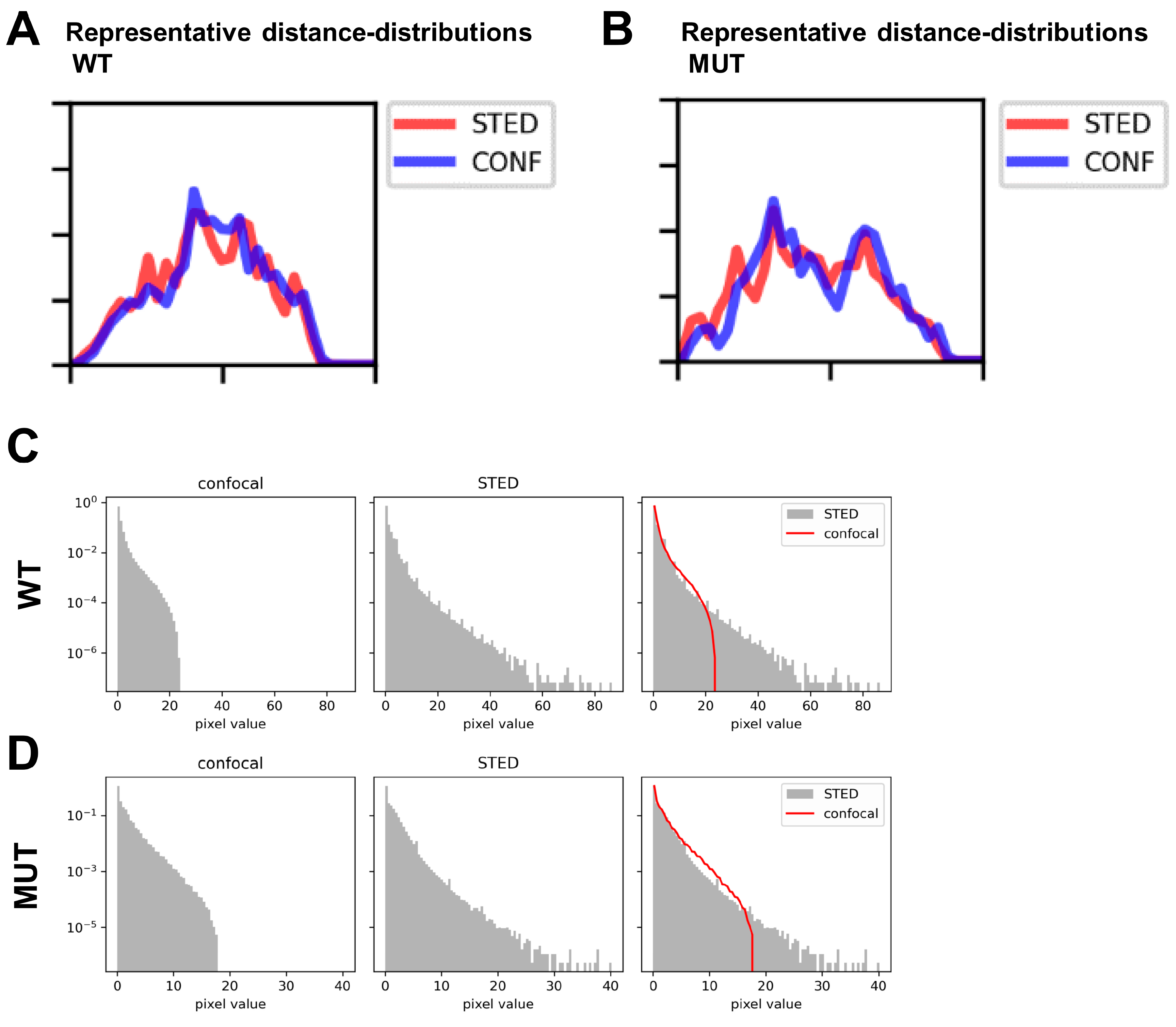
3.7. STED Super-Resolution Microscopy Combined with 3D Spherical Modeling Enables More Accurate Quantification of Vesicular Content and Distribution
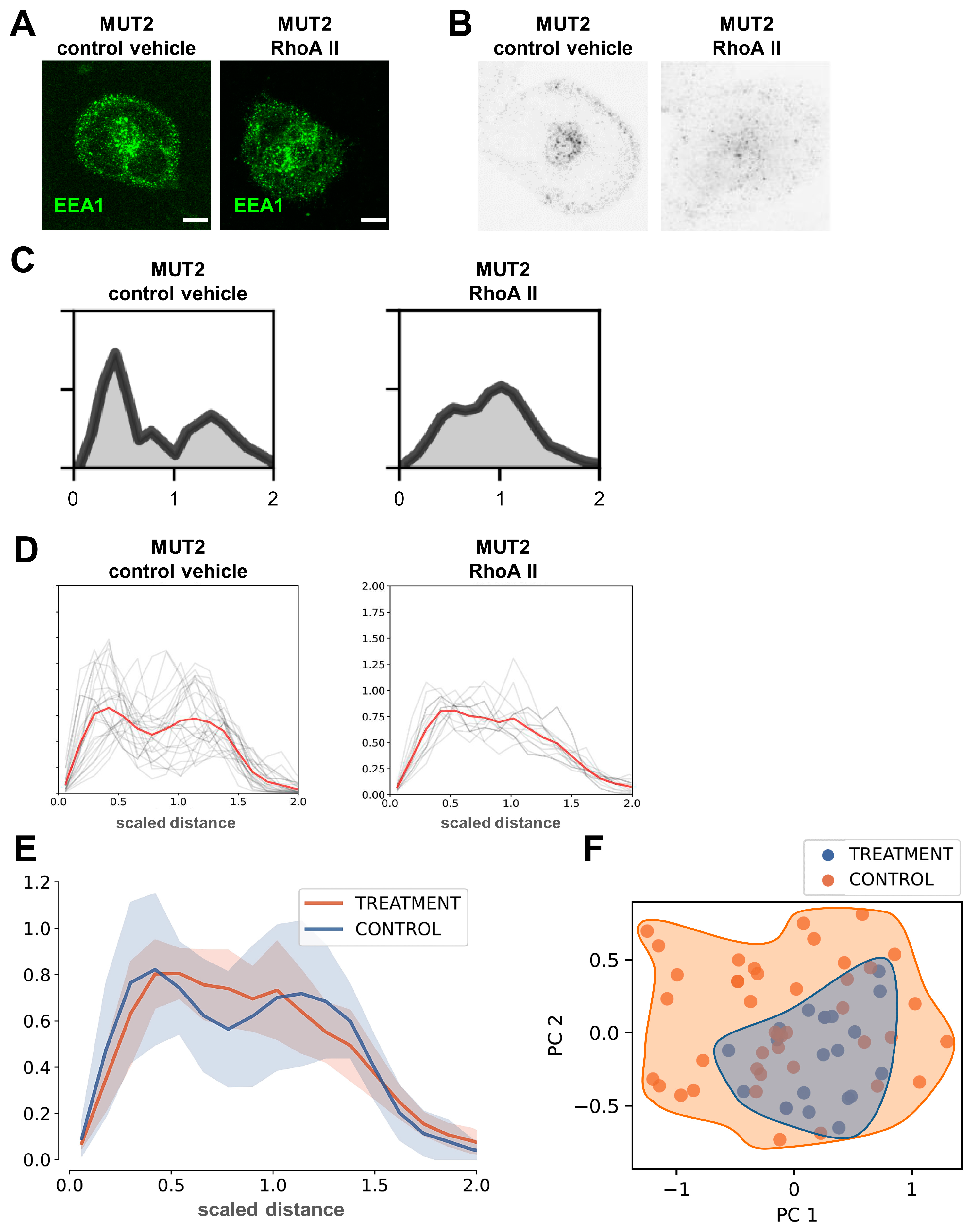
3.8. 3D Spherical Modeling Confirms Recovery of Vesicular Distributions in MUT iPSC-CMs following RhoA II Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yamanaka, S.; Takahashi, K. [Induction of pluripotent stem cells from mouse fibroblast cultures]. Tanpakushitsu Kakusan Koso 2006, 51, 2346–2351. [Google Scholar] [PubMed]
- Chen, G.; Gulbranson, D.R.; Hou, Z.; Bolin, J.M.; Ruotti, V.; Probasco, M.D.; Smuga-Otto, K.; Howden, S.E.; Diol, N.R.; Propson, N.E.; et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 2011, 8, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Vergara, X.; Sevilla, A.; D’Souza, S.L.; Ang, Y.S.; Schaniel, C.; Lee, D.F.; Yang, L.; Kaplan, A.D.; Adler, E.D.; Rozov, R.; et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature 2010, 465, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Yazawa, M.; Hsueh, B.; Jia, X.; Pasca, A.M.; Bernstein, J.A.; Hallmayer, J.; Dolmetsch, R.E. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature 2011, 471, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Moretti, A.; Bellin, M.; Welling, A.; Jung, C.B.; Lam, J.T.; Bott-Flugel, L.; Dorn, T.; Goedel, A.; Hohnke, C.; Hofmann, F.; et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 2010, 363, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Itzhaki, I.; Maizels, L.; Huber, I.; Zwi-Dantsis, L.; Caspi, O.; Winterstern, A.; Feldman, O.; Gepstein, A.; Arbel, G.; Hammerman, H.; et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature 2011, 471, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, P.; Lan, F.; Wu, H.; Lisowski, L.; Gu, M.; Hu, S.; Kay, M.A.; Urnov, F.D.; Shinnawi, R.; et al. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J. Am. Coll. Cardiol. 2014, 64, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Lee, A.S.; Liang, P.; Sanchez-Freire, V.; Nguyen, P.K.; Wang, L.; Han, L.; Yen, M.; Wang, Y.; Sun, N.; et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 2013, 12, 101–113. [Google Scholar] [CrossRef]
- Sun, N.; Yazawa, M.; Liu, J.; Han, L.; Sanchez-Freire, V.; Abilez, O.J.; Navarrete, E.G.; Hu, S.; Wang, L.; Lee, A.; et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 2012, 4, 130ra147. [Google Scholar] [CrossRef]
- Dai, Y.; Amenov, A.; Ignatyeva, N.; Koschinski, A.; Xu, H.; Soong, P.L.; Tiburcy, M.; Linke, W.A.; Zaccolo, M.; Hasenfuss, G.; et al. Troponin destabilization impairs sarcomere-cytoskeleton interactions in iPSC-derived cardiomyocytes from dilated cardiomyopathy patients. Sci. Rep. 2020, 10, 209. [Google Scholar] [CrossRef]
- Dai, Y.; Ignatyeva, N.; Xu, H.; Wali, R.; Toischer, K.; Brandenburg, S.; Lenz, C.; Pronto, J.; Fakuade, F.E.; Sossalla, S.; et al. An Alternative Mechanism of Subcellular Iron Uptake Deficiency in Cardiomyocytes. Circ. Res. 2023, 133, e19–e46. [Google Scholar] [CrossRef] [PubMed]
- Zerial, M.; McBride, H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001, 2, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Wandinger-Ness, A.; Zerial, M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 2014, 6, a022616. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.; Spannl, S.; Ferguson, C.; Hyman, A.A.; Parton, R.G.; Zerial, M. Rab5 and Alsin regulate stress-activated cytoprotective signaling on mitochondria. Elife 2018, 7, e32282. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wali, R.; Cheruiyot, C.; Bodenschatz, J.; Hasenfuss, G.; Janshoff, A.; Habeck, M.; Ebert, A. Non-negative blind deconvolution for signal processing in a CRISPR-edited iPSC-cardiomyocyte model of dilated cardiomyopathy. FEBS Lett. 2021, 595, 2544–2557. [Google Scholar] [CrossRef] [PubMed]
- Ebert, A.D.; Kodo, K.; Liang, P.; Wu, H.; Huber, B.C.; Riegler, J.; Churko, J.; Lee, J.; de Almeida, P.; Lan, F.; et al. Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system. Sci. Transl. Med. 2014, 6, 255ra130. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Hsiao, C.; Wilson, G.; Zhu, K.; Hazeltine, L.B.; Azarin, S.M.; Raval, K.K.; Zhang, J.; Kamp, T.J.; Palecek, S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E1848–E1857. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Zhang, J.; Azarin, S.M.; Zhu, K.; Hazeltine, L.B.; Bao, X.; Hsiao, C.; Kamp, T.J.; Palecek, S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat. Protoc. 2013, 8, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Ebert, A.; Joshi, A.U.; Andorf, S.; Dai, Y.; Sampathkumar, S.; Chen, H.; Li, Y.; Garg, P.; Toischer, K.; Hasenfuss, G.; et al. Proteasome-Dependent Regulation of Distinct Metabolic States During Long-Term Culture of Human iPSC-Derived Cardiomyocytes. Circ. Res. 2019, 125, 90–103. [Google Scholar] [CrossRef]
- Matsa, E.; Burridge, P.W.; Yu, K.H.; Ahrens, J.H.; Termglinchan, V.; Wu, H.; Liu, C.; Shukla, P.; Sayed, N.; Churko, J.M.; et al. Transcriptome Profiling of Patient-Specific Human iPSC-Cardiomyocytes Predicts Individual Drug Safety and Efficacy Responses In Vitro. Cell Stem Cell 2016, 19, 311–325. [Google Scholar] [CrossRef]
- Del Conte-Zerial, P.; Brusch, L.; Rink, J.C.; Collinet, C.; Kalaidzidis, Y.; Zerial, M.; Deutsch, A. Membrane identity and GTPase cascades regulated by toggle and cut-out switches. Mol. Syst. Biol. 2008, 4, 206. [Google Scholar] [CrossRef] [PubMed]
- Foret, L.; Dawson, J.E.; Villaseñor, R.; Collinet, C.; Deutsch, A.; Brusch, L.; Zerial, M.; Kalaidzidis, Y.; Jülicher, F. A general theoretical framework to infer endosomal network dynamics from quantitative image analysis. Curr. Biol. 2012, 22, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Eschenhagen, T.; Mummery, C.; Knollmann, B.C. Modelling sarcomeric cardiomyopathies in the dish: From human heart samples to iPSC cardiomyocytes. Cardiovasc. Res. 2015, 105, 424–438. [Google Scholar] [CrossRef] [PubMed]
- Klar, T.A.; Jakobs, S.; Dyba, M.; Egner, A.; Hell, S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. USA 2000, 97, 8206–8210. [Google Scholar] [CrossRef] [PubMed]
- Sahl, S.J.; Hell, S.W.; Jakobs, S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 2017, 18, 685–701. [Google Scholar] [CrossRef]
- Stephan, T.; Bruser, C.; Deckers, M.; Steyer, A.M.; Balzarotti, F.; Barbot, M.; Behr, T.S.; Heim, G.; Hubner, W.; Ilgen, P.; et al. MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J. 2020, 39, e104105. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saleem, H.N.; Ignatyeva, N.; Stuut, C.; Jakobs, S.; Habeck, M.; Ebert, A. 3D Computational Modeling of Defective Early Endosome Distribution in Human iPSC-Based Cardiomyopathy Models. Cells 2024, 13, 923. https://doi.org/10.3390/cells13110923
Saleem HN, Ignatyeva N, Stuut C, Jakobs S, Habeck M, Ebert A. 3D Computational Modeling of Defective Early Endosome Distribution in Human iPSC-Based Cardiomyopathy Models. Cells. 2024; 13(11):923. https://doi.org/10.3390/cells13110923
Chicago/Turabian StyleSaleem, Hafiza Nosheen, Nadezda Ignatyeva, Christiaan Stuut, Stefan Jakobs, Michael Habeck, and Antje Ebert. 2024. "3D Computational Modeling of Defective Early Endosome Distribution in Human iPSC-Based Cardiomyopathy Models" Cells 13, no. 11: 923. https://doi.org/10.3390/cells13110923
APA StyleSaleem, H. N., Ignatyeva, N., Stuut, C., Jakobs, S., Habeck, M., & Ebert, A. (2024). 3D Computational Modeling of Defective Early Endosome Distribution in Human iPSC-Based Cardiomyopathy Models. Cells, 13(11), 923. https://doi.org/10.3390/cells13110923






