Cardiac Development Long Non-Coding RNA (CARDEL) Is Activated during Human Heart Development and Contributes to Cardiac Specification and Homeostasis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Cardiomyocyte Differentiation
2.2. Lines with Doxycycline-Inducible Expression and Knock-Out Lines
2.3. Karyotype
2.4. Flow Cytometry
2.5. RNA Isolation and RT-qPCR
2.6. Whole Cell Patch Clamp
2.7. RNA-seq and Data Analysis
2.8. Data and Statistical Analysis
3. Results
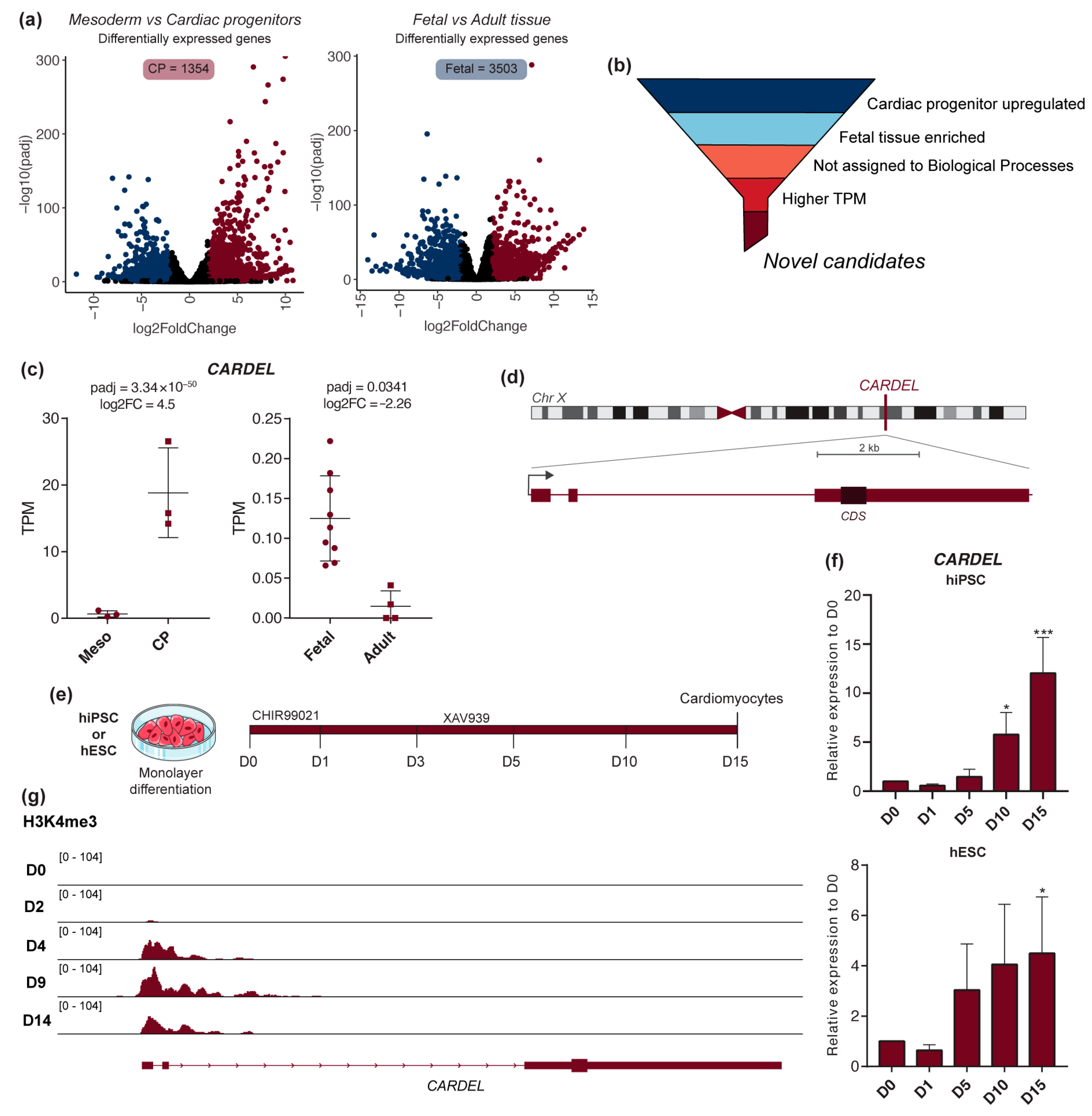
3.1. CARDEL Is Expressed during the Human Cardiac-Lineage Specification
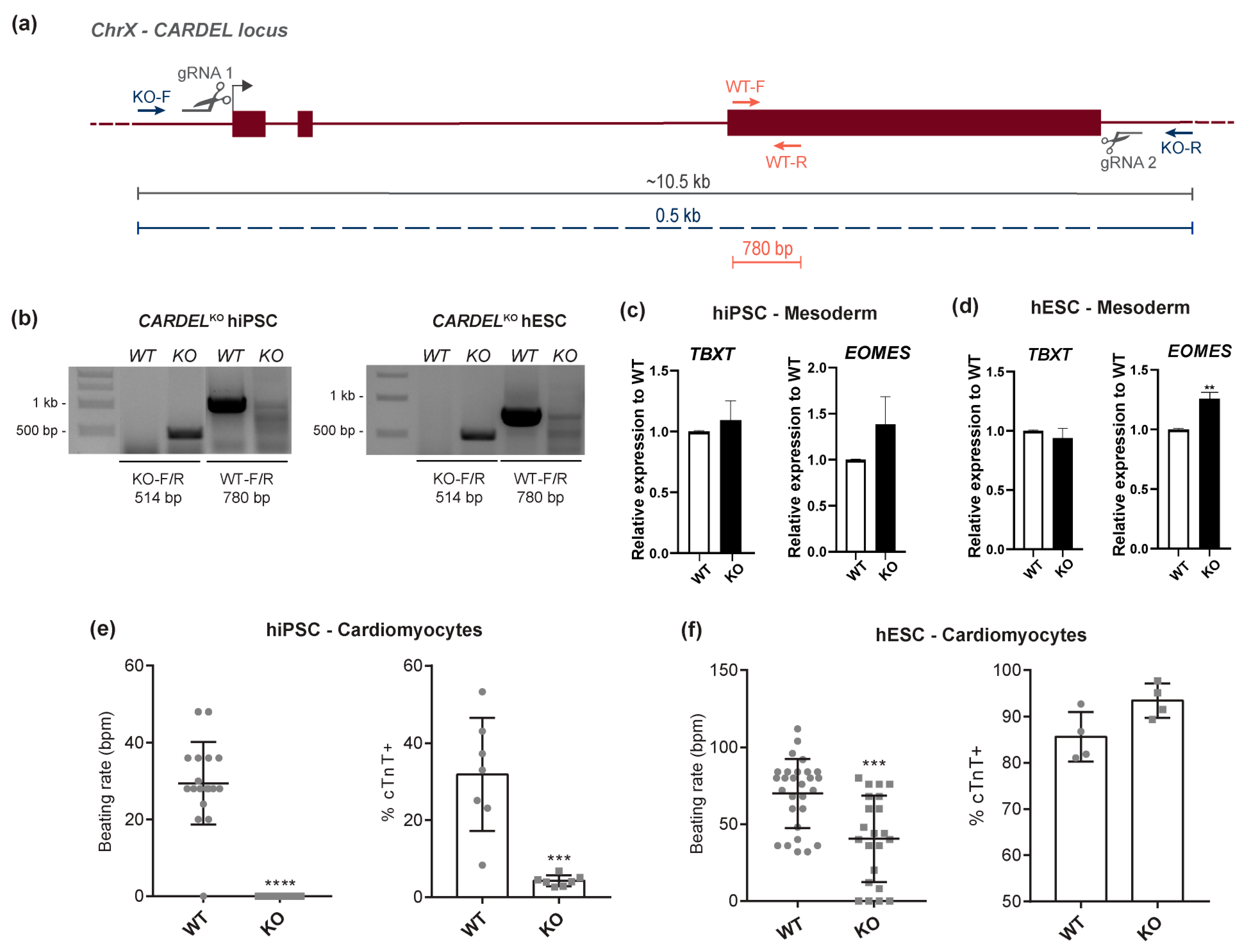
3.2. Cardiomyocyte Differentiation and Homeostasis Were Impaired in CARDELKO Cells
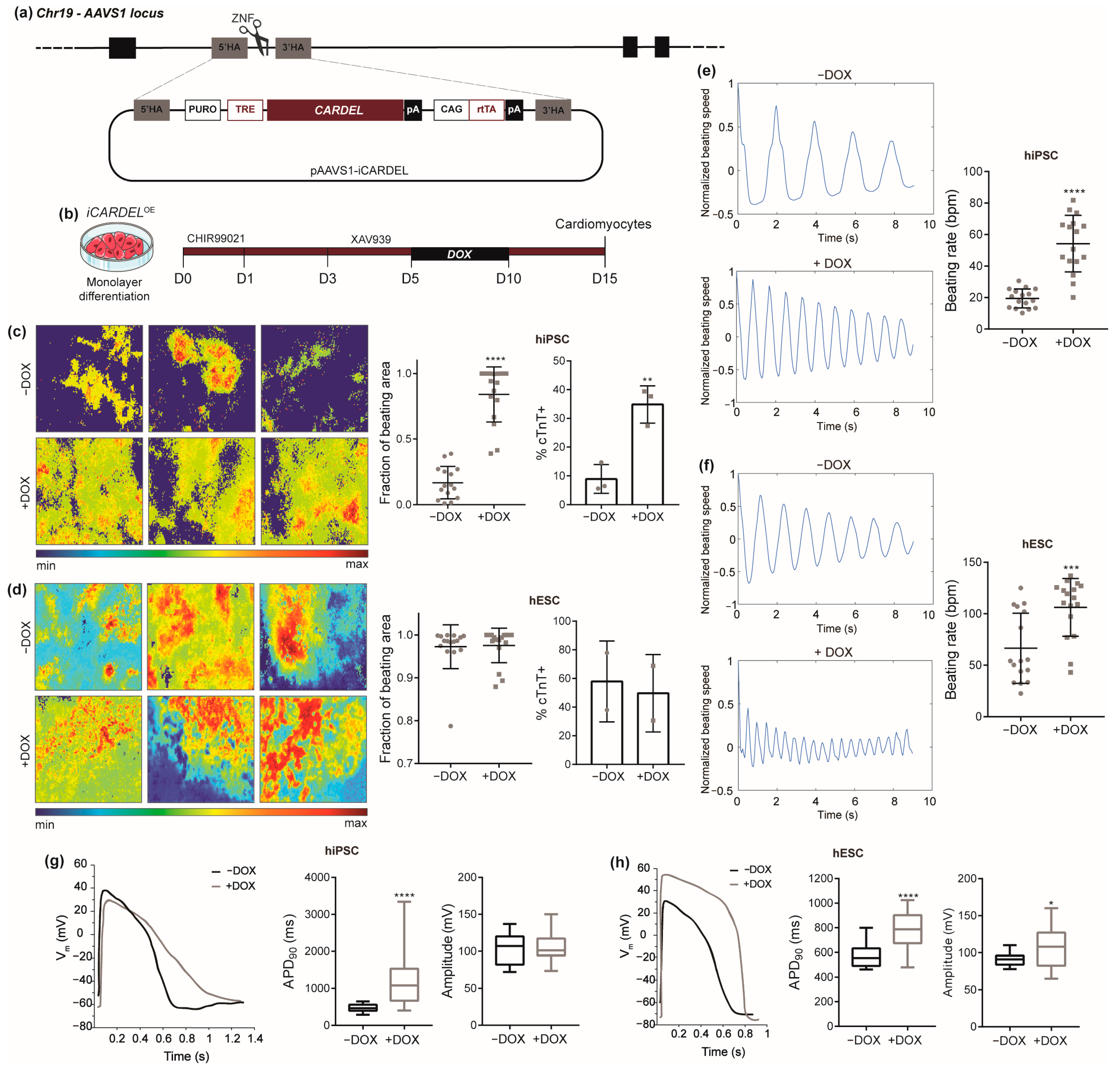
3.3. Overexpression of CARDEL Improves hPSC Cardiomyogenic Differentiation and Alters Cardiomyocyte Functional Properties
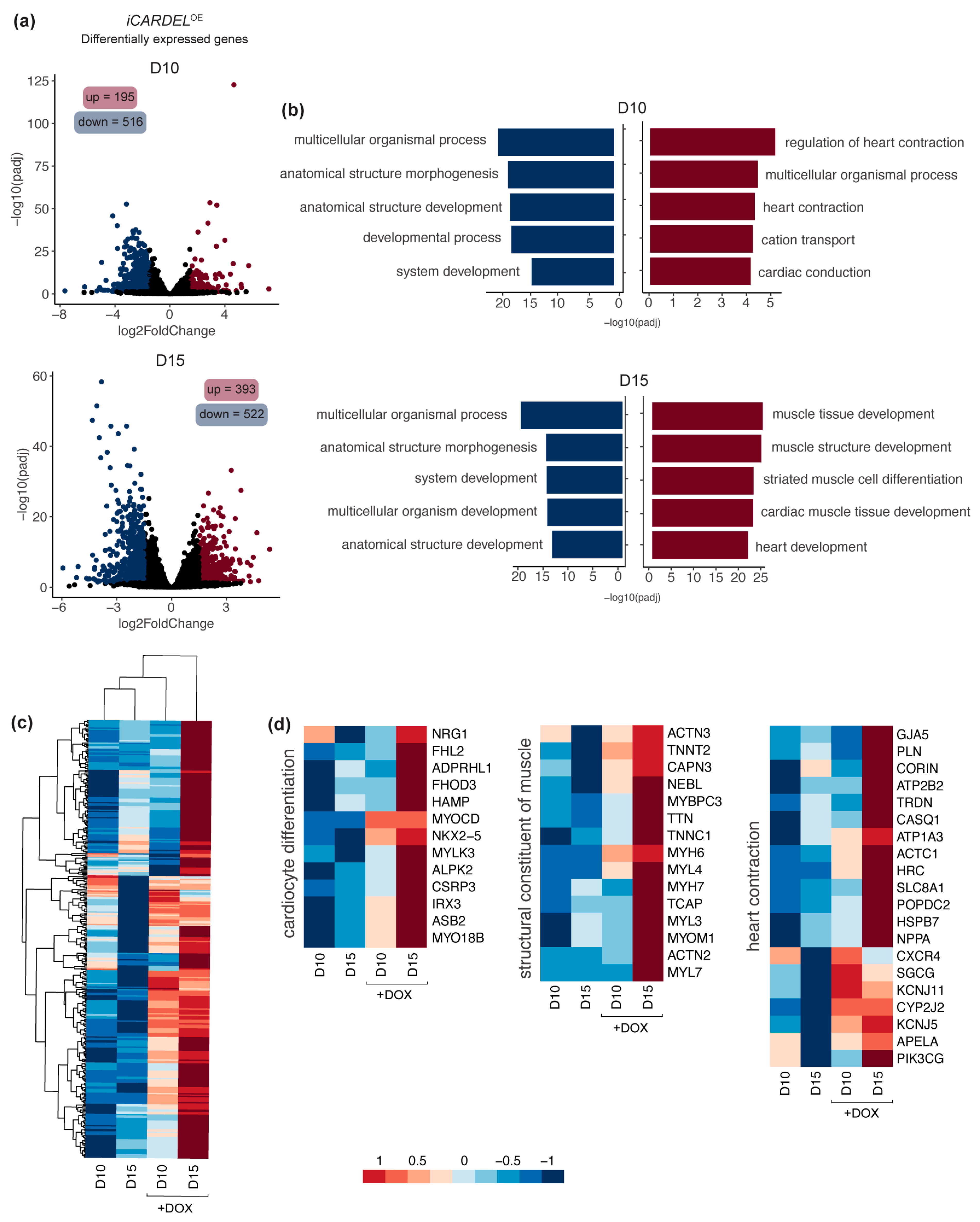
3.4. CARDEL Expression Contributes to Physiological and Molecular Changes during Cardiomyocyte Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meilhac, S.M.; Buckingham, M.E. The Deployment of Cell Lineages That Form the Mammalian Heart. Nat. Rev. Cardiol. 2018, 15, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Spater, D.; Hansson, E.M.; Zangi, L.; Chien, K.R. How to Make a Cardiomyocyte. Development 2014, 141, 4418–4431. [Google Scholar] [CrossRef]
- Olson, E.N. Gene Regulatory Networks in the Evolution and Development of the Heart. Science 2006, 313, 1922–1927. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, P.; Wu, S.M.; Zaffran, S.; Pucéat, M. Early Cardiac Development: A View from Stem Cells to Embryos. Cardiovasc. Res. 2012, 96, 352–362. [Google Scholar] [CrossRef] [PubMed]
- Protze, S.I.; Lee, J.H.; Keller, G.M. Human Pluripotent Stem Cell-Derived Cardiovascular Cells: From Developmental Biology to Therapeutic Applications. Cell Stem Cell 2019, 25, 311–327. [Google Scholar] [CrossRef]
- Evans, S.M.; Yelon, D.; Conlon, F.L.; Kirby, M.L. Myocardial Lineage Development. Circ. Res. 2010, 107, 1428–1444. [Google Scholar] [CrossRef]
- Feng, H.Z.; Jin, J.P. A Protocol to Study Ex Vivo Mouse Working Heart at Human-like Heart Rate. J. Mol. Cell. Cardiol. 2018, 114, 175–184. [Google Scholar] [CrossRef]
- Su, Z.; Li, F.; Spitzer, K.W.; Yao, A.; Ritter, M.; Barry, W.H. Comparison of Sarcoplasmic Reticulum Ca2+-ATPase Function in Human, Dog, Rabbit, and Mouse Ventricular Myocytes. J. Mol. Cell. Cardiol. 2003, 35, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Sadayappan, S.; Gulick, J.; Klevitsky, R.; Lorenz, J.N.; Sargent, M.; Molkentin, J.D.; Robbins, J. Cardiac Myosin Binding Protein-C Phosphorylation in a β-Myosin Heavy Chain Background. Circulation 2009, 119, 1253–1262. [Google Scholar] [CrossRef]
- Devalla, H.D.; Passier, R. Cardiac Differentiation of Pluripotent Stem Cells and Implications for Modeling the Heart in Health and Disease. Sci. Transl. Med. 2018, 10, eaah5457. [Google Scholar] [CrossRef]
- Hofbauer, P.; Jahnel, S.M.; Mendjan, S. In Vitro Models of the Human Heart. Development 2021, 148, dev199672. [Google Scholar] [CrossRef]
- Leitolis, A.; Robert, A.W.; Pereira, I.T.; Correa, A.; Stimamiglio, M.A.; Carr, C.; Robert, A.W. Cardiomyogenesis Modeling Using Pluripotent Stem Cells: The Role of Microenvironmental Signaling. Front. Cell Dev. Biol. 2019, 7, 164. [Google Scholar] [CrossRef] [PubMed]
- Buijtendijk, M.F.J.; Barnett, P.; Hoff, M.J.B. Development of the Human Heart. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Darabi, R.; Arpke, R.W.; Irion, S.; Dimos, J.T.; Grskovic, M.; Kyba, M.; Perlingeiro, R.C.R. Human ES- and IPS-Derived Myogenic Progenitors Restore DYSTROPHIN and Improve Contractility upon Transplantation in Dystrophic Mice. Cell Stem Cell 2012, 10, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Zhang, J.; Azarin, S.M.; Zhu, K.; Hazeltine, L.B.; Bao, X.; Hsiao, C.; Kamp, T.J.; Palecek, S.P. Directed Cardiomyocyte Differentiation from Human Pluripotent Stem Cells by Modulating Wnt/β-Catenin Signaling under Fully Defined Conditions. Nat. Protoc. 2012, 8, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Huebsch, N.; Loskill, P.; Mandegar, M.A.; Marks, N.C.; Sheehan, A.S.; Ma, Z.; Mathur, A.; Nguyen, T.N.; Yoo, J.C.; Judge, L.M.; et al. Automated Video-Based Analysis of Contractility and Calcium Flux in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Cultured over Different Spatial Scales. Tissue Eng. Part. C Methods 2015, 21, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Pereira, I.T.; Spangenberg, L.; Robert, A.W.; Amorín, R.; Stimamiglio, M.A.; Naya, H.; Dallagiovanna, B. Polysome Profiling Followed by RNA-Seq of Cardiac Differentiation Stages in HESCs. Sci. Data 2018, 5, 180287. [Google Scholar] [CrossRef] [PubMed]
- Pervolaraki, E.; Dachtler, J.; Anderson, R.A.; Holden, A.V. The Developmental Transcriptome of the Human Heart. Sci. Rep. 2018, 8, 15362. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R.; Osborne, C.S. Large Scale Loss of Data in Low-Diversity Illumina Sequencing Libraries Can Be Recovered by Deferred Cluster Calling. PLoS ONE 2011, 6, e16607. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Paige, S.L.; Thomas, S.; Stoick-Cooper, C.L.; Wang, H.; Maves, L.; Sandstrom, R.; Pabon, L.; Reinecke, H.; Pratt, G.; Keller, G.; et al. A Temporal Chromatin Signature in Human Embryonic Stem Cells Identifies Regulators of Cardiac Development. Cell 2012, 151, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Bedada, F.B.; Chan, S.S.K.; Metzger, S.K.; Zhang, L.; Zhang, J.; Garry, D.J.; Kamp, T.J.; Kyba, M.; Metzger, J.M. Acquisition of a Quantitative, Stoichiometrically Conserved Ratiometric Marker of Maturation Status in Stem Cell-Derived Cardiac Myocytes. Stem Cell Rep. 2014, 3, 594–605. [Google Scholar] [CrossRef]
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. Lncipedia 5: Towards a Reference Set of Human Long Non-Coding Rnas. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef] [PubMed]
- D’Antonio, M.; Nguyen, J.P.; Arthur, T.D.; Matsui, H.; Donovan, M.K.R.; D’Antonio-Chronowska, A.; Frazer, K.A. In Heart Failure Reactivation of RNA-Binding Proteins Is Associated with the Expression of 1523 Fetal-Specific Isoforms. PLoS Comput. Biol. 2022, 18, e1009918. [Google Scholar] [CrossRef] [PubMed]
- Hartman, M.E.; Dai, D.F.; Laflamme, M.A. Human Pluripotent Stem Cells: Prospects and Challenges as a Source of Cardiomyocytes for in Vitro Modeling and Cell-Based Cardiac Repair. Adv. Drug Deliv. Rev. 2016, 96, 3–17. [Google Scholar] [CrossRef]
- den Hartogh, S.C.; Wolstencroft, K.; Mummery, C.L.; Passier, R. A Comprehensive Gene Expression Analysis at Sequential Stages of in Vitro Cardiac Differentiation from Isolated MESP1-Expressing-Mesoderm Progenitors. Sci. Rep. 2016, 6, 19386. [Google Scholar] [CrossRef]
- Fu, K.; Nakano, H.; Morselli, M.; Chen, T.; Pappoe, H.; Nakano, A.; Pellegrini, M. A Temporal Transcriptome and Methylome in Human Embryonic Stem Cell-Derived Cardiomyocytes Identifies Novel Regulators of Early Cardiac Development. Epigenetics 2018, 13, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Kurian, L.; Aguirre, A.; Sancho-Martinez, I.; Benner, C.; Hishida, T.; Nguyen, T.B.; Reddy, P.; Nivet, E.; Krause, M.N.; Nelles, D.A.; et al. Identification of Novel Long Noncoding RNAs Underlying Vertebrate Cardiovascular Development. Circulation 2015, 131, 1278–1290. [Google Scholar] [CrossRef]
- Li, Y.; Lin, B.; Yang, L. Comparative Transcriptomic Analysis of Multiple Cardiovascular Fates from Embryonic Stem Cells Predicts Novel Regulators in Human Cardiogenesis. Sci. Rep. 2015, 5, 9758. [Google Scholar] [CrossRef] [PubMed]
- Pereira, I.T.; Spangenberg, L.; Robert, A.W.; Amorín, R.; Stimamiglio, M.A.; Naya, H.; Dallagiovanna, B. Cardiomyogenic Differentiation Is Fine-Tuned by Differential MRNA Association with Polysomes. BMC Genom. 2019, 20, 219. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, J.D.; Jung, M.; Chen, C.Y.; Lin, Z.; Ye, J.; Godatha, S.; Lizhar, E.; Wu, X.; Hsu, D.; Couture, L.A.; et al. Mapping Human Pluripotent-to-Cardiomyocyte Differentiation: Methylomes, Transcriptomes, and Exon DNA Methylation “Memories”. eBioMedicine 2016, 4, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Q.; Soo, S.Y.; Sun, W.; Zweigerdt, R. Global Expression Profile of Highly Enriched Cardiomyocytes Derived from Human Embryonic Stem Cells. Stem Cells 2009, 27, 2163–2174. [Google Scholar] [CrossRef]
- Uchida, S.; Schneider, A.; Wiesnet, M.; Jungblut, B.; Zarjitskaya, P.; Jenniches, K.; Kreymborg, K.G.; Seeger, W.; Braun, T. An Integrated Approach for the Systematic Identification and Characterization of Heart-Enriched Genes with Unknown Functions. BMC Genom. 2009, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-Specific Expression by Genome-Wide Integration of Transcriptomics and Antibody-Based Proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.-D.; Khorram, O. Expression Profiling of IncRNAs, MiRNAs, and MRNAs and Their Differential Expression in Leiomyoma Using Next-Generation RNA Sequencing. Reprod. Sci. 2018, 25, 246–255. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Nakagawa, S. Lessons from Reverse-Genetic Studies of LncRNAs. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Hartford, C.C.R.; Lal, A. When Long Noncoding Becomes Protein Coding. Mol. Cell. Biol. 2020, 40, e00528-19. [Google Scholar] [CrossRef] [PubMed]
- Yeasmin, F.; Yada, T.; Akimitsu, N. Micropeptides Encoded in Transcripts Previously Identified as Long Noncoding RNAs: A New Chapter in Transcriptomics and Proteomics. Front. Genet. 2018, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Acosta, R.; Addleman, N.J.; Adrian, J.; Afzal, V.; Aken, B.; Akiyama, J.A.; Al Jammal, O.; Amrhein, H.; Anderson, S.M.; et al. Expanded Encyclopaedias of DNA Elements in the Human and Mouse Genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Ulitsky, I. Methods for Distinguishing between Protein-Coding and Long Noncoding RNAs and the Elusive Biological Purpose of Translation of Long Noncoding RNAs. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Mackowiak, S.D.; Zauber, H.; Bielow, C.; Thiel, D.; Kutz, K.; Calviello, L.; Mastrobuoni, G.; Rajewsky, N.; Kempa, S.; Selbach, M.; et al. Extensive Identification and Analysis of Conserved Small ORFs in Animals. Genome Biol. 2015, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260.e29. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Samuels, D.C.; Zhao, S.; Xiang, Y.; Zhao, Y.Y.; Guo, Y. Current Research on Non-Coding Ribonucleic Acid (RNA). Genes 2017, 8, 366. [Google Scholar] [CrossRef] [PubMed]
- Gaertner, B.; van Heesch, S.; Schneider-Lunitz, V.; Schulz, J.F.; Witte, F.; Blachut, S.; Nguyen, S.; Wong, R.; Matta, I.; Hübner, N.; et al. A Human ESC-Based Screen Identifies a Role for the Translated LncRNA LINC00261 in Pancreatic Endocrine Differentiation. eLife 2020, 9, e58659. [Google Scholar] [CrossRef]
- Broadwell, L.J.; Smallegan, M.J.; Rigby, K.M.; Navarro-Arriola, J.S.; Montgomery, R.L.; Rinn, J.L.; Leinwand, L.A. Myosin 7b Is a Regulatory Long Noncoding RNA (LncMYH7b) in the Human Heart. J. Biol. Chem. 2021, 296, 100694. [Google Scholar] [CrossRef]
- Devaux, Y.; Zangrando, J.; Schroen, B.; Creemers, E.E.; Pedrazzini, T.; Chang, C.P.; Dorn, G.W.; Thum, T.; Heymans, S. Long Noncoding RNAs in Cardiac Development and Ageing. Nat. Rev. Cardiol. 2015, 12, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Huo, C.; Ding, N.; Li, J.; Xiao, J.; Lin, X.; Cai, B.; Zhang, Y.; Xu, J. Dynamic Organization of LncRNA and Circular RNA Regulators Collectively Controlled Cardiac Differentiation in Humans. eBioMedicine 2017, 24, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Pajares, V. Long Non-Coding RNA Regulation of Gene Expression during Differentiation. Pflug. Arch. 2016, 468, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Pereira, I.T.; Spangenberg, L.; Cabrera, G.; Dallagiovanna, B. Polysome-Associated LncRNAs during Cardiomyogenesis of HESCs. Mol. Cell. Biochem. 2020, 468, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The Tissue-Specific LncRNA Fendrr Is an Essential Regulator of Heart and Body Wall Development in the Mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef]
- Klattenhoff, C.A.; Scheuermann, J.C.; Surface, L.E.; Bradley, R.K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a Long Noncoding RNA Required for Cardiovascular Lineage Commitment. Cell 2013, 152, 570–583. [Google Scholar] [CrossRef]
- Ounzain, S.; Micheletti, R.; Beckmann, T.; Schroen, B.; Alexanian, M.; Pezzuto, I.; Crippa, S.; Nemir, M.; Sarre, A.; Johnson, R.; et al. Genome-Wide Profiling of the Cardiac Transcriptome after Myocardial Infarction Identifies Novel Heart-Specific Long Non-Coding RNAs. Eur. Heart J. 2015, 36, 353–368. [Google Scholar] [CrossRef]
- Dirkx, E.; da Costa Martins, P.A.; De Windt, L.J. Regulation of Fetal Gene Expression in Heart Failure. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2013, 1832, 2414–2424. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Collyer, J.; Wang, M.; Sun, F.; Xu, F. Genetic Dissection of Hypertrophic Cardiomyopathy with Myocardial RNA-Seq. Int. J. Mol. Sci. 2020, 21, 3040. [Google Scholar] [CrossRef]
- Caruso, M.; Meurant, S.; Detraux, D.; Mathieu, A.; Gilson, M.; Dieu, M.; Fattaccioli, A.; Demazy, C.; Najimi, M.; Sokal, E.; et al. The Global Downregulation of Protein Synthesis Observed during Hepatogenic Maturation Is Associated with a Decrease in TOP MRNA Translation. Stem Cell Rep. 2023, 18, 254–268. [Google Scholar] [CrossRef]
- Senichkin, V.V.; Prokhorova, E.A.; Zhivotovsky, B.; Kopeina, G.S. Simple and Efficient Protocol for Subcellular Fractionation of Normal and Apoptotic Cells. Cells 2021, 10, 852. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, I.T.; Gomes-Júnior, R.; Hansel-Frose, A.; França, R.S.V.; Liu, M.; Soliman, H.A.N.; Chan, S.S.K.; Dudley, S.C., Jr.; Kyba, M.; Dallagiovanna, B. Cardiac Development Long Non-Coding RNA (CARDEL) Is Activated during Human Heart Development and Contributes to Cardiac Specification and Homeostasis. Cells 2024, 13, 1050. https://doi.org/10.3390/cells13121050
Pereira IT, Gomes-Júnior R, Hansel-Frose A, França RSV, Liu M, Soliman HAN, Chan SSK, Dudley SC Jr., Kyba M, Dallagiovanna B. Cardiac Development Long Non-Coding RNA (CARDEL) Is Activated during Human Heart Development and Contributes to Cardiac Specification and Homeostasis. Cells. 2024; 13(12):1050. https://doi.org/10.3390/cells13121050
Chicago/Turabian StylePereira, Isabela T., Rubens Gomes-Júnior, Aruana Hansel-Frose, Rhaíza S. V. França, Man Liu, Hossam A. N. Soliman, Sunny S. K. Chan, Samuel C. Dudley, Jr., Michael Kyba, and Bruno Dallagiovanna. 2024. "Cardiac Development Long Non-Coding RNA (CARDEL) Is Activated during Human Heart Development and Contributes to Cardiac Specification and Homeostasis" Cells 13, no. 12: 1050. https://doi.org/10.3390/cells13121050
APA StylePereira, I. T., Gomes-Júnior, R., Hansel-Frose, A., França, R. S. V., Liu, M., Soliman, H. A. N., Chan, S. S. K., Dudley, S. C., Jr., Kyba, M., & Dallagiovanna, B. (2024). Cardiac Development Long Non-Coding RNA (CARDEL) Is Activated during Human Heart Development and Contributes to Cardiac Specification and Homeostasis. Cells, 13(12), 1050. https://doi.org/10.3390/cells13121050








