Immune Control of Human Cytomegalovirus (HCMV) Infection in HCMV-Seropositive Solid Organ Transplant Recipients: The Predictive Role of Different Immunological Assays
Abstract
1. Introduction
2. Material and Methods
2.1. Study Subjects
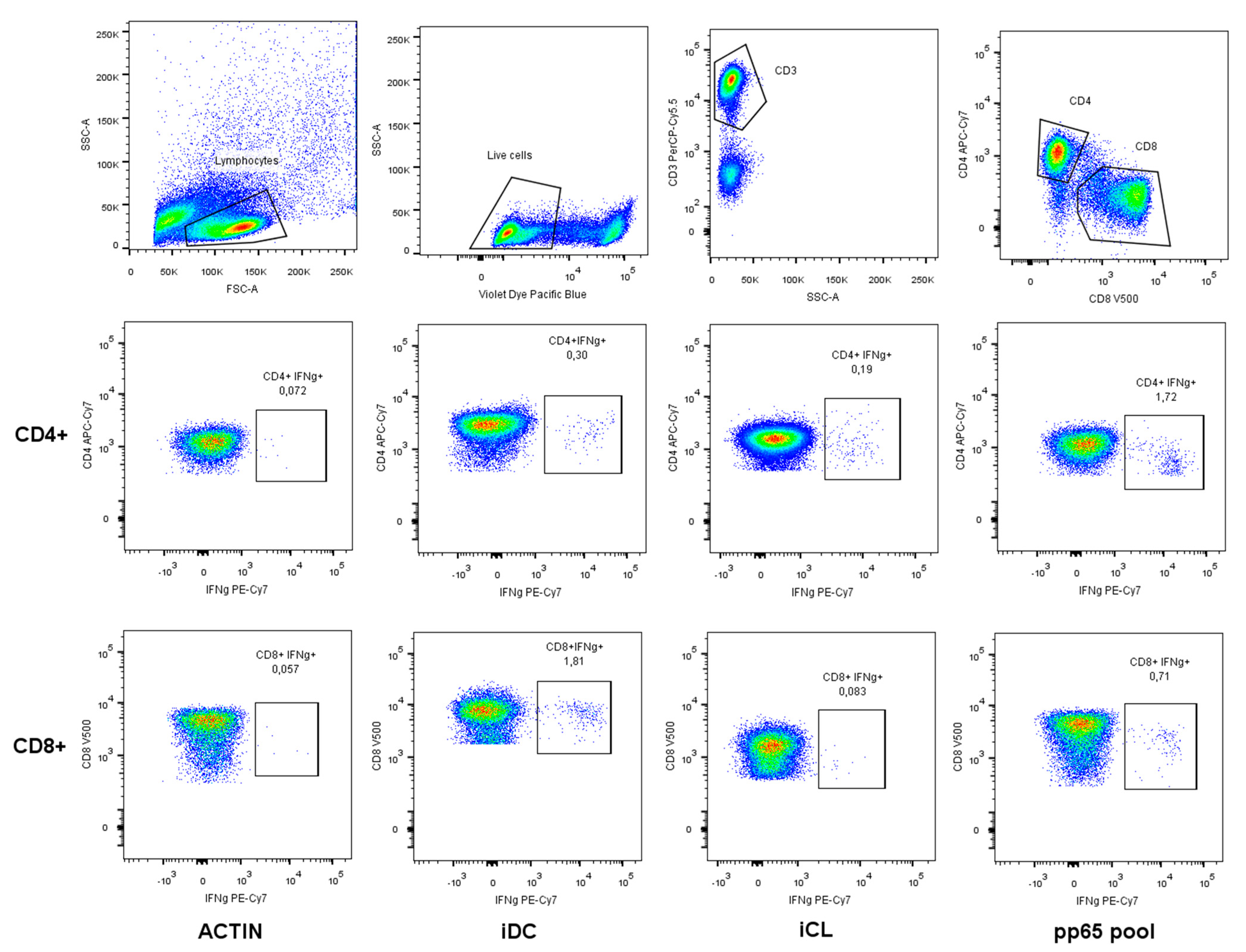
2.2. CFC-iDC, CFC-iCL and CFC-pp65 Pool Assays and Cytokine Flow Cytometry Staining
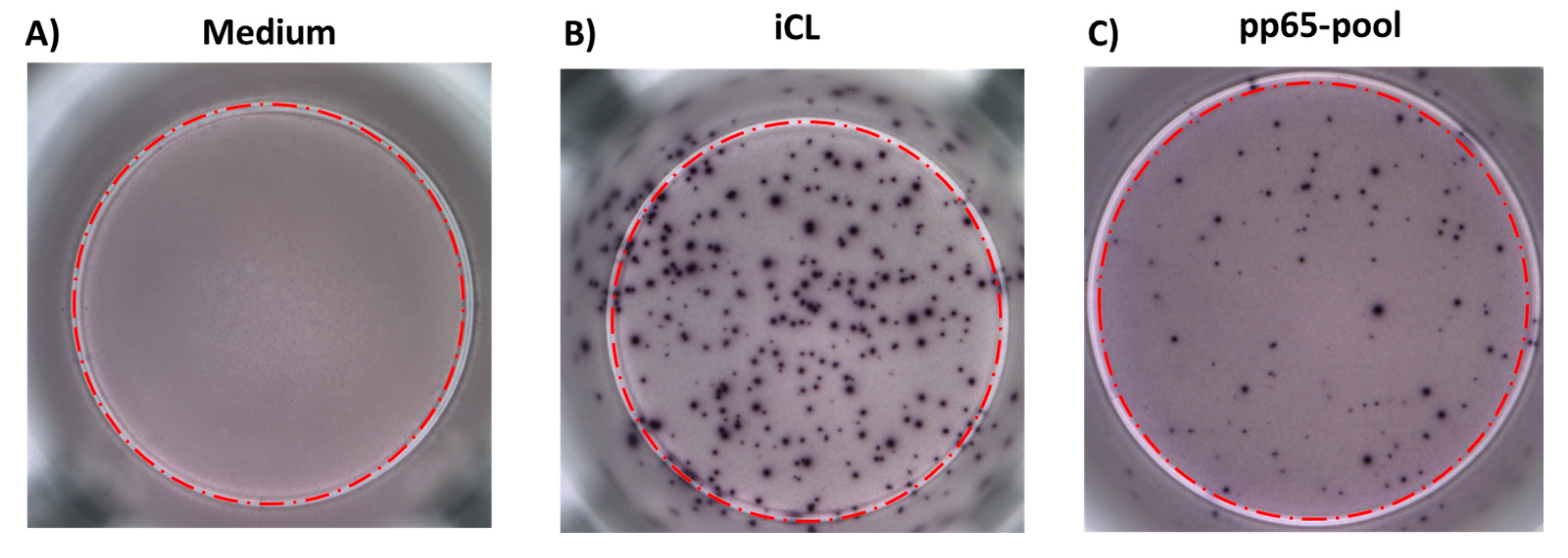
2.3. ELISPOT-iCL
2.4. ELISPOT-pp65 Peptide Pool
2.5. Statistical Analysis
3. Results
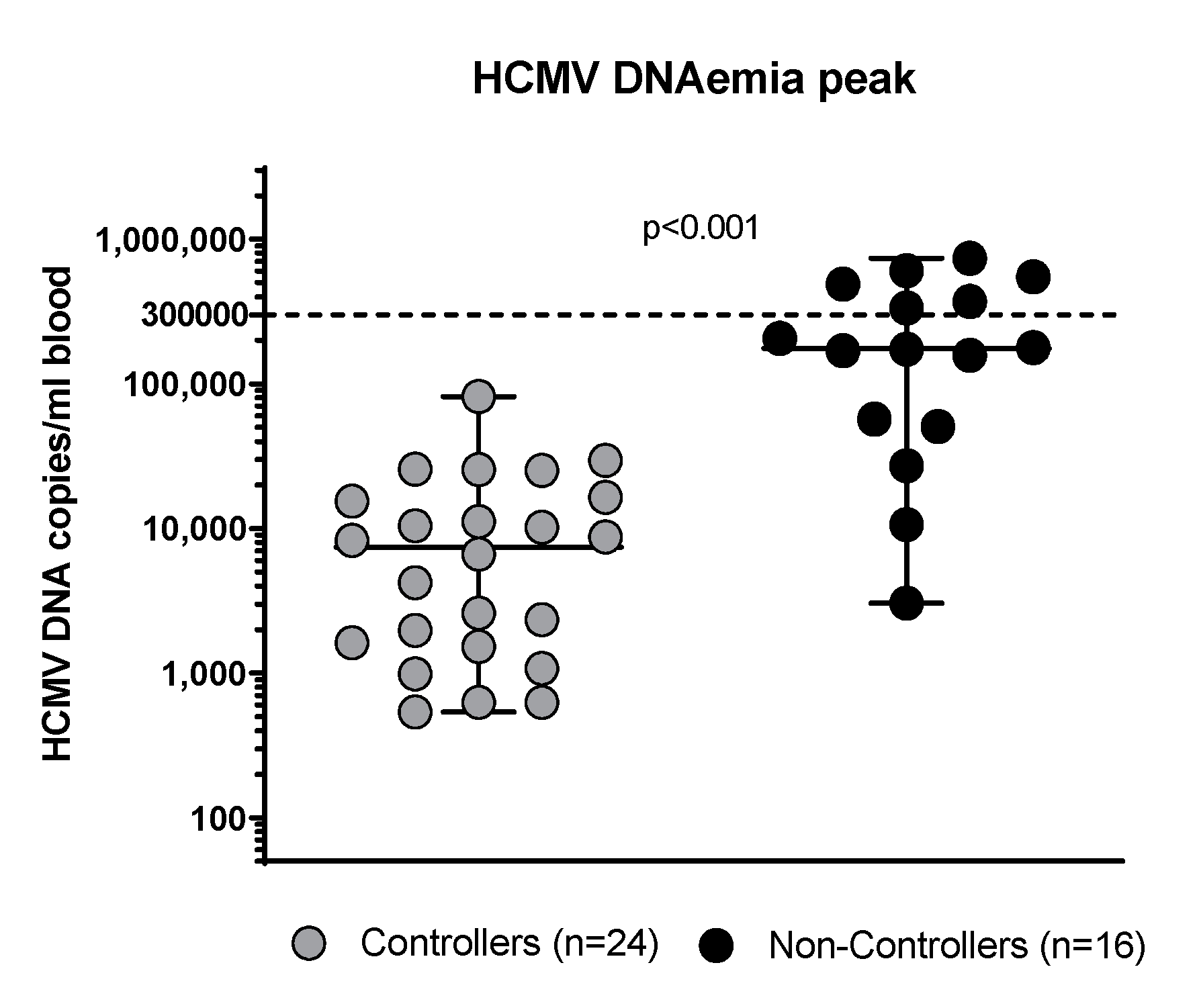
3.1. Clinical Characteristics of the Patients Enrolled
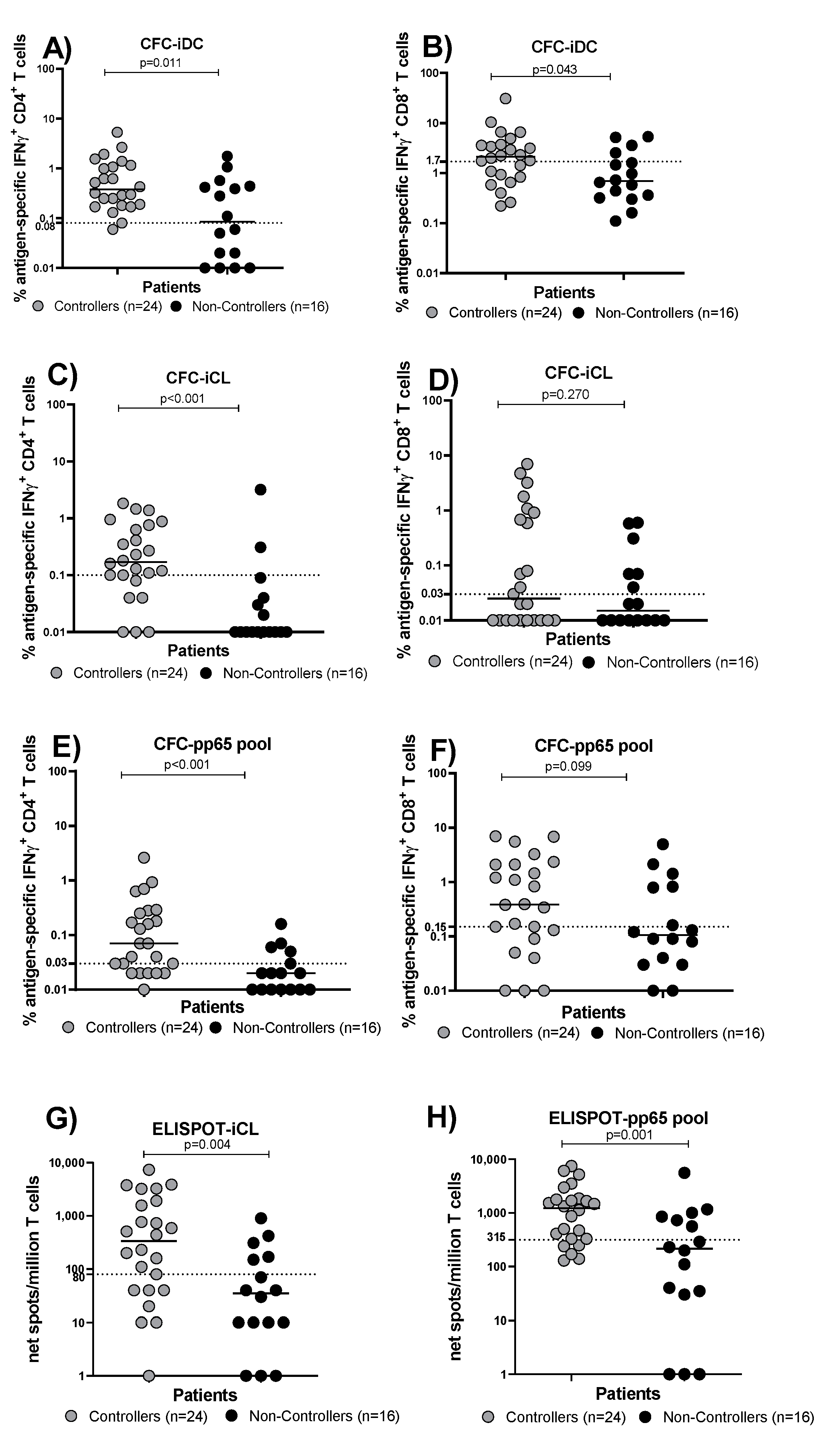
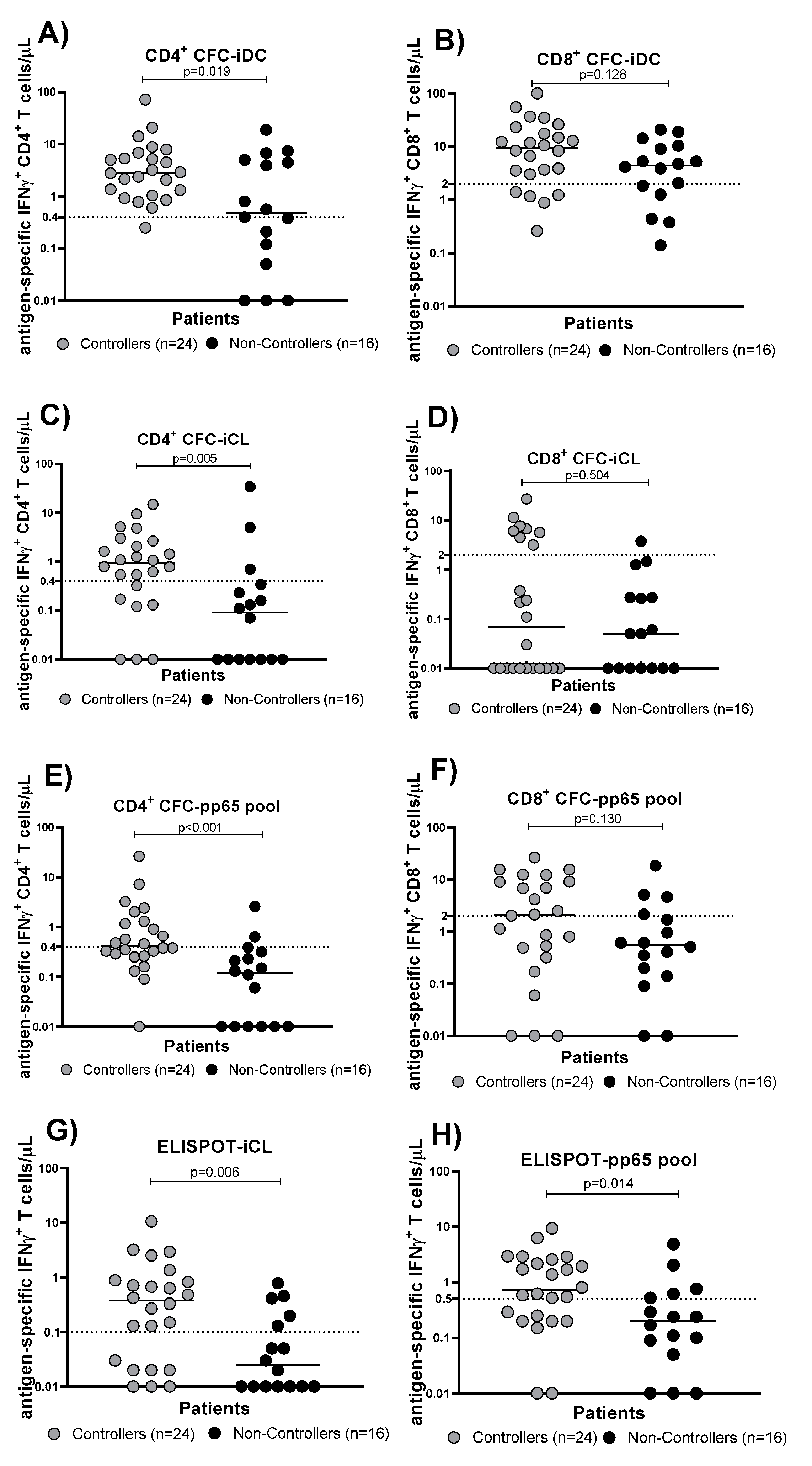
3.2. Evaluation of HCMV-Specific T Cell Response in Controllers and Non-Controllers Using Five Different Immunological Assays
3.3. Comparison of Prognostic Performance of Five Different Immunological Assays and Determination of Cut-off Values for Protective T Cell Response
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Razonable, R.R.; Humar, A. Cytomegalovirus in solid organ transplant recipients-Guidelines of the American Society of Transplantation Infectious Diseases Community of Practice. Clin. Transplant. 2019, 33, e13512. [Google Scholar] [CrossRef] [PubMed]
- Maffini, E.; Giaccone, L.; Festuccia, M.; Brunello, L.; Busca, A.; Bruno, B. Treatment of CMV infection after allogeneic hematopoietic stem cell transplantation. Expert Rev. Hematol. 2016, 9, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Kotton, C.N.; Kumar, D.; Caliendo, A.M.; Huprikar, S.; Chou, S.; Danziger-Isakov, L.; Humar, A. The Transplantation Society International CMV Consensus Group. The Third International Consensus Guidelines on the Management of Cytomegalovirus in Solid-Organ Transplantation. Transplantation 2018, 102, 900–931. [Google Scholar] [CrossRef] [PubMed]
- Fishman, J.A.; Emery, V.; Freeman, R.; Pascual, M.; Rostaing, L.; Schlitt, H.J.; Sgarabotto, D.; Torre-Cisneros, J.; E Uknis, M. Cytomegalovirus in transplantation—Challenging the status quo. Clin. Transplant. 2007, 21, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Bruminhent, J.; Razonable, R.R. Management of cytomegalovirus infection and disease in liver transplant recipients. World J. Hepatol. 2014, 6, 370–383. [Google Scholar] [CrossRef] [PubMed]
- Avetisyan, G.; Aschan, J.; Hägglund, H.; Ringdén, O.; Ljungman, P. Evaluation of intervention strategy based on CMV-specific immune responses after allogeneic SCT. Bone Marrow Transplant. 2007, 40, 865–869. [Google Scholar] [CrossRef] [PubMed]
- Gerna, G.; Lilleri, D.; Chiesa, A.; Zelini, P.; Furione, M.; Comolli, G.; Pellegrini, C.; Sarchi, E.; Migotto, C.; Bonora, M.R.; et al. Virologic and immunologic monitoring of cytomegalovirus to guide preemptive therapy in solid-organ transplantation. Am. J. Transplant. 2011, 11, 2463–2471. [Google Scholar] [CrossRef] [PubMed]
- Avetisyan, G.; Larsson, K.; Aschan, J.; Nilsson, C.; Hassan, M.; Ljungman, P. Impact on the cytomegalovirus (CMV) viral load by CMV-specific T-cell immunity in recipients of allogeneic stem cell transplantation. Bone Marrow Transplant. 2006, 38, 687–692. [Google Scholar] [CrossRef]
- Sood, S.; Haifer, C.; Yu, L.; Pavlovic, J.; Gow, P.J.; Jones, R.M.; Visvanathan, K.; Angus, P.W.; Testro, A.G. Targeted individual prophylaxis offers superior risk stratification for cytomegalovirus reactivation after liver transplantation. Liver Transpl. 2015, 21, 1478–1485. [Google Scholar] [CrossRef]
- Ruan, Y.; Guo, W.; Liang, S.; Xu, Z.; Niu, T. Diagnostic performance of cytomegalovirus (CMV) immune monitoring with ELISPOT and QuantiFERON-CMV assay in kidney transplantation: A PRISMA-compliant article. Medicine 2019, 98, e15228. [Google Scholar] [CrossRef]
- Sester, U.; Gärtner, B.C.; Wilkens, H.; Schwaab, B.; Wössner, R.; Kindermann, I.; Girndt, M.; Meyerhans, A.; Mueller-Lantzsch, N.; Schäferse, H.-J.; et al. Differences in CMV-specific T-cell levels and long-term susceptibility to CMV infection after kidney, heart and lung transplantation. Am. J. Transplant. 2005, 5, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Rogers, R.; Saharia, K.; Chandorkar, A.; Weiss, Z.F.; Vieira, K.; Koo, S.; Farmakiotis, D. Clinical experience with a novel assay measuring cytomegalovirus (CMV)-specific CD4+ and CD8+ T-cell immunity by flow cytometry and intracellular cytokine staining to predict clinically significant CMV events. BMC Infect. Dis. 2020, 20, 58, Erratum in BMC Infect. Dis. 2020, 20, 122. [Google Scholar] [CrossRef]
- Gliga, S.; Fiedler, M.; Dornieden, T.; Achterfeld, A.; Paul, A.; Horn, P.A.; Herzer, K.; Lindemann, M. Comparison of Three Cellular Assays to Predict the Course of CMV Infection in Liver Transplant Recipients. Vaccines 2021, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Zavaglio, F.; Rivela, F.; Cassaniti, I.; Arena, F.; Gabanti, E.; Asti, A.L.; Lilleri, D.; Rampino, T.; Baldanti, F.; Gregorini, M. ELISPOT assays with pp65 peptides or whole HCMV antigen are reliable predictors of immune control of HCMV infection in seropositive kidney transplant recipients. J. Med. Virol. 2023, 95, e28507. [Google Scholar] [CrossRef] [PubMed]
- Banas, B.; Steubl, D.; Renders, L.; Chittka, D.; Banas, M.C.; Wekerle, T.; Koch, M.; Witzke, O.; Mühlfeld, A.; Sommerer, C.; et al. Clinical validation of a novel enzyme-linked immunosorbent spot assay-based in vitro diagnostic assay to monitor cytomegalovirus-specific cell-mediated immunity in kidney transplant recipients: A multicenter, longitudinal, prospective, observational study. Transpl. Int. 2018, 31, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Chanouzas, D.; Small, A.; Borrows, R.; Ball, S. Assessment of the T-SPOT. CMV interferon-gamma release assay in renal transplant recipients: A single center cohort study. PLoS ONE 2018, 13, e0193968. [Google Scholar] [CrossRef]
- Shin, K.H.; Lee, H.J.; Chang, C.L.; Kim, E.J.; Lim, S.; Lee, S.J.; Ryu, J.H.; Yang, K.; Choi, B.H.; Lee, T.B.; et al. CMV specific T cell immunity predicts early viremia after liver transplantation. Transpl. Immunol. 2018, 51, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Thompson, G.; Boan, P.; Baumwol, J.; Chakera, A.; MacQuillan, G.; Swaminathan, S.; Lavender, M.; Flexman, J.; James, I.; John, M. Analysis of the QuantiFERON-CMV assay, CMV viraemia and antiviral treatment following solid organ transplantation in Western Australia. Pathology 2018, 50, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Mian, M.; Singer, L.; Humar, A. An Interventional Study Using Cell-Mediated Immunity to Personalize Therapy for Cytomegalovirus Infection after Transplantation. Am. J. Transplant. 2017, 17, 2468–2473. [Google Scholar] [CrossRef]
- Manuel, O.; Husain, S.; Kumar, D.; Zayas, C.; Mawhorter, S.; Levi, M.E.; Kalpoe, J.; Lisboa, L.; Ely, L.; Kaul, D.R.; et al. Assessment of cytomegalovirus-specific cell-mediated immunity for the prediction of cytomegalovirus disease in high-risk solid-organ transplant recipients: A multicenter cohort study. Clin. Infect. Dis. 2013, 56, 817–824. [Google Scholar] [CrossRef]
- Kumar, D.; Chin-Hong, P.; Kayler, L.; Wojciechowski, D.; Limaye, A.P.; Osama Gaber, A.; Ball, S.; Mehta, A.K.; Cooper, M.; Blanchard, T.; et al. A prospective multicenter observational study of cell-mediated immunity as a predictor for cytomegalovirus infection in kidney transplant recipients. Am. J. Transplant. 2019, 19, 2505–2516. [Google Scholar] [CrossRef] [PubMed]
- Lisboa, L.F.; Kumar, D.; Wilson, L.E.; Humar, A. Clinical utility of cytomegalovirus cell-mediated immunity in transplant recipients with cytomegalovirus viremia. Transplantation 2012, 93, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Chiereghin, A.; Potena, L.; Borgese, L.; Gibertoni, D.; Squarzoni, D.; Turello, G.; Petrisli, E.; Piccirilli, G.; Gabrielli, L.; Grigioni, F.; et al. Monitoring of Cytomegalovirus (CMV)-Specific Cell-Mediated Immunity in Heart Transplant Recipients: Clinical Utility of the QuantiFERON-CMV Assay for Management of Posttransplant CMV Infection. J. Clin. Microbiol. 2018, 56, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Sidoti, F.; Piralla, A.; Costa, C.; Scarasciulli, M.L.; Calvario, A.; Conaldi, P.G.; Paba, P.; Perno, C.F.; Gaeta, A.; Antonelli, G.; et al. Collaborative national multicenter for the identification of conversion factors from copies/mL to international units/mL for the normalization of HCMV DNA load. Diagn. Microbiol. Infect. Dis. 2019, 95, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Lozza, L.; Lilleri, D.; Percivalle, E.; Fornara, C.; Comolli, G.; Revello, M.G.; Gerna, G. Simultaneous quantification of human cytomegalovirus (HCMV)-specific CD4+ and CD8+ T cells by a novel method using monocyte-derived HCMV-infected immature dendritic cells. Eur. J. Immunol. 2005, 35, 1795–1804. [Google Scholar] [CrossRef] [PubMed]
- Zelini, P.; Lilleri, D.; Comolli, G.; Rognoni, V.; Chiesa, A.; Fornara, C.; Locatelli, F.; Meloni, F.; Gerna, G. Human cytomegalovirus-specific CD4(+) and CD8(+) T-cell response determination: Comparison of short-term (24 h) assays vs long-term (7-day) infected dendritic cell assay in the immunocompetent and the immunocompromised host. Clin. Immunol. 2010, 136, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Cassaniti, I.; Cavagna, L.; Calarota, S.A.; Adzasehoun, K.M.G.; Comolli, G.; Montecucco, C. Evaluation of EBV- and HCMV-Specific T Cell Responses in Systemic Lupus Erythematosus (SLE) Patients Using a Normalized Enzyme-Linked Immunospot (ELISPOT) Assay. J. Immunol. Res. 2019, 2019, 4236503. [Google Scholar] [CrossRef] [PubMed]
- Gabanti, E.; Bruno, F.; Scaramuzzi, L.; Mangione, F.; Zelini, P.; Gerna, G.; Lilleri, D. Predictive value of human cytomegalovirus (HCMV) T-cell response in the control of HCMV infection by seropositive solid-organ transplant recipients according to different assays and stimuli. New Microbiol. 2016, 39, 247–258. [Google Scholar] [PubMed]
- Kumar, D.; Chernenko, S.; Moussa, G.; Cobas, I.; Manuel, O.; Preiksaitis, J.; Venkataraman, S.; Humar, A. Cell mediated immunity to predict cytomegalovirus disease in high-risk solid organ transplant recipients. Am. J. Transplant. 2009, 9, 1214–1222. [Google Scholar] [CrossRef]
- Deborska-Materkowska, D.; Perkowska-Ptasinska, A.; Sadowska, A.; Gozdowska, J.; Ciszek, M.; Serwanska-Swietek, M.; Domagala, P.; Miszewska-Szyszkowska, D.; Sitarek, E.; Jozwik, A.; et al. Diagnostic utility of monitoring cytomegalovirus-specific immunity by QuantiFERON-cytomegalovirus assay in kidney transplant recipients. BMC Infect. Dis. 2018, 18, 179. [Google Scholar] [CrossRef] [PubMed]
- Abate, D.; Saldan, A.; Mengoli, C.; Fiscon, M.; Silvestre, C.; Fallico, L.; Peracchi, M.; Furian, L.; Cusinato, R.; Bonfante, L.; et al. Comparison of cytomegalovirus (CMV) enzyme-linked immunosorbent spot and CMV quantiferon gamma interferon-releasing assays in assessing risk of CMV infection in kidney transplant recipients. J. Clin. Microbiol. 2013, 51, 2501–2507. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gayowski, T.; Wagener, M.M.; Zeevi, A. T-helper cell responses in liver transplant recipients: Correlation with cytomegalovirus and other major infections. Transpl. Infect. Dis. 2004, 6, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Reusser, P.; Cathomas, G.; Attenhofer, R.; Tamm, M.; Thiel, G. Cytomegalovirus (CMV)-specific T cell immunity after renal transplantation mediates protection from CMV disease by limiting the systemic virus load. J. Infect. Dis. 1999, 180, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Egli, A.; Binet, I.; Binggeli, S.; Jäger, C.; Dumoulin, A.; Schaub, S.; Steiger, J.; Sester, U.; Sester, M.; Hirsch, H.H. Cytomegalovirus-specific T-cell responses and viral replication in kidney transplant recipients. J. Transl. Med. 2008, 6, 29. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Radha, R.; Jordan, S.; Puliyanda, D.; Bunnapradist, S.; Petrosyan, A.; Amet, N.; Toyoda, M. Cellular immune responses to cytomegalovirus in renal transplant recipients. Am. J. Transplant. 2005, 5, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Bunde, T.; Kirchner, A.; Hoffmeister, B.; Habedank, D.; Hetzer, R.; Cherepnev, G.; Proesch, S.; Reinke, P.; Volk, H.D.; Lehmkuhl, H.; et al. Protection from cytomegalovirus after transplantation is correlated with immediate early 1-specific CD8 T cells. J. Exp. Med. 2005, 201, 1031–1036. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gabanti, E.; Bruno, F.; Lilleri, D.; Fornara, C.; Zelini, P.; Cane, I.; Migotto, C.; Sarchi, E.; Furione, M.; Gerna, G. Human cytomegalovirus (HCMV)-specific CD4+ and CD8+ T cells are both required for prevention of HCMV disease in seropositive solid-organ transplant recipients. PLoS ONE 2014, 9, e106044. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Barabas, S.; Spindler, T.; Kiener, R.; Tonar, C.; Lugner, T.; Batzilla, J.; Bendfeldt, H.; Rascle, A.; Asbach, B.; Wagner, R.; et al. An optimized IFN-γ ELISpot assay for the sensitive and standardized monitoring of CMV protein-reactive effector cells of cell-mediated immunity. BMC Immunol. 2017, 18, 14. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lilleri, D.; Zelini, P.; Fornara, C.; Comolli, G.; Gerna, G. Inconsistent responses of cytomegalovirus-specific T cells to pp65 and IE-1 versus infected dendritic cells in organ transplant recipients. Am. J. Transplant. 2007, 7, 1997–2005. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | All Patients (n = 40) | Controllers (n = 24) | Non-Controllers (n = 16) | p Value |
|---|---|---|---|---|
| Age, median [IQR] | 51 [45–58] | 51 [43–58] | 53 [45–59] | 0.401 |
| Gender, n (%): | ||||
| Male | 28 (70) | 15 (62) | 13 (81) | 0.290 |
| Female | 12 (30) | 9 (38) | 3 (19) | |
| Kind of Transplant, n (%): | ||||
| KTR | 27 (67) | 17 (71) | 10 (63) | 0.683 |
| LTR | 6 (15) | 4 (17) | 2 (12) | 0.684 |
| HTR | 7 (18) | 3 (12) | 4 (25) | 0.591 |
| Induction Therapy, n (%): | ||||
| Basiliximab | 29 (72) | 17 (71) | 12 (75) | 0.457 |
| ATG | 7 (17) | 5 (21) | 2 (12) | 0.418 |
| Steroids | 40 (100) | 24 (100) | 16 (100) | 0.365 |
| Maintenance Therapy, n (%): | ||||
| FK506, MMF, MPRE | 33 (83) | 20 (83) | 13 (81) | 0.380 |
| CyA, MMF, MPRE | 4 (10) | 3 (13) | 1 (7) | 0.458 |
| FK506, Everolimus, MPRE | 3 (7) | 1 (4) | 2 (12) | 0.561 |
| CFC-iDC CD4 | CFC-iDC CD8 | CFC-iCL CD4 | CFC-iCL CD8 | CFC-pp65 CD4 | CFC-pp65 CD8 | ELISPOT- iCL | ELISPOT-pp65 | |
|---|---|---|---|---|---|---|---|---|
| AUC | 0.73 | 0.69 | 0.81 | 0.60 | 0.80 | 0.65 | 0.78 | 0.76 |
| CUT-OFF (%) | 0.08 | 1.70 | 0.10 | 0.03 | 0.03 | 0.15 | 80 | 315 |
| sensitivity (%) | 95 | 62 | 75 | 50 | 75 | 70 | 70 | 79 |
| specificity (%) | 40 | 75 | 87 | 62 | 68 | 62 | 68 | 64 |
| PPV (%) | 74 | 79 | 90 | 67 | 78 | 74 | 77 | 76 |
| NPV (%) | 89 | 57 | 70 | 45 | 65 | 59 | 61 | 67 |
| CFC-iDC CD4 | CFC-iDC CD8 | CFC-iCL CD4 | CFC-iCL CD8 | CFC-pp65 CD4 | CFC-pp65 CD8 | ELISPOT- iCL | ELISPOT-pp65 | |
|---|---|---|---|---|---|---|---|---|
| AUC | 0.72 | 0.64 | 0.76 | 0.56 | 0.81 | 0.64 | 0.75 | 0.73 |
| CUT-OFF | 0.4 | 2 | 0.4 | 2 | 0.4 | 2 | 0.1 | 0.5 |
| sensitivity (%) | 95 | 76 | 71 | 33 | 50 | 50 | 71 | 62 |
| specificity (%) | 44 | 31 | 75 | 94 | 81 | 75 | 69 | 75 |
| PPV (%) | 72 | 66 | 85 | 89 | 86 | 76 | 77 | 76 |
| NPV (%) | 85 | 50 | 65 | 48 | 54 | 52 | 61 | 58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zavaglio, F.; Cassaniti, I.; d’Angelo, P.; Zelini, P.; Comolli, G.; Gregorini, M.; Rampino, T.; Del Frate, L.; Meloni, F.; Pellegrini, C.; et al. Immune Control of Human Cytomegalovirus (HCMV) Infection in HCMV-Seropositive Solid Organ Transplant Recipients: The Predictive Role of Different Immunological Assays. Cells 2024, 13, 1325. https://doi.org/10.3390/cells13161325
Zavaglio F, Cassaniti I, d’Angelo P, Zelini P, Comolli G, Gregorini M, Rampino T, Del Frate L, Meloni F, Pellegrini C, et al. Immune Control of Human Cytomegalovirus (HCMV) Infection in HCMV-Seropositive Solid Organ Transplant Recipients: The Predictive Role of Different Immunological Assays. Cells. 2024; 13(16):1325. https://doi.org/10.3390/cells13161325
Chicago/Turabian StyleZavaglio, Federica, Irene Cassaniti, Piera d’Angelo, Paola Zelini, Giuditta Comolli, Marilena Gregorini, Teresa Rampino, Lucia Del Frate, Federica Meloni, Carlo Pellegrini, and et al. 2024. "Immune Control of Human Cytomegalovirus (HCMV) Infection in HCMV-Seropositive Solid Organ Transplant Recipients: The Predictive Role of Different Immunological Assays" Cells 13, no. 16: 1325. https://doi.org/10.3390/cells13161325
APA StyleZavaglio, F., Cassaniti, I., d’Angelo, P., Zelini, P., Comolli, G., Gregorini, M., Rampino, T., Del Frate, L., Meloni, F., Pellegrini, C., Abelli, M., Ticozzelli, E., Lilleri, D., & Baldanti, F. (2024). Immune Control of Human Cytomegalovirus (HCMV) Infection in HCMV-Seropositive Solid Organ Transplant Recipients: The Predictive Role of Different Immunological Assays. Cells, 13(16), 1325. https://doi.org/10.3390/cells13161325








