Involvement of Cyclooxygenase-2 in Establishing an Immunosuppressive Microenvironment in Tumorspheres Derived from TMZ-Resistant Glioblastoma Cell Lines and Primary Cultures
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. GBM Primary Cultures
2.3. Tumorsphere Formation Assay
2.4. Reagents and Treatments
2.5. Proliferation Assay
2.6. Macrophage Infiltration into Tumorspheres
2.7. Western Blot
2.8. ELISA Kit
2.9. Flow Cytometry Analysis
2.10. Statistics Analysis
3. Results
3.1. Effect of CXB, TMZ, and Their Combination on Tumorsphere Formation and Macrophage Infiltration
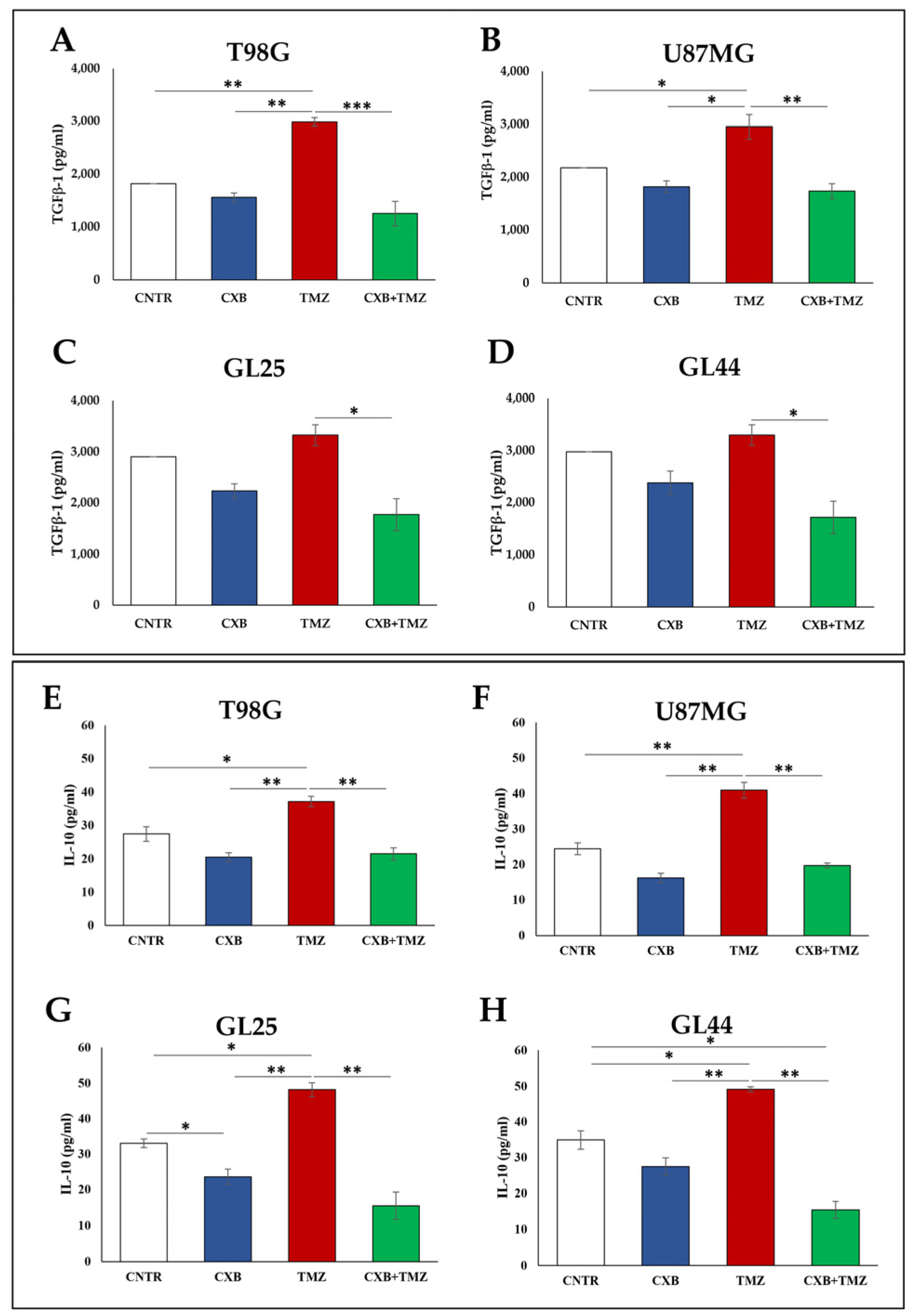
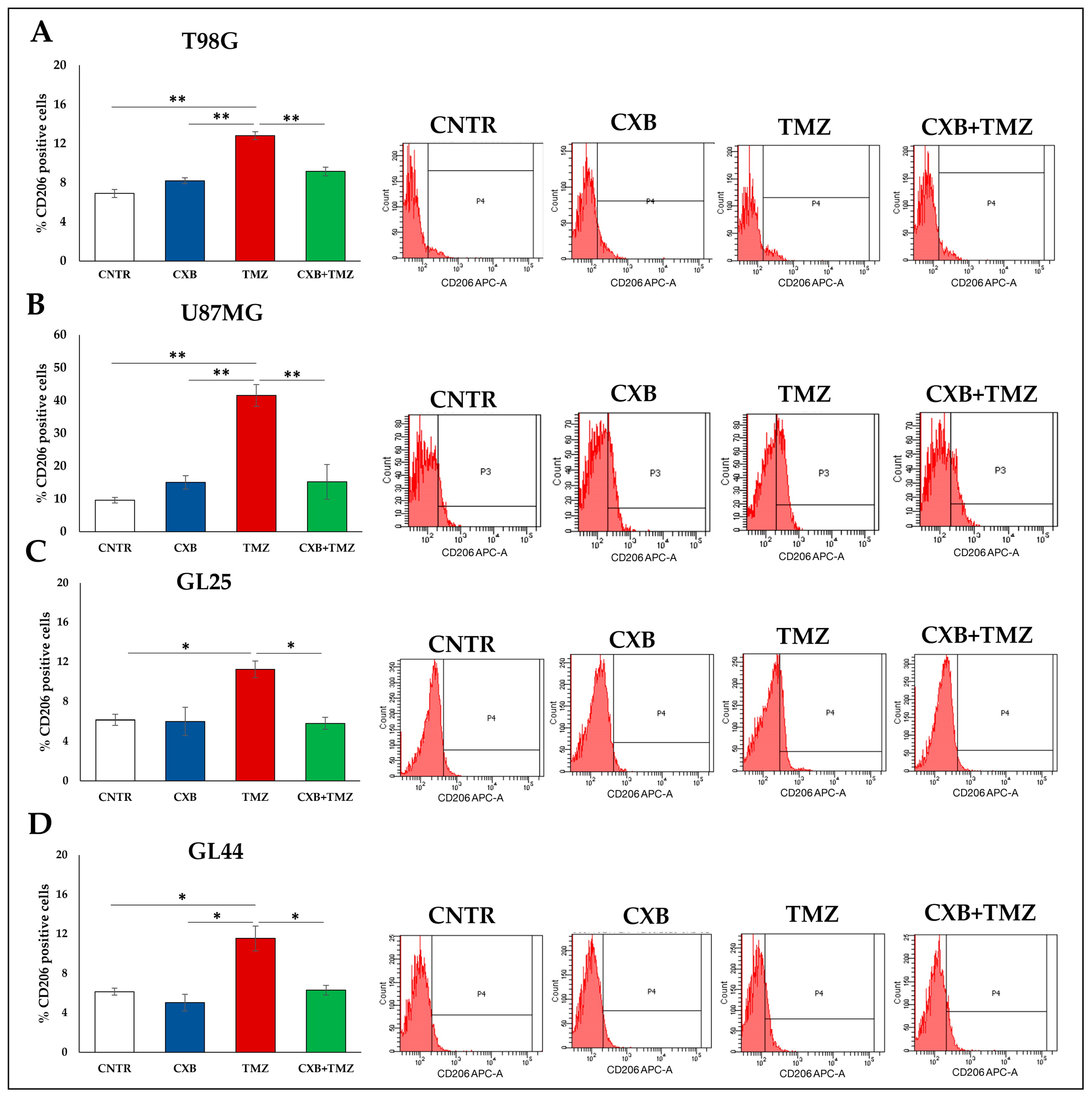
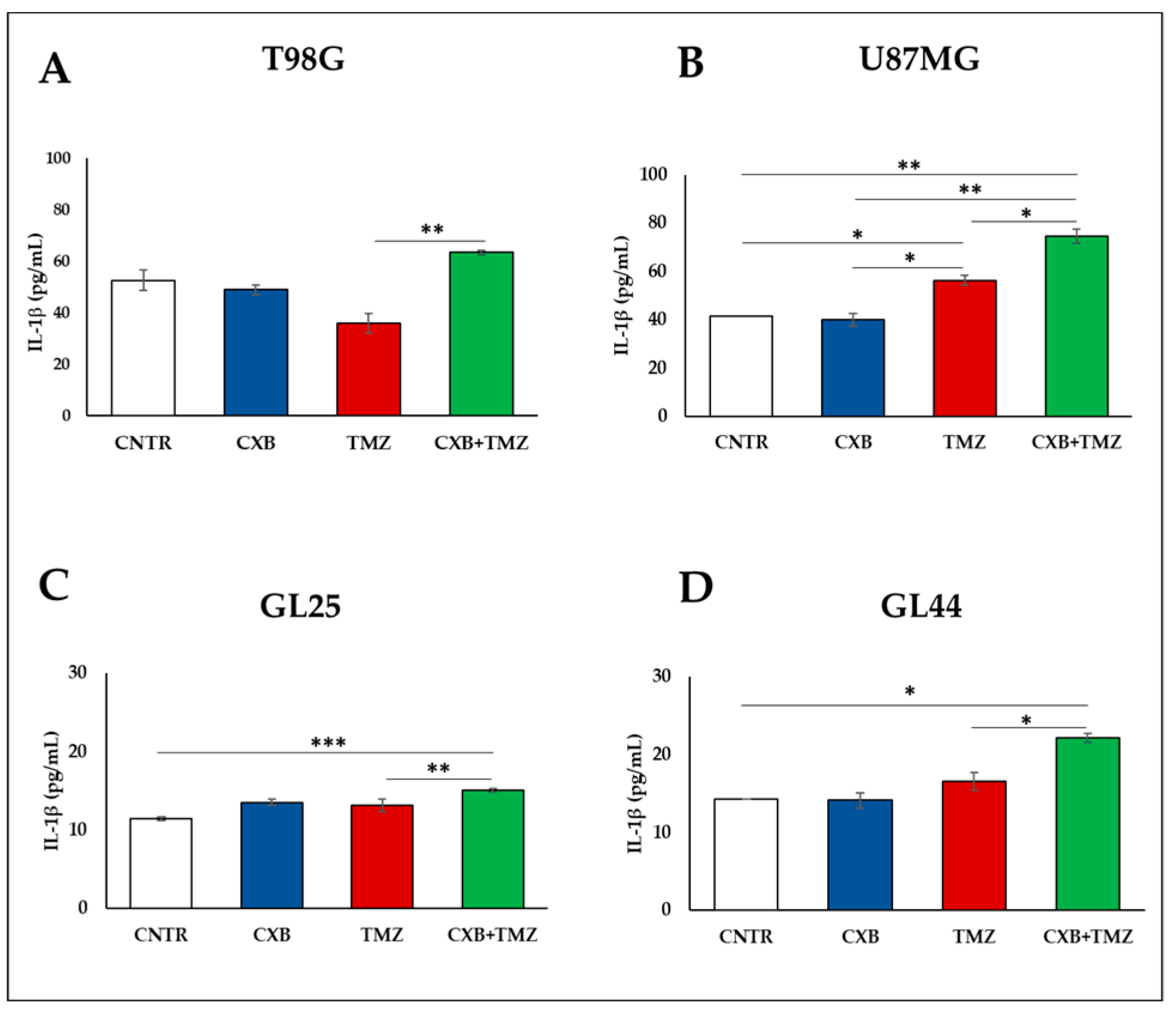
3.2. COX-2 Inhibition Affects the Immunosuppressive Macrophage M2 Phenotype
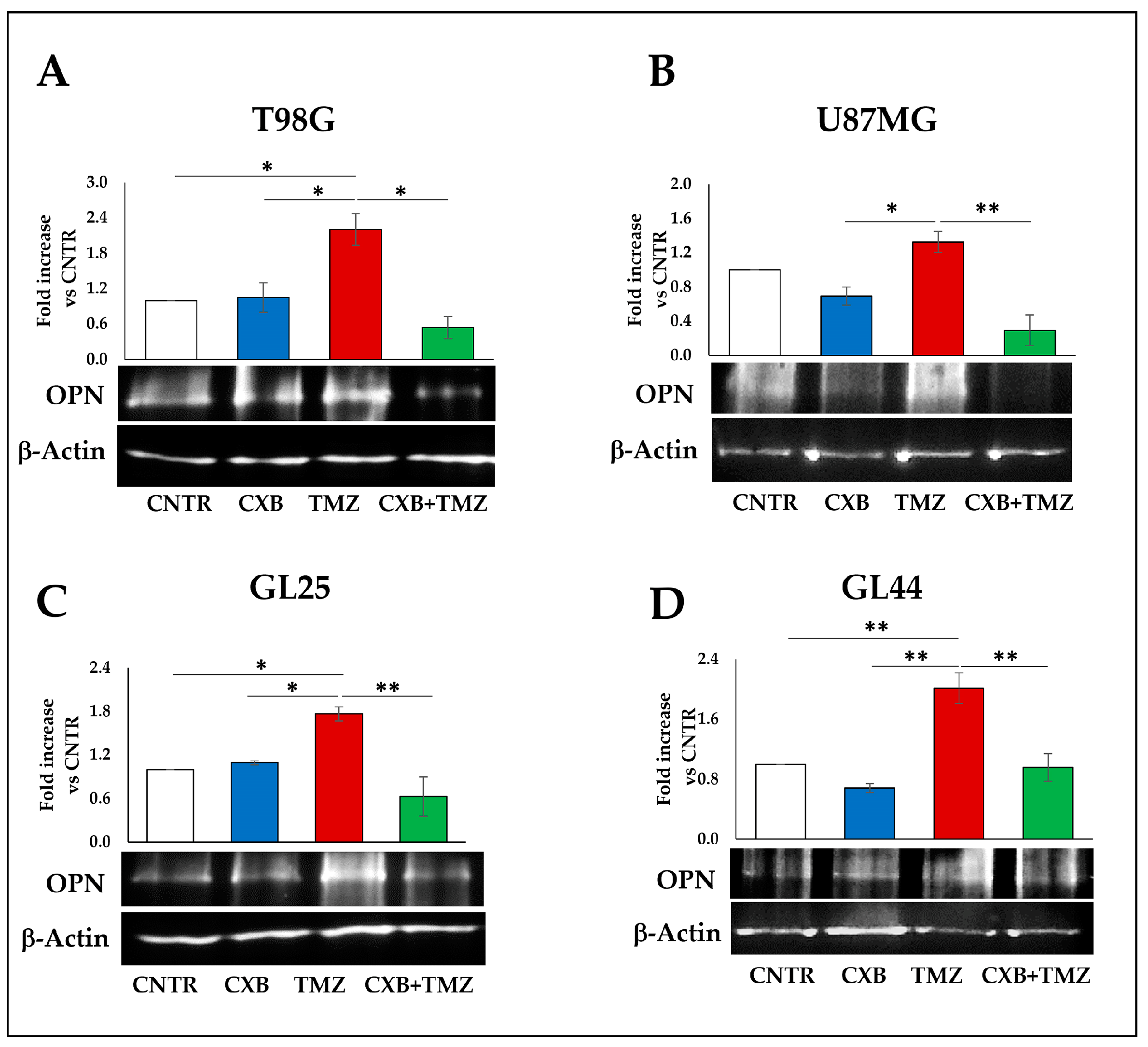
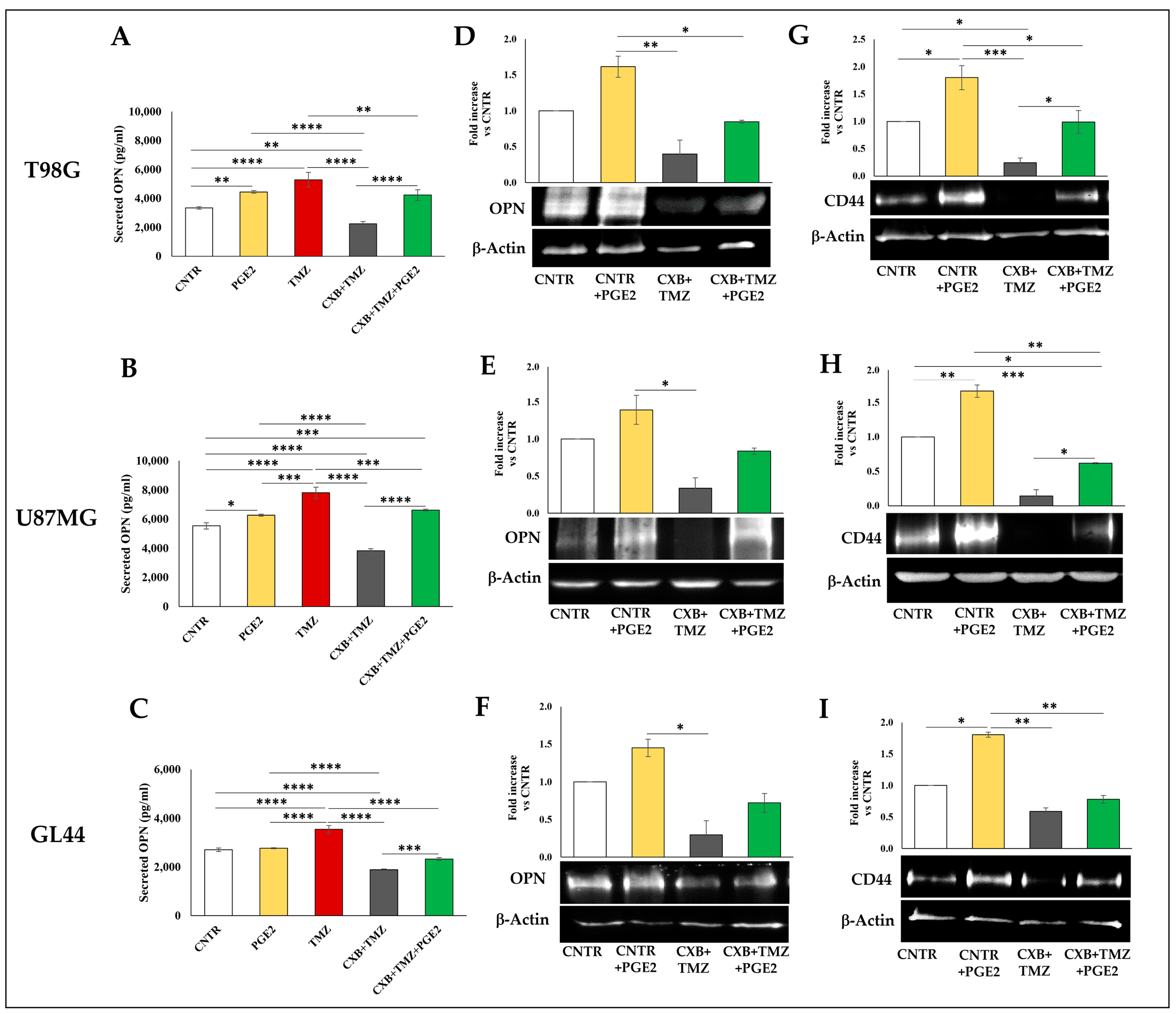
3.3. COX Inhibition Counteracted TMZ-Induced OPN Overexpression
3.4. COX-2 Inhibition Counteracted the TMZ-Induced CD44 Upregulation
3.5. Effect of Exogenous PGE2 on TMZ-Induced OPN in GBM Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Di Ianni, N.; Maffezzini, M.; Eoli, M.; Pellegatta, S. Revisiting the Immunological Aspects of Temozolomide Considering the Genetic Landscape and the Immune Microenvironment Composition of Glioblastoma. Front. Oncol. 2021, 11, 747690. [Google Scholar] [CrossRef] [PubMed]
- Burster, T.; Traut, R.; Yermekkyzy, Z.; Mayer, K.; Westhoff, M.A.; Bischof, J.; Knippschild, U. Critical View of Novel Treatment Strategies for Glioblastoma: Failure and Success of Resistance Mechanisms by Glioblastoma Cells. Front. Cell Dev. Biol. 2021, 9, 695325. [Google Scholar] [CrossRef] [PubMed]
- Fabro, F.; Lamfers, M.L.M.; Leenstra, S. Advancements, Challenges, and Future Directions in Tackling Glioblastoma Resistance to Small Kinase Inhibitors. Cancers 2022, 14, 600. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ye, Z.; Song, F.; He, Y.; Liu, J. The Role of TAMs in Tumor Microenvironment and New Research Progress. Stem Cells Int. 2022, 2022, 5775696. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Xiao, M.; Li, X.; Xin, L.; Song, J.; Zhan, Q.; Wang, C.; Zhang, Q.; Yuan, X.; Tan, Y.; et al. Origin, activation, and targeted therapy of glioma-associated macrophages. Front. Immunol. 2022, 13, 974996. [Google Scholar] [CrossRef] [PubMed]
- Serpe, C.; Monaco, L.; Relucenti, M.; Iovino, L.; Familiari, P.; Scavizzi, F.; Raspa, M.; Familiari, G.; Civiero, L.; D’Agnano, I.; et al. Microglia-Derived Small Extracellular Vesicles Reduce Glioma Growth by Modifying Tumor Cell Metabolism and Enhancing Glutamate Clearance through miR-124. Cells 2021, 10, 2066. [Google Scholar] [CrossRef]
- Wang, G.; Zhong, K.; Wang, Z.; Zhang, Z.; Tang, X.; Tong, A.; Zhou, L. Tumor-associated microglia and macrophages in glioblastoma: From basic insights to therapeutic opportunities. Front. Immunol. 2022, 13, 964898. [Google Scholar] [CrossRef]
- Wei, J.; Chen, P.; Gupta, P.; Ott, M.; Zamler, D.; Kassab, C.; Bhat, K.P.; Curran, M.A.; de Groot, J.F.; Heimberger, A.B. Immune biology of glioma-associated macrophages and microglia: Functional and therapeutic implications. Neuro Oncol. 2020, 22, 180–194. [Google Scholar] [CrossRef]
- Mattei, V.; Santilli, F.; Martellucci, S.; Delle Monache, S.; Fabrizi, J.; Colapietro, A.; Angelucci, A.; Festuccia, C. The Importance of Tumor Stem Cells in Glioblastoma Resistance to Therapy. Int. J. Mol. Sci. 2021, 22, 3863. [Google Scholar] [CrossRef]
- Luo, S.; Yang, G.; Ye, P.; Cao, N.; Chi, X.; Yang, W.H.; Yan, X. Macrophages Are a Double-Edged Sword: Molecular Crosstalk between Tumor-Associated Macrophages and Cancer Stem Cells. Biomolecules 2022, 12, 850. [Google Scholar] [CrossRef]
- Wei, J.; Marisetty, A.; Schrand, B.; Gabrusiewicz, K.; Hashimoto, Y.; Ott, M.; Grami, Z.; Kong, L.Y.; Ling, X.Y.; Caruso, H.; et al. Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J. Clin. Investig. 2019, 129, 137–149. [Google Scholar] [CrossRef]
- Chen, P.W.; Zhao, D.; Li, J.; Liang, X.; Li, J.X.; Chang, A.; Henry, V.K.; Lan, Z.D.; Spring, D.J.; Rao, G.; et al. Symbiotic Macrophage-Glioma Cell Interactions Reveal Synthetic Lethality in PTEN-Null Glioma. Cancer Cell 2019, 35, 868–884. [Google Scholar] [CrossRef]
- Kariya, Y.; Kariya, Y. Osteopontin in Cancer: Mechanisms and Therapeutic Targets. Int. J. Transl. Med. 2022, 2, 419–447. [Google Scholar] [CrossRef]
- Tan, Y.Y.; Zhao, L.; Yang, Y.G.; Liu, W.T. The Role of Osteopontin in Tumor Progression through Tumor-Associated Macrophages. Front. Oncol. 2022, 12, 953283. [Google Scholar] [CrossRef] [PubMed]
- Cantor, H.; Shinohara, M.L. Regulation of T-helper-cell lineage development by osteopontin: The inside story. Nat. Rev. Immunol. 2009, 9, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Renkl, A.C.; Wussler, J.; Ahrens, T.; Thoma, K.; Kon, S.; Uede, T.; Martin, S.F.; Simon, J.C.; Weiss, J.M. Osteopontin functionally activates dendritic cells and induces their differentiation toward a Th1-polarizing phenotype. Blood 2005, 106, 946–955. [Google Scholar] [CrossRef] [PubMed]
- Atai, N.A.; Bansal, M.; Lo, C.; Bosman, J.; Tigchelaar, W.; Bosch, K.S.; Jonker, A.; De Witt Hamer, P.C.; Troost, D.; McCulloch, C.A.; et al. Osteopontin is up-regulated and associated with neutrophil and macrophage infiltration in glioblastoma. Immunology 2011, 132, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Sreekanthreddy, P.; Srinivasan, H.; Kumar, D.M.; Nijaguna, M.B.; Sridevi, S.; Vrinda, M.; Arivazhagan, A.; Balasubramaniam, A.; Hegde, A.S.; Chandramouli, B.A.; et al. Identification of potential serum biomarkers of glioblastoma: Serum osteopontin levels correlate with poor prognosis. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1409–1422. [Google Scholar] [CrossRef]
- Toy, H.; Yavas, O.; Eren, O.; Genc, M.; Yavas, C. Correlation between osteopontin protein expression and histological grade of astrocytomas. Pathol. Oncol. Res. 2009, 15, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Kijewska, M.; Kocyk, M.; Kloss, M.; Stepniak, K.; Korwek, Z.; Polakowska, R.; Dabrowski, M.; Gieryng, A.; Wojtas, B.; Ciechomska, I.A.; et al. The embryonic type of SPP1 transcriptional regulation is re-activated in glioblastoma. Oncotarget 2017, 8, 16340–16355. [Google Scholar] [CrossRef]
- Lamour, V.; Henry, A.; Kroonen, J.; Nokin, M.J.; von Marschall, Z.; Fisher, L.W.; Chau, T.L.; Chariot, A.; Sanson, M.; Delattre, J.Y.; et al. Targeting osteopontin suppresses glioblastoma stem-like cell character and tumorigenicity in vivo. Int. J. Cancer 2015, 137, 1047–1057. [Google Scholar] [CrossRef]
- Kolliopoulos, C.; Ali, M.M.; Castillejo-Lopez, C.; Heldin, C.H.; Heldin, P. CD44 Depletion in Glioblastoma Cells Suppresses Growth and Stemness and Induces Senescence. Cancers 2022, 14, 3747. [Google Scholar] [CrossRef]
- Johansson, E.; Grassi, E.S.; Pantazopoulou, V.; Tong, B.; Lindgren, D.; Berg, T.J.; Pietras, E.J.; Axelson, H.; Pietras, A. CD44 Interacts with HIF-2 alpha to Modulate the Hypoxic Phenotype of Perinecrotic and Perivascular Glioma Cells. Cell Rep. 2017, 20, 1641–1653. [Google Scholar] [CrossRef]
- Polat, B.; Wohlleben, G.; Kosmala, R.; Lisowski, D.; Mantel, F.; Lewitzki, V.; Lohr, M.; Blum, R.; Herud, P.; Flentje, M.; et al. Differences in stem cell marker and osteopontin expression in primary and recurrent glioblastoma. Cancer Cell Int. 2022, 22, 87. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Li, P.; Yan, W.; Shi, L.; Zhang, J.; Wang, Y.; Liu, H.; You, Y. Downregulation of osteopontin enhances the sensitivity of glioma U251 cells to temozolomide and cisplatin by targeting the NF-kappaB/Bcl-2 pathway. Mol. Med. Rep. 2015, 11, 1951–1955. [Google Scholar] [CrossRef]
- Lombardi, F.; Augello, F.R.; Artone, S.; Gugu, M.K.; Cifone, M.G.; Cinque, B.; Palumbo, P. Up-Regulation of Cyclooxygenase-2 (COX-2) Expression by Temozolomide (TMZ) in Human Glioblastoma (GBM) Cell Lines. Int. J. Mol. Sci. 2022, 23, 1545. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, F.; Augello, F.R.; Artone, S.; Ayroldi, E.; Giusti, I.; Dolo, V.; Cifone, M.G.; Cinque, B.; Palumbo, P. Cyclooxygenase-2 Upregulated by Temozolomide in Glioblastoma Cells Is Shuttled In Extracellular Vesicles Modifying Recipient Cell Phenotype. Front. Oncol. 2022, 12, 933746. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, R.; Rizzarelli, E.; Copani, A. Role for Metallothionein-3 in the Resistance of Human U87 Glioblastoma Cells to Temozolomide. ACS Omega 2020, 5, 17900–17907. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.M.; Kim, J.Y.; Cho, H.J.; Lee, W.J.; Nguyen, D.; Kim, S.S.; Oh, Y.T.; Kim, H.J.; Hubert, C.G.; Jung, C.W.; et al. Tissue factor is a critical regulator of radiation therapy-induced glioblastoma remodeling. Cancer Cell 2023, 41, 1480. [Google Scholar] [CrossRef]
- Peng, P.; Zhu, H.T.; Liu, D.; Chen, Z.R.; Zhang, X.L.; Guo, Z.Y.; Dong, M.H.; Wan, L.J.; Zhang, P.; Liu, G.H.; et al. TGFBI secreted by tumor-associated macrophages promotes glioblastoma stem cell-driven tumor growth via integrin αvβ5-Src-Stat3 signaling. Theranostics 2022, 12, 4221–4236. [Google Scholar] [CrossRef]
- Vidal, V.; Gutierrez, O.; Talamillo, A.; Velasquez, C.; Fernandez-Luna, J.L. Glioblastoma invasion factor ODZ1 is induced by microenvironmental signals through activation of a Stat3-dependent transcriptional pathway. Sci. Rep. 2021, 11, 16196. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, J.; Zhang, Z.; Gao, Z.; Qi, Y.; Qiu, W.; Pan, Z.; Guo, Q.; Li, B.; Zhao, S.; et al. Hypoxic glioma-derived exosomes promote M2-like macrophage polarization by enhancing autophagy induction. Cell Death Dis. 2021, 12, 373. [Google Scholar] [CrossRef] [PubMed]
- Miconi, G.; Palumbo, P.; Dehcordi, S.R.; La Torre, C.; Lombardi, F.; Evtoski, Z.; Cimini, A.M.; Galzio, R.; Cifone, M.G.; Cinque, B. Immunophenotypic characterization of human glioblastoma stem cells: Correlation with clinical outcome. J. Cell Biochem. 2015, 116, 864–876. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, P.; Miconi, G.; Cinque, B.; Lombardi, F.; La Torre, C.; Dehcordi, S.R.; Galzio, R.; Cimini, A.; Giordano, A.; Cifone, M.G. NOS2 expression in glioma cell lines and glioma primary cell cultures: Correlation with neurosphere generation and SOX-2 expression. Oncotarget 2017, 8, 25582–25598. [Google Scholar] [CrossRef]
- Majchrzak-Celinska, A.; Misiorek, J.O.; Kruhlenia, N.; Przybyl, L.; Kleszcz, R.; Rolle, K.; Krajka-Kuzniak, V. COXIBs and 2,5-dimethylcelecoxib counteract the hyperactivated Wnt/beta-catenin pathway and COX-2/PGE2/EP4 signaling in glioblastoma cells. BMC Cancer 2021, 21, 493. [Google Scholar] [CrossRef]
- Wu, M.G.; Guan, J.; Li, C.; Gunter, S.; Nusrat, L.; Ng, S.; Dhand, K.; Morshead, C.; Kim, A.; Das, S. Aberrantly activated Cox-2 and Wnt signaling interact to maintain cancer stem cells in glioblastoma. Oncotarget 2017, 8, 82217–82230. [Google Scholar] [CrossRef]
- Ferreira, M.T.; Miyake, J.A.; Gomes, R.N.; Feitoza, F.; Stevannato, P.B.; da Cunha, A.S.; Serachi, F.O.; Panagopoulos, A.T.; Colquhoun, A. Cyclooxygenase Inhibition Alters Proliferative, Migratory, and Invasive Properties of Human Glioblastoma Cells In Vitro. Int. J. Mol. Sci. 2021, 22, 4297. [Google Scholar] [CrossRef]
- Xu, Z.J.; Gu, Y.; Wang, C.Z.; Jin, Y.; Wen, X.M.; Ma, J.C.; Tang, L.J.; Mao, Z.W.; Qian, J.; Lin, J. The M2 macrophage marker CD206: A novel prognostic indicator for acute myeloid leukemia. Oncoimmunology 2020, 9, 1683347. [Google Scholar] [CrossRef]
- Palumbo, P.; Lombardi, F.; Augello, F.R.; Giusti, I.; Dolo, V.; Leocata, P.; Cifone, M.G.; Cinque, B. Biological effects of selective COX-2 inhibitor NS398 on human glioblastoma cell lines. Cancer Cell Int. 2020, 20, 167. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Y.; Zang, J.; Zheng, M.H.; Zhang, Y.F.; Yue, K.Y.; Cao, X.L.; Cao, Y.; Li, X.X.; Han, H.; Jiang, X.F.; et al. Temozolomide Treatment Induces HMGB1 to Promote the Formation of Glioma Stem Cells via the TLR2/NEAT1/Wnt Pathway in Glioblastoma. Front. Cell Dev. Biol. 2021, 9, 620883. [Google Scholar] [CrossRef] [PubMed]
- Pietras, A.; Katz, A.M.; Ekstrom, E.J.; Wee, B.; Halliday, J.J.; Pitter, K.L.; Werbeck, J.L.; Amankulor, N.M.; Huse, J.T.; Holland, E.C. Osteopontin-CD44 Signaling in the Glioma Perivascular Niche Enhances Cancer Stem Cell Phenotypes and Promotes Aggressive Tumor Growth. Cell Stem Cell 2014, 14, 357–369. [Google Scholar] [CrossRef]
- Qi, L.; Yu, H.Q.; Zhang, Y.; Zhao, D.H.; Lv, P.; Zhong, Y.; Xu, Y. IL-10 secreted by M2 macrophage promoted tumorigenesis through interaction with JAK2 in glioma. Oncotarget 2016, 7, 71673–71685. [Google Scholar] [CrossRef]
- Dean, P.T.; Hooks, S.B. Pleiotropic effects of the COX-2/PGE2 axis in the glioblastoma tumor microenvironment. Front. Oncol. 2022, 12, 1116014. [Google Scholar] [CrossRef] [PubMed]
- Zahner, G.; Schaper, M.; Panzer, U.; Kluger, M.; Stahl, R.A.; Thaiss, F.; Schneider, A. Prostaglandin EP2 and EP4 receptors modulate expression of the chemokine CCL2 (MCP-1) in response to LPS-induced renal glomerular inflammation. Biochem. J. 2009, 422, 563–570. [Google Scholar] [CrossRef]
- Jin, K.; Qian, C.; Lin, J.; Liu, B. Cyclooxygenase-2-Prostaglandin E2 pathway: A key player in tumor-associated immune cells. Front. Oncol. 2023, 13, 1099811. [Google Scholar] [CrossRef] [PubMed]
- Ching, M.M.; Reader, J.; Fulton, A.M. Eicosanoids in Cancer: Prostaglandin E(2) Receptor 4 in Cancer Therapeutics and Immunotherapy. Front. Pharmacol. 2020, 11, 819. [Google Scholar] [CrossRef]
- Vergadi, E.; Ieronymaki, E.; Lyroni, K.; Vaporidi, K.; Tsatsanis, C. Akt Signaling Pathway in Macrophage Activation and M1/M2 Polarization. J. Immunol. 2017, 198, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.; Qian, B.Z.; Rowan, C.; Muthana, M.; Keklikoglou, I.; Olson, O.C.; Tazzyman, S.; Danson, S.; Addison, C.; Clemons, M.; et al. Perivascular M2 Macrophages Stimulate Tumor Relapse after Chemotherapy. Cancer Res. 2015, 75, 3479–3491. [Google Scholar] [CrossRef]
- Gimba, E.R.P.; Brum, M.C.M.; De Moraes, G.N. Full-length osteopontin and its splice variants as modulators of chemoresistance and radioresistance (Review). Int. J. Oncol. 2019, 54, 420–430. [Google Scholar] [CrossRef]
- Hao, C.C.; Lane, J.; Jiang, W.G. Osteopontin and Cancer: Insights into Its Role in Drug Resistance. Biomedicines 2023, 11, 197. [Google Scholar] [CrossRef]
- Kale, S.; Raja, R.; Thorat, D.; Soundararajan, G.; Patil, T.V.; Kundu, G.C. Osteopontin signaling upregulates cyclooxygenase-2 expression in tumor-associated macrophages leading to enhanced angiogenesis and melanoma growth via alpha 9 beta 1 integrin. Oncogene 2014, 33, 2295–2306. [Google Scholar] [CrossRef]
- Jain, S.; Chakraborty, G.; Kundu, G.C. The crucial role of cyclooxygenase-2 in osteopontin-induced protein kinase C alpha/c-Src/IkappaB kinase alpha/beta-dependent prostate tumor progression and angiogenesis. Cancer Res. 2006, 66, 6638–6648. [Google Scholar] [CrossRef]
- Amilca-Seba, K.; Sabbah, M.; Larsen, A.K.; Denis, J.A. Osteopontin as a Regulator of Colorectal Cancer Progression and Its Clinical Applications. Cancers 2021, 13, 3793. [Google Scholar] [CrossRef]
- Zagani, R.; Hamzaoui, N.; Cacheux, W.; De Reyniès, A.; Terris, B.; Chaussade, S.; Romagnolo, B.; Perret, C.; Lamarque, D. Cyclooxygenase-2 Inhibitors Down-regulate Osteopontin and Nr4a2-New Therapeutic Targets for Colorectal Cancers. Gastroenterology 2009, 137, 1358–1366. [Google Scholar] [CrossRef]
- Arora, A.; Somasundaram, K. Glioblastoma vs temozolomide: Can the red queen race be won? Cancer Biol. Ther. 2019, 20, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016, 3, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Miner, A.; Hennis, L.; Mittal, S. Mechanisms of temozolomide resistance in glioblastoma—A comprehensive review. Cancer Drug Resist. 2021, 4, 17–43. [Google Scholar] [CrossRef] [PubMed]
- Yi, G.Z.; Huang, G.; Guo, M.; Zhang, X.; Wang, H.; Deng, S.; Li, Y.; Xiang, W.; Chen, Z.; Pan, J.; et al. Acquired temozolomide resistance in MGMT-deficient glioblastoma cells is associated with regulation of DNA repair by DHC2. Brain 2019, 142, 2352–2366. [Google Scholar] [CrossRef] [PubMed]
- Almeida Lima, K.; Osawa, I.Y.A.; Ramalho, M.C.C.; de Souza, I.; Guedes, C.B.; Souza Filho, C.H.D.; Monteiro, L.K.S.; Latancia, M.T.; Rocha, C.R.R. Temozolomide Resistance in Glioblastoma by NRF2: Protecting the Evil. Biomedicines 2023, 11, 1081. [Google Scholar] [CrossRef] [PubMed]



| Primary Antibody | Dilution | Company |
|---|---|---|
| rabbit monoclonal anti-COX-2 | 1:1000 | Cell Signaling Technology, Danvers, MA, USA |
| rabbit monoclonal anti-osteopontin | 1:1000 | Boster Biological Technology, Pleasanton, CA, USA |
| mouse monoclonal anti-CD44 | 1:1000 | Cell Signaling Technology, Danvers, MA, USA |
| mouse monoclonal anti-β-actin | 1:1000 | Bio-Rad, Hercules, CA, USA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lombardi, F.; Augello, F.R.; Artone, S.; Ciafarone, A.; Topi, S.; Cifone, M.G.; Cinque, B.; Palumbo, P. Involvement of Cyclooxygenase-2 in Establishing an Immunosuppressive Microenvironment in Tumorspheres Derived from TMZ-Resistant Glioblastoma Cell Lines and Primary Cultures. Cells 2024, 13, 258. https://doi.org/10.3390/cells13030258
Lombardi F, Augello FR, Artone S, Ciafarone A, Topi S, Cifone MG, Cinque B, Palumbo P. Involvement of Cyclooxygenase-2 in Establishing an Immunosuppressive Microenvironment in Tumorspheres Derived from TMZ-Resistant Glioblastoma Cell Lines and Primary Cultures. Cells. 2024; 13(3):258. https://doi.org/10.3390/cells13030258
Chicago/Turabian StyleLombardi, Francesca, Francesca Rosaria Augello, Serena Artone, Alessia Ciafarone, Skender Topi, Maria Grazia Cifone, Benedetta Cinque, and Paola Palumbo. 2024. "Involvement of Cyclooxygenase-2 in Establishing an Immunosuppressive Microenvironment in Tumorspheres Derived from TMZ-Resistant Glioblastoma Cell Lines and Primary Cultures" Cells 13, no. 3: 258. https://doi.org/10.3390/cells13030258
APA StyleLombardi, F., Augello, F. R., Artone, S., Ciafarone, A., Topi, S., Cifone, M. G., Cinque, B., & Palumbo, P. (2024). Involvement of Cyclooxygenase-2 in Establishing an Immunosuppressive Microenvironment in Tumorspheres Derived from TMZ-Resistant Glioblastoma Cell Lines and Primary Cultures. Cells, 13(3), 258. https://doi.org/10.3390/cells13030258









