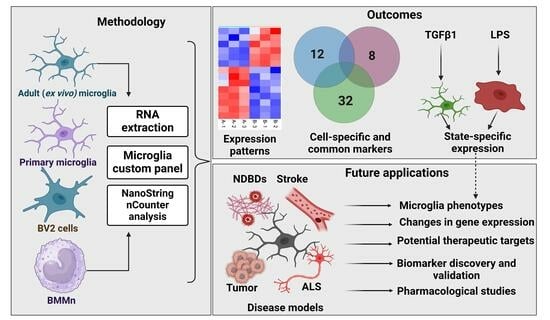
A Custom Panel for Profiling Microglia Gene Expression
Abstract
1. Introduction
2. Materials and Methods
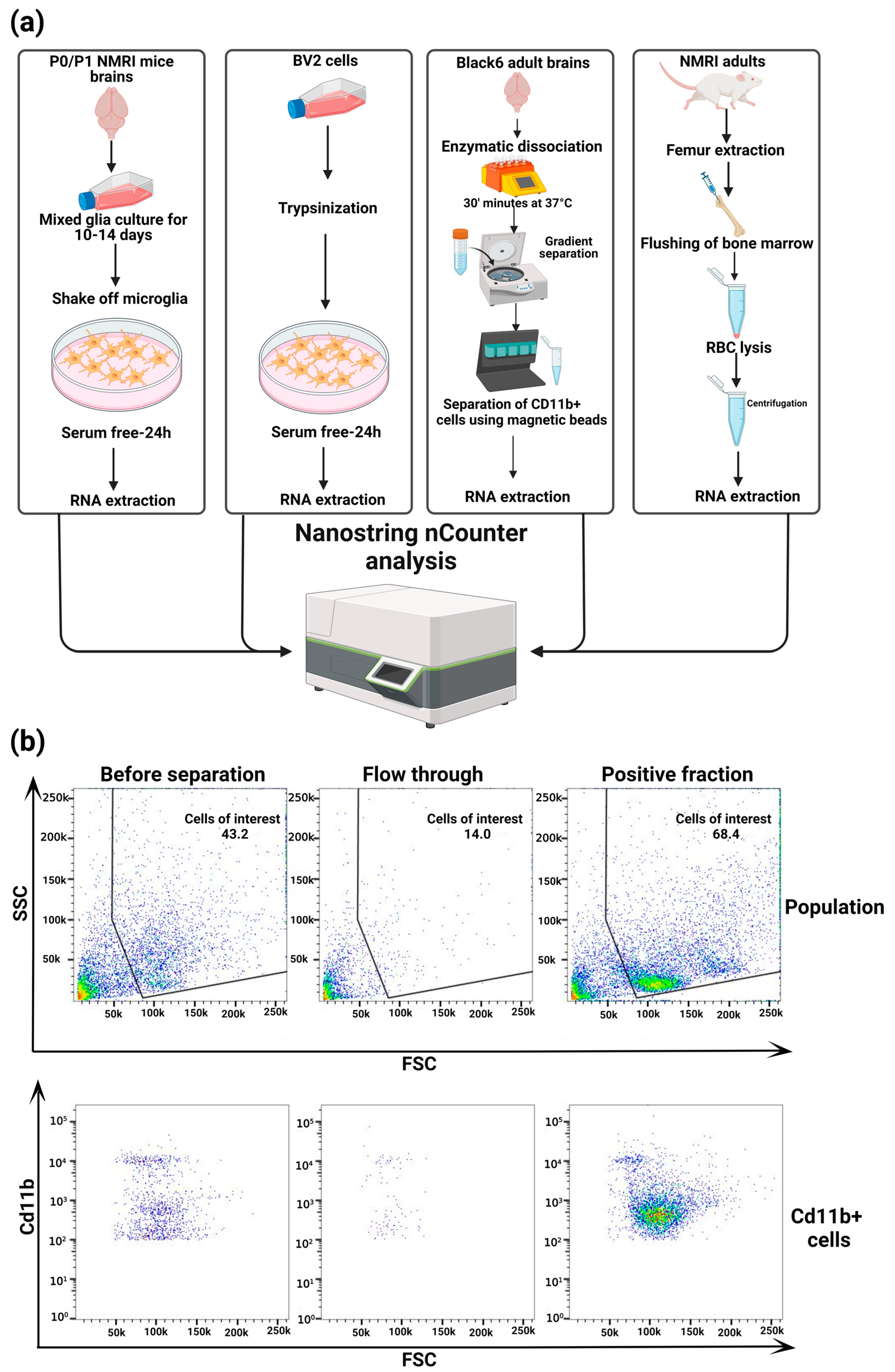
2.1. Animals
2.2. Primary Microglia Culture
2.3. BV2 Culture
2.4. Bone Marrow Monocyte Extraction
2.5. Ex Vivo Microglia Isolation and Flow Cytometry
2.6. RNA Extraction
2.7. NanoString nCounter Analysis
2.8. Data Availability
3. Results
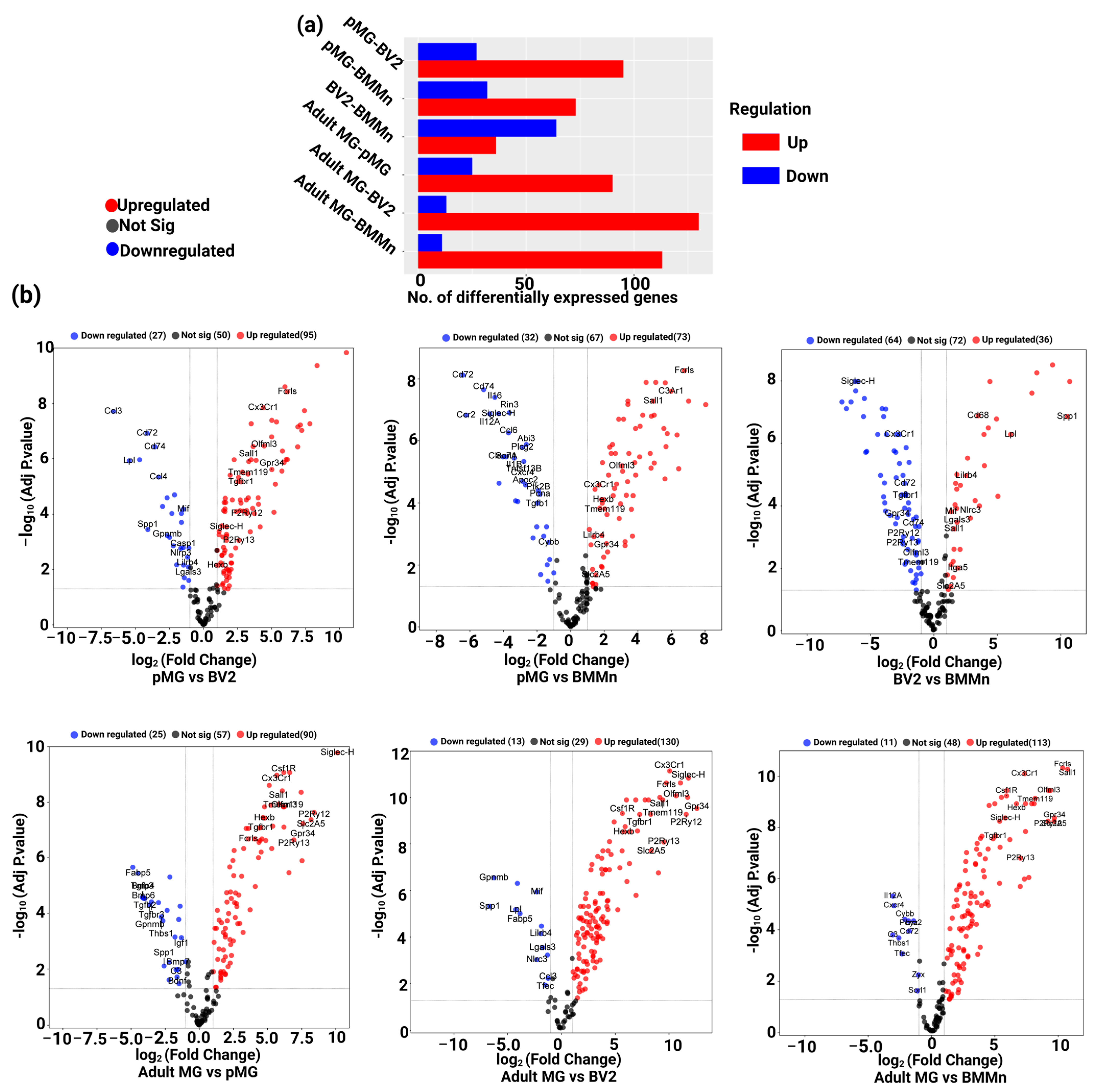
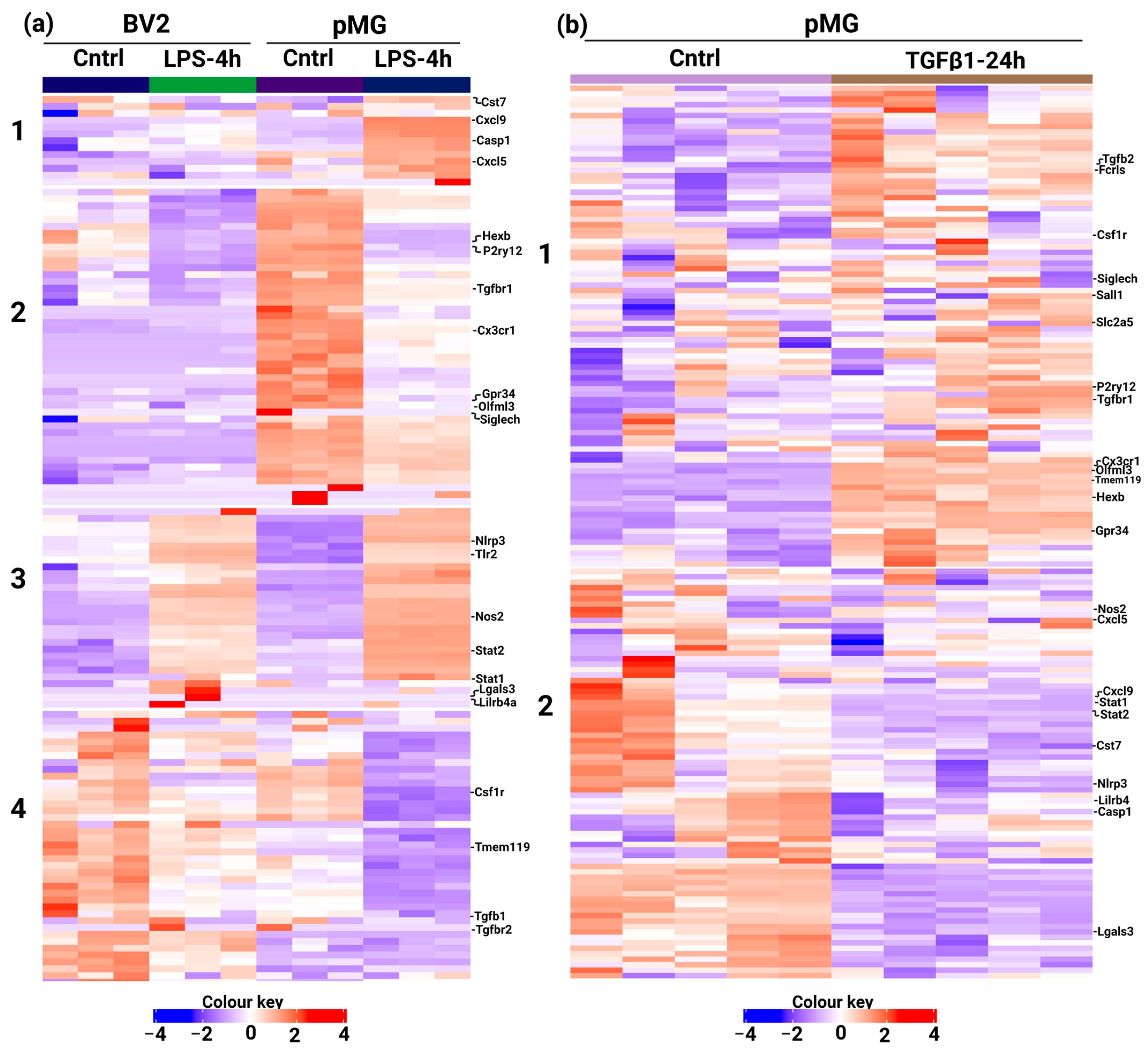
3.1. Custom Panel Identifies Distinct Gene Expression Patterns in Microglia Cells
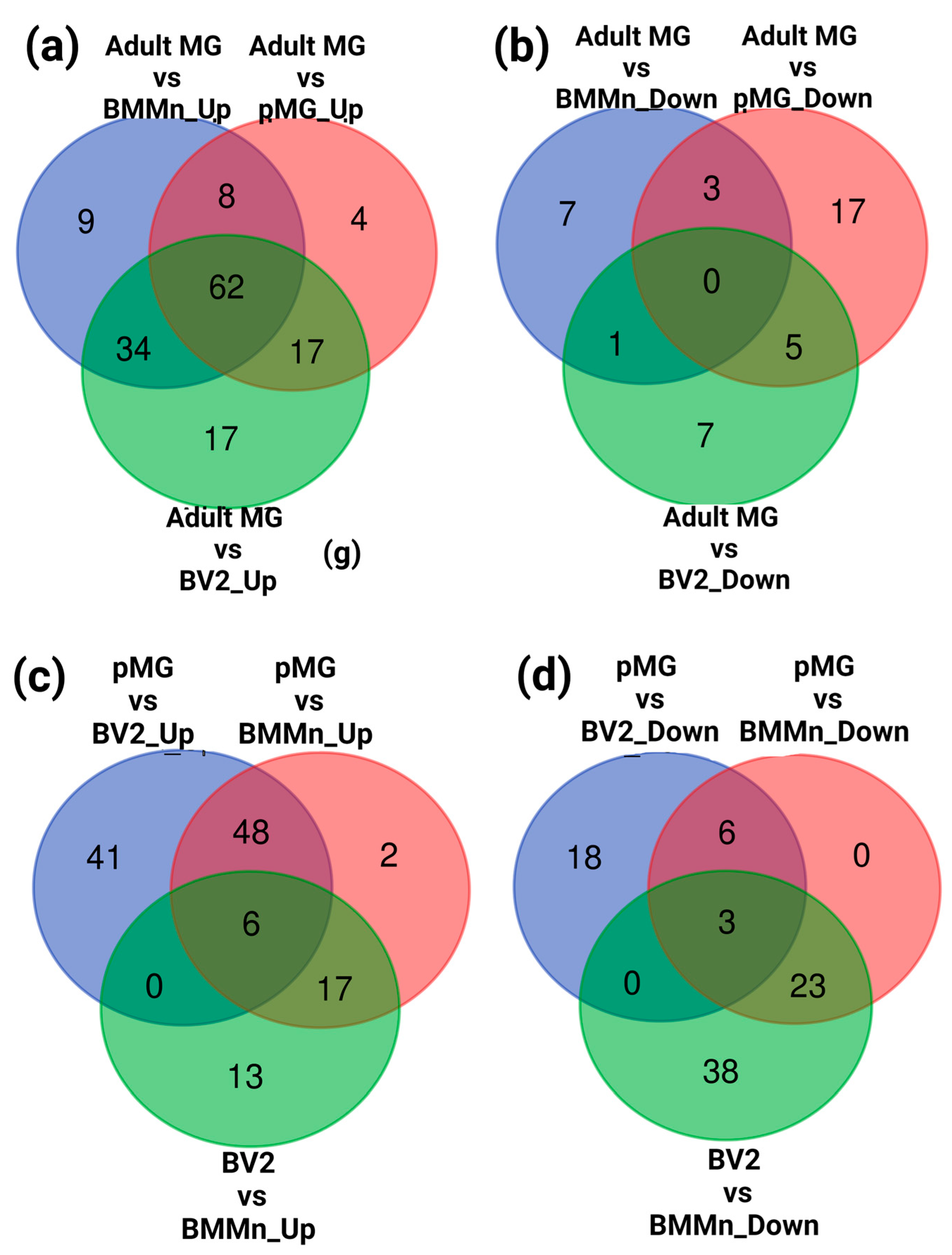
3.2. Differential Gene Expression across Microglia Cell Types and BMMns
3.3. Custom Panels Can Account for Functional Changes in Microglia
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nimmerjahn, A.; Kirchhoff, F.; Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005, 308, 1314–1318. [Google Scholar] [CrossRef] [PubMed]
- Davalos, D.; Grutzendler, J.; Yang, G.; Kim, J.V.; Zuo, Y.; Jung, S.; Littman, D.R.; Dustin, M.L.; Gan, W.-B. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 2005, 8, 752–758. [Google Scholar] [CrossRef]
- Schafer, D.P.; Lehrman, E.K.; Kautzman, A.G.; Koyama, R.; Mardinly, A.R.; Yamasaki, R.; Ransohoff, R.M.; Greenberg, M.E.; Barres, B.A.; Stevens, B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 2012, 74, 691–705. [Google Scholar] [CrossRef] [PubMed]
- Spittau, B.; Dokalis, N.; Prinz, M. The Role of TGFβ Signaling in Microglia Maturation and Activation. Trends Immunol. 2020, 41, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Prinz, M.; Priller, J. Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nat. Rev. Neurosci. 2014, 15, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Prinz, M.; Jung, S.; Priller, J. Microglia Biology: One Century of Evolving Concepts. Cell 2019, 179, 292–311. [Google Scholar] [CrossRef] [PubMed]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a Unique TGF-β Dependent Molecular and Functional Signature in Microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Amann, L.; Sankowski, R.; Staszewski, O.; Lenz, M.; d’Errico, P.; Snaidero, N.; Jordão, M.J.C.; Böttcher, C.; Kierdorf, K.; et al. Novel Hexb-based tools for studying microglia in the CNS. Nat. Immunol. 2020, 21, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Boche, D.; Gordon, M.N. Diversity of transcriptomic microglial phenotypes in aging and Alzheimer’s disease. Alzheimers Dement. 2022, 18, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Butovsky, O.; Weiner, H.L. Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 2018, 19, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zöller, T.; Krieglstein, K.; Spittau, B. TGFβ1 inhibits IFNγ-mediated microglia activation and protects mDA neurons from IFNγ-driven neurotoxicity. J. Neurochem. 2015, 134, 125–134. [Google Scholar] [CrossRef]
- Wagner, M.; Koester, H.; Deffge, C.; Weinert, S.; Lauf, J.; Francke, A.; Lee, J.; Dullaeus, R.C.B.; Herold, J. Isolation and intravenous injection of murine bone marrow derived monocytes. J. Vis. Exp. JoVE 2014, 27, e52347. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Kim, S.H.; Arifuzzaman, S.; Yoon, T.; Chai, J.C.; Lee, Y.S.; Park, K.S.; Jung, K.H.; Chai, Y.G. Transcriptome sequencing reveals that LPS-triggered transcriptional responses in established microglia BV2 cell lines are poorly representative of primary microglia. J. Neuroinflamm. 2016, 13, 182. [Google Scholar] [CrossRef] [PubMed]
- Zöller, T.; Schneider, A.; Kleimeyer, C.; Masuda, T.; Potru, P.S.; Pfeifer, D.; Blank, T.; Prinz, M.; Spittau, B. Silencing of TGFβ signalling in microglia results in impaired homeostasis. Nat. Commun. 2018, 9, 4011. [Google Scholar] [CrossRef] [PubMed]
- Ayata, P.; Badimon, A.; Strasburger, H.J.; Duff, M.K.; Montgomery, S.E.; Loh, Y.-H.E.; Ebert, A.; Pimenova, A.A.; Ramirez, B.R.; Chan, A.T.; et al. Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci. 2018, 21, 1049–1060. [Google Scholar] [CrossRef]
- Grubman, A.; Choo, X.Y.; Chew, G.; Ouyang, J.F.; Sun, G.; Croft, N.P.; Rossello, F.J.; Simmons, R.; Buckberry, S.; Landin, D.V.; et al. Transcriptional signature in microglia associated with Aβ plaque phagocytosis. Nat. Commun. 2021, 12, 3015. [Google Scholar] [CrossRef] [PubMed]
- Haage, V.; Semtner, M.; Vidal, R.O.; Hernandez, D.P.; Pong, W.W.; Chen, Z.; Hambardzumyan, D.; Magrini, V.; Ly, A.; Walker, J.; et al. Comprehensive gene expression meta-analysis identifies signature genes that distinguish microglia from peripheral monocytes/macrophages in health and glioma. Acta Neuropathol. Commun. 2019, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. [Google Scholar] [CrossRef] [PubMed]
- Krasemann, S.; Madore, C.; Cialic, R.; Baufeld, C.; Calcagno, N.; El Fatimy, R.; Beckers, L.; O’Loughlin, E.; Xu, Y.; Fanek, Z.; et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 2017, 47, 566–581.e9. [Google Scholar] [CrossRef] [PubMed]
- Zrzavy, T.; Hametner, S.; Wimmer, I.; Butovsky, O.; Weiner, H.L.; Lassmann, H. Loss of “homeostatic” microglia and patterns of their activation in active multiple sclerosis. Brain J. Neurol. 2017, 140, 1900–1913. [Google Scholar] [CrossRef] [PubMed]
- Goytain, A.; Ng, T. NanoString nCounter Technology: High-Throughput RNA Validation. In Chimeric RNA: Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2079, pp. 125–139. [Google Scholar] [CrossRef]
- Cadiz, M.P.; Jensen, T.D.; Sens, J.P.; Zhu, K.; Song, W.-M.; Zhang, B.; Ebbert, M.; Chang, R.; Fryer, J.D. Culture shock: Microglial heterogeneity, activation, and disrupted single-cell microglial networks in vitro. Mol. Neurodegener. 2022, 17, 26. [Google Scholar] [CrossRef]
- Kretzschmar, F.; Piecha, R.; Jahn, J.; Potru, P.S.; Spittau, B. Characterization of the Leucocyte Immunoglobulin-like Receptor B4 (Lilrb4) Expression in Microglia. Biology 2021, 10, 1300. [Google Scholar] [CrossRef] [PubMed]
- Von Ehr, A.; Attaai, A.; Neidert, N.; Potru, P.S.; Ruß, T.; Zöller, T.; Spittau, B. Inhibition of Microglial TGFβ Signaling Increases Expression of Mrc1. Front. Cell. Neurosci. 2020, 14, 66. Available online: https://www.frontiersin.org/articles/10.3389/fncel.2020.00066 (accessed on 25 January 2024). [CrossRef] [PubMed]
- Zhao, Y.; Ma, C.; Chen, C.; Li, S.; Wang, Y.; Yang, T.; Stetler, R.A.; Bennett, M.V.L.; Dixon, C.E.; Chen, J.; et al. STAT1 Contributes to Microglial/Macrophage Inflammation and Neurological Dysfunction in a Mouse Model of Traumatic Brain Injury. J. Neurosci. 2022, 42, 7466–7481. [Google Scholar] [CrossRef] [PubMed]
- Hirbec, H.; Marmai, C.; Duroux-Richard, I.; Roubert, C.; Esclangon, A.; Croze, S.; Lachuer, J.; Peyroutou, R.; Rassendren, F. The microglial reaction signature revealed by RNAseq from individual mice. Glia 2018, 66, 971–986. [Google Scholar] [CrossRef] [PubMed]
- Lively, S.; Schlichter, L.C. Microglia Responses to Pro-inflammatory Stimuli (LPS, IFNγ+TNFα) and Reprogramming by Resolving Cytokines (IL-4, IL-10). Front. Cell. Neurosci. 2018, 12, 215. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.; Golebiewska, A.; Poovathingal, S.K.; Kaoma, T.; Pires-Afonso, Y.; Martina, S.; Coowar, D.; Azuaje, F.; Skupin, A.; Balling, R.; et al. Single-cell transcriptomics reveals distinct inflammation-induced microglia signatures. EMBO Rep. 2018, 19, e46171. [Google Scholar] [CrossRef] [PubMed]
- Paolicelli, R.C.; Sierra, A.; Stevens, B.; Tremblay, M.-E.; Aguzzi, A.; Ajami, B.; Amit, I.; Audinat, E.; Bechmann, I.; Bennett, M.; et al. Microglia states and nomenclature: A field at its crossroads. Neuron 2022, 110, 3458–3483. [Google Scholar] [CrossRef] [PubMed]
- Attaai, A.; Neidert, N.; von Ehr, A.; Potru, P.S.; Zöller, T.; Spittau, B. Postnatal maturation of microglia is associated with alternative activation and activated TGFβ signaling. Glia 2018, 66, 1695–1708. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Potru, P.S.; Vidovic, N.; Wiemann, S.; Russ, T.; Trautmann, M.; Spittau, B. A Custom Panel for Profiling Microglia Gene Expression. Cells 2024, 13, 630. https://doi.org/10.3390/cells13070630
Potru PS, Vidovic N, Wiemann S, Russ T, Trautmann M, Spittau B. A Custom Panel for Profiling Microglia Gene Expression. Cells. 2024; 13(7):630. https://doi.org/10.3390/cells13070630
Chicago/Turabian StylePotru, Phani Sankar, Natascha Vidovic, Susanne Wiemann, Tamara Russ, Marcel Trautmann, and Björn Spittau. 2024. "A Custom Panel for Profiling Microglia Gene Expression" Cells 13, no. 7: 630. https://doi.org/10.3390/cells13070630
APA StylePotru, P. S., Vidovic, N., Wiemann, S., Russ, T., Trautmann, M., & Spittau, B. (2024). A Custom Panel for Profiling Microglia Gene Expression. Cells, 13(7), 630. https://doi.org/10.3390/cells13070630