Enhanced In Vitro Recapitulation of In Vivo Liver Regeneration by Co-Culturing Hepatocyte Organoids with Adipose-Derived Mesenchymal Stem Cells, Alleviating Steatosis and Apoptosis in Acute Alcoholic Liver Injury
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statements
2.2. Cultivation of HOs
2.3. Induction of an ALI Model
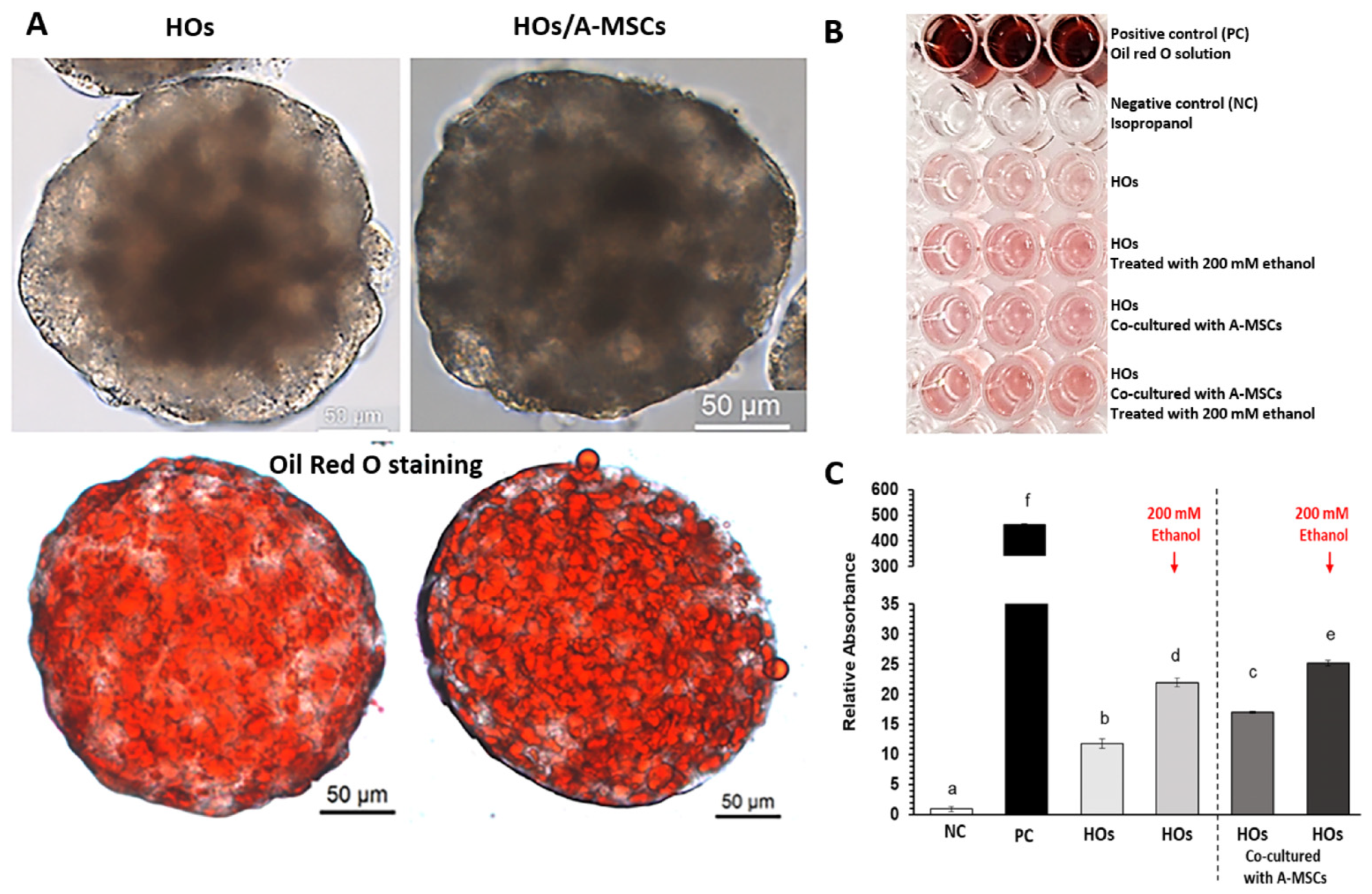
2.4. Oil Red O Staining
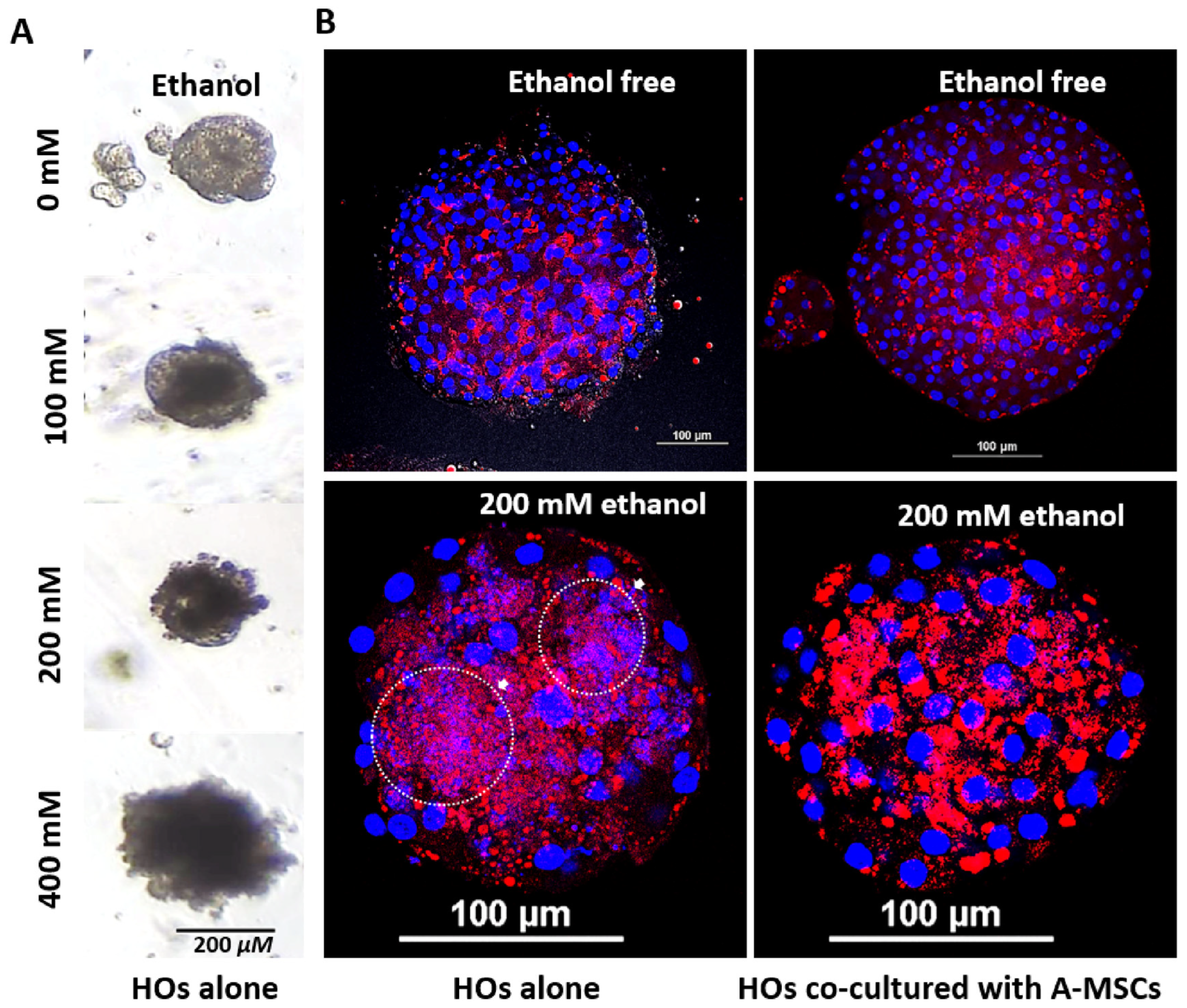
2.5. Nile Red Staining
2.6. Transcriptome and Differential Gene Expression Analysis
2.7. Real-Time Reverse-Transcription Quantitative PCR (Real-Time qPCR)
2.8. Statistical Analyses
3. Results
3.1. Transcriptome Analysis in HOs
3.2. Comprehensive Transcriptomic Analysis of Liver Factors and Metabolism in HOs
3.3. Transcriptomic Analysis of Key Factors in the 3D Tissue Architecture Formation of HOs
3.4. Effect of A-MSCs Co-Culture on Ethanol-Induced Hepatic Lipid Accumulation in HOs
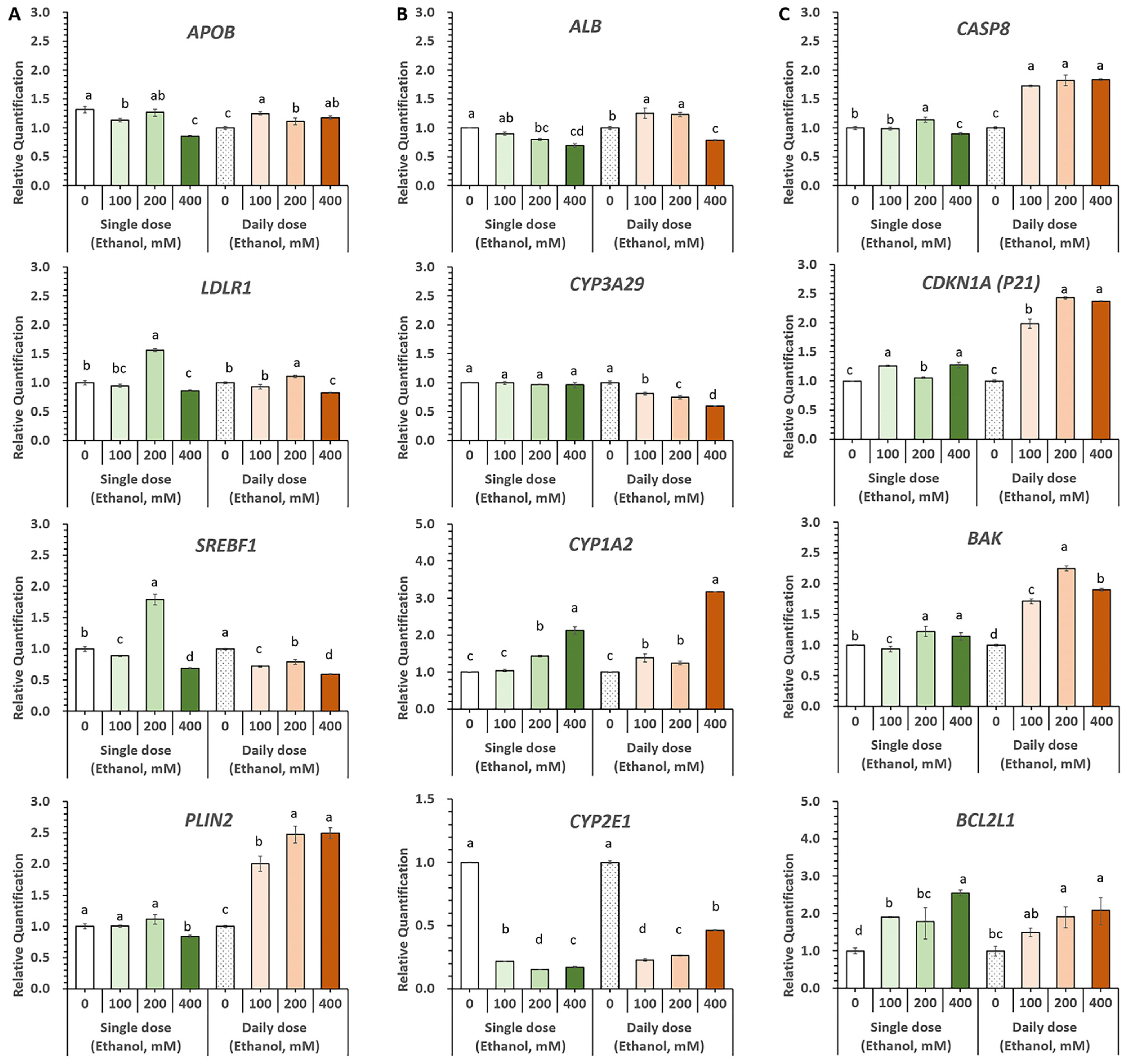
3.5. Evaluation of Alcohol-Induced Liver Injury Models Using Diverse Ethanol Exposure Methods in HOs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Levy, G.; Bomze, D.; Heinz, S.; Ramachandran, S.D.; Noerenberg, A.; Cohen, M.; Shibolet, O.; Sklan, E.; Braspenning, J.; Nahmias, Y. Long-term culture and expansion of primary human hepatocytes. Nat. Biotechnol. 2015, 33, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Huch, M.; Gehart, H.; van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.; Ellis, E.; van Wenum, M.; Fuchs, S.A.; de Ligt, J.; et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Gehart, H.; Artegiani, B.; LÖpez-Iglesias, C.; Dekkers, F.; Basak, O.; van Es, J.; Chuva de Sousa Lopes, S.M.; Begthel, H.; Korving, J.; et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 2018, 175, 1591–1606.e19. [Google Scholar] [CrossRef]
- Ock, S.A.; Kim, S.Y.; Ju, W.S.; Kim, Y.I.; Wi, H.Y.; Lee, P. Adipose tissue-derived mesenchymal stem cells extend the lifespan and enhance liver function in hepatocyte organoids. Int. J. Mol. Sci. 2023, 24, 15429. [Google Scholar] [CrossRef] [PubMed]
- Asai, A.; Aihara, E.; Watson, C.; Mourya, R.; Mizuochi, T.; Shivakumar, P.; Phelan, K.; Mayhew, C.; Helmrath, M.; Takebe, T.; et al. Paracrine signals regulate human liver organoid maturation from induced pluripotent stem cells. Development 2017, 144, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- He, Y.T.; Zhu, X.L.; Li, S.F.; Zhang, B.Q.; Li, Y.; Wu, Q.; Zhang, Y.L.; Zhou, Y.Y.; Li, L.; Qi, Y.N.; et al. Creating rat hepatocyte organoid as an in vitro model for drug testing. World J. Stem Cells 2020, 12, 1184–1195. [Google Scholar] [CrossRef]
- Ceni, E.; Mello, T.; Galli, A. Pathogenesis of alcoholic liver disease: Role of oxidative metabolism. World J. Gastroenterol. 2014, 20, 17756–17772. [Google Scholar] [CrossRef]
- Ghosh Dastidar, S.; Warner, J.B.; Warner, D.R.; McClain, C.J.; Kirpich, I.A. Rodent models of alcoholic liver disease: Role of binge ethanol administration. Biomolecules 2018, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Testino, G.; Burra, P.; Bonino, F.; Piani, F.; Sumberaz, A.; Peressutti, R.; Giannelli Castiglione, A.; Patussi, V.; Fanucchi, T.; Ancarani, O.; et al. Acute alcoholic hepatitis, end stage alcoholic liver disease and liver transplantation: An Italian position statement. World J. Gastroenterol. 2014, 20, 14642–14651. [Google Scholar] [CrossRef]
- Hendriks, D.; Brouwers, J.F.; Hamer, K.; Geurts, M.H.; Luciana, L.; Massalini, S.; López-Iglesias, C.; Peters, P.J.; Rodríguez-Colman, M.J.; Chuva de Sousa Lopes, S.; et al. Engineered human hepatocyte organoids enable CRISPR-based target discovery and drug screening for steatosis. Nat. Biotechnol. 2023, 41, 1567–1581. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Huang, C.; Meng, X.; Li, J. CYP1A2 contributes to alcohol-induced abnormal lipid metabolism through the PTEN/AKT/SREBP-1c pathway. Biochem. Biophys. Res. Commun. 2019, 513, 509–514. [Google Scholar] [CrossRef]
- Angireddy, R.; Chowdhury, A.R.; Zielonka, J.; Ruthel, G.; Kalyanaraman, B.; Avadhani, N.G. Alcohol-induced CYP2E1, mitochondrial dynamics and retrograde signaling in human hepatic 3D organoids. Free Radic. Biol. Med. 2020, 159, 1–14. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Perincheri, S.; Dingle, R.W.; Peterson, M.L.; Spear, B.T. Hereditary persistence of alpha-fetoprotein and H19 expression in liver of BALB/cJ mice is due to a retrovirus insertion in the Zhx2 gene. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, J.; Zhang, H.; Wang, J.; Cai, H.; Li, C.; Li, K.; Liu, J.; Guo, X.; Zou, G.; et al. Integrated transcriptional and proteomic analysis with in vitro biochemical assay reveal the important role of CYP3A46 in T-2 toxin hydroxylation in porcine primary hepatocytes. Mol. Cell. Proteomics 2011, 10, M111.008748. [Google Scholar] [CrossRef]
- Wu, J.; Chen, R.; Zhang, C.; Li, K.; Xu, W.; Wang, L.; Chen, Q.; Mu, P.; Jiang, J.; Wen, J.; et al. Bioactivation and regioselectivity of pig cytochrome P450 3A29 towards aflatoxin b1. Toxins 2016, 8, 267. [Google Scholar] [CrossRef] [PubMed]
- Huch, M.; Dorrell, C.; Boj, S.F.; van Es, J.H.; Li, V.S.; van de Wetering, M.; Sato, T.; Hamer, K.; Sasaki, N.; Finegold, M.J.; et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013, 494, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Nyström, H. Extracellular matrix proteins in metastases to the liver – Composition, function and potential applications. Semin. Cancer Biol. 2021, 71, 134–142. [Google Scholar] [CrossRef] [PubMed]
- McQuitty, C.E.; Williams, R.; Chokshi, S.; Urbani, L. Immunomodulatory role of the extracellular matrix within the liver disease microenvironment. Front. Immunol. 2020, 11, 574276. [Google Scholar] [CrossRef]
- Korkida, F.; Stamatopoulou, A.; Roubelakis, M.G. Recent advances in mesenchymal stem/stromal cell-based therapy for alcohol-associated liver disease and non-alcoholic fatty liver disease. Stem Cells Transl. Med. 2024, 13, 107–115. [Google Scholar] [CrossRef]
- Wan, Y.M.; Li, Z.Q.; Liu, C.; He, Y.F.; Wang, M.J.; Wu, X.N.; Zhang, Y.; Li, Y.H. Mesenchymal stem cells reduce alcoholic hepatitis in mice via suppression of hepatic neutrophil and macrophage infiltration, and of oxidative stress. PLOS ONE 2020, 15, e0228889. [Google Scholar] [CrossRef]
- Wan, Y.M.; Li, Z.Q.; Zhou, Q.; Liu, C.; Wang, M.J.; Wu, H.X.; Mu, Y.Z.; He, Y.F.; Zhang, Y.; Wu, X.N.; et al. Mesenchymal stem cells alleviate liver injury induced by chronic-binge ethanol feeding in mice via release of TSG6 and suppression of STAT3 activation. Stem Cell Res. Ther. 2020, 11, 24. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, D.; Wang, X.; Ward, S.C.; Cederbaum, A.I. Chronic alcohol-induced liver injury and oxidant stress are decreased in cytochrome P4502E1 knockout mice and restored in humanized cytochrome P4502E1 knock-in mice. Free Radic. Biol. Med. 2010, 49, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
- Kono, H.; Bradford, B.U.; Yin, M.; Sulik, K.K.; Koop, D.R.; Peters, J.M.; Gonzalez, F.J.; McDonald, T.; Dikalova, A.; Kadiiska, M.B.; et al. CYP2E1 is not involved in early alcohol-induced liver injury. Am. J. Physiol. 1999, 277, G1259–G1267. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.G.; Davies, R.; Dalton, T.P.; Miller, M.L.; Judah, D.; Riley, J.; Gant, T.; Nebert, D.W. Intrinsic hepatic phenotype associated with the Cyp1a2 gene as shown by cDNA expression microarray analysis of the knockout mouse. EHP Toxicogenom. 2003, 111, 45–51. [Google Scholar] [CrossRef]

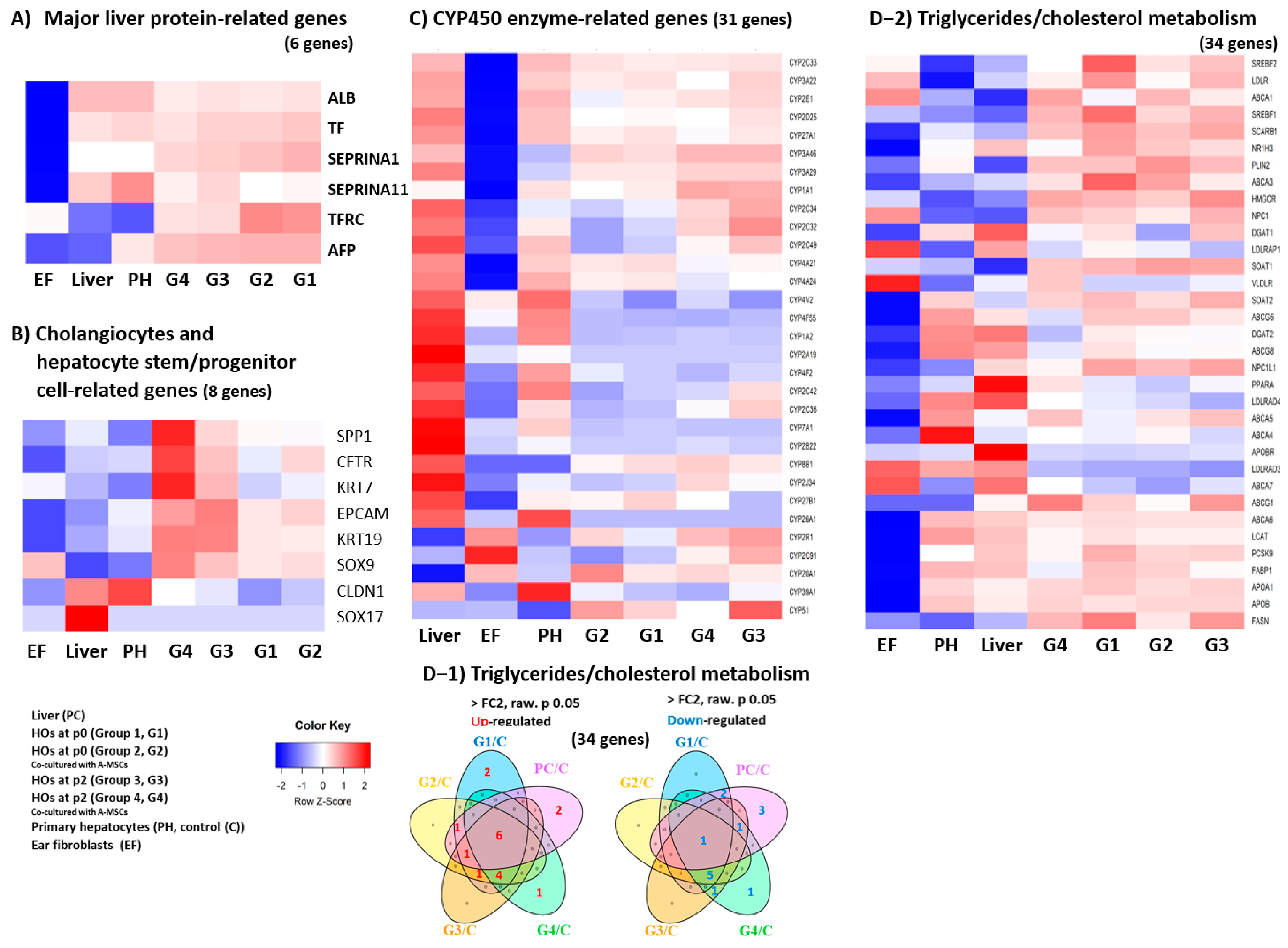


| Genes | * Fold-Change Values [log2 Each Group/Primary Hepatocytes, (PH)] | |||||
|---|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | Liver | ||
| 1 | AFP | 8.71 | 8.82 | 6.74 | 4.65 | −6173.24 |
| 2 | ALB | −4.01 | −4.27 | −3.89 | −5.98 | 1.07 |
| 3 | SERPINA1 | 10.77 | 7.20 | 5.20 | 3.55 | 1.02 |
| 4 | SERPINA11 | −5.96 | −8.12 | −3.59 | −5.63 | −2.85 |
| 5 | TF | 1.69 | 1.20 | 1.19 | −1.45 | −1.43 |
| 6 | TFRC | 5.31 | 5.64 | 3.46 | 3.36 | 1.19 |
| Genes | * Fold-Change Values [log2 Each Group/Primary Hepatocytes, (PH)] | |||||
|---|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | Liver | ||
| 1 | CFTR | 1.38 | 3.30 | 4.27 | 19.89 | −1.13 |
| 2 | CLDN1 | −6.74 | −4.77 | −3.81 | −3.18 | −1.49 |
| 3 | EPCAM | 2.02 | 2.77 | 9.60 | 5.84 | −5.47 |
| 4 | KRT19 | 2.28 | 2.27 | 9.15 | 9.72 | −2.83 |
| 5 | KRT7 | 28.32 | 47.55 | 190.98 | 1222.52 | 15.77 |
| 6 | SOX9 | 6.63 | 7.46 | 9.67 | 18.48 | −2.05 |
| 7 | SOX17 | 1.00 | 1.00 | 1.00 | 1.00 | 350.65 |
| 8 | SPP1 | 15.74 | 12.83 | 27.11 | 254.03 | 10.20 |
| 9 | LGR5 | −1.38 | −2.27 | 1.19 | 4.36 | −2.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ock, S.A.; Kim, S.-Y.; Kim, Y.-I.; Ju, W.S.; Lee, P. Enhanced In Vitro Recapitulation of In Vivo Liver Regeneration by Co-Culturing Hepatocyte Organoids with Adipose-Derived Mesenchymal Stem Cells, Alleviating Steatosis and Apoptosis in Acute Alcoholic Liver Injury. Cells 2024, 13, 1303. https://doi.org/10.3390/cells13151303
Ock SA, Kim S-Y, Kim Y-I, Ju WS, Lee P. Enhanced In Vitro Recapitulation of In Vivo Liver Regeneration by Co-Culturing Hepatocyte Organoids with Adipose-Derived Mesenchymal Stem Cells, Alleviating Steatosis and Apoptosis in Acute Alcoholic Liver Injury. Cells. 2024; 13(15):1303. https://doi.org/10.3390/cells13151303
Chicago/Turabian StyleOck, Sun A, Seo-Yeon Kim, Young-Im Kim, Won Seok Ju, and Poongyeon Lee. 2024. "Enhanced In Vitro Recapitulation of In Vivo Liver Regeneration by Co-Culturing Hepatocyte Organoids with Adipose-Derived Mesenchymal Stem Cells, Alleviating Steatosis and Apoptosis in Acute Alcoholic Liver Injury" Cells 13, no. 15: 1303. https://doi.org/10.3390/cells13151303
APA StyleOck, S. A., Kim, S.-Y., Kim, Y.-I., Ju, W. S., & Lee, P. (2024). Enhanced In Vitro Recapitulation of In Vivo Liver Regeneration by Co-Culturing Hepatocyte Organoids with Adipose-Derived Mesenchymal Stem Cells, Alleviating Steatosis and Apoptosis in Acute Alcoholic Liver Injury. Cells, 13(15), 1303. https://doi.org/10.3390/cells13151303








