Miro GTPases at the Crossroads of Cytoskeletal Dynamics and Mitochondrial Trafficking
Abstract
:1. Introduction

2. Domain Organization of Miro GTPases
3. Miro GTPases Are Involved in Organelle Homeostasis
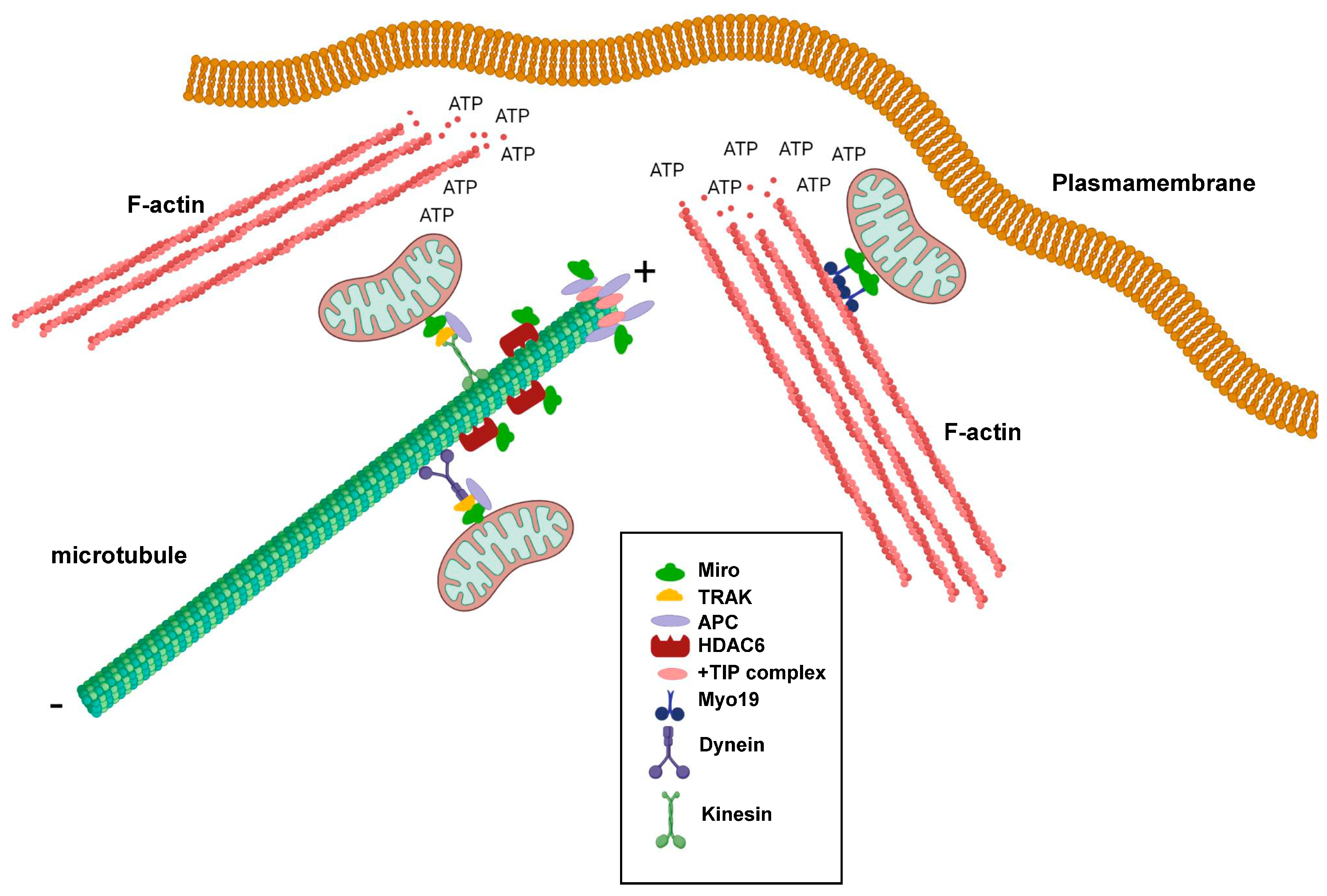
4. Miro GTPases in Microtubule Dynamics
5. Miro GTPases Are Involved in Actin Dynamics
6. Miro in Neuropathologies
7. Conclusions
Funding
Conflicts of Interest
References
- Aspenström, P.; Fransson, Å.; Saras, J. The Rho GTPases have diverse effects on the organization of the actin filament system. Biochem. J. 2004, 377, 327–337. [Google Scholar] [CrossRef]
- Fransson, Å.; Ruusala, A.; Aspenström, P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem. 2003, 278, 6495–6502. [Google Scholar] [CrossRef]
- Klosowiak, J.L.; Focia, P.J.; Chakravarthy, S.; Landahl, E.C.; Freymann, D.M.; Rice, S.E. Structural coupling of the EF hand and C-terminal GTPase domains in the mitochondrial protein Miro. EMBO Rep. 2013, 14, 968–974. [Google Scholar] [CrossRef]
- Wennerberg, K.; Der, C.J. Rho-family GTPases: It’s not only Rac and Rho (and I like it). J. Cell Sci. 2004, 117, 1301–1312. [Google Scholar] [CrossRef]
- Fransson, Å.; Ruusala, A.; Aspenström, P. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem. Biophys. Res. Commun. 2006, 344, 500–510. [Google Scholar] [CrossRef]
- Boureux, A.; Vignal, E.; Faure, S.; Fort, P. Evolution of the Rho family of Ras-like GTPases in eukaryotes. Mol. Biol. Evol. 2007, 24, 203–216. [Google Scholar] [CrossRef]
- Cai, Q.; Davis, M.L.; Sheng, Z.H. Regulation of axonal mitochondrial transport and its impact on synaptic transmission. Neurosci. Res. 2011, 70, 9–15. [Google Scholar] [CrossRef]
- Tang, B.L. MIRO GTPases in Mitochondrial Transport, Homeostasis and Pathology. Cells 2015, 5, 1. [Google Scholar] [CrossRef]
- Kay, L.; Pienaar, I.S.; Cooray, R.; Black, G.; Soundararajan, M. Understanding Miro GTPases: Implications in the Treatment of Neurodegenerative Disorders. Mol. Neurobiol. 2018, 55, 7352–7365. [Google Scholar] [CrossRef]
- Eberhardt, E.L.; Ludlam, A.V.; Tan, Z.; Cianfrocco, M.A. Miro: A molecular switch at the center of mitochondrial regulation. Protein Sci. 2020, 29, 1269–1284. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Frederick, R.L.; McCaffery, J.M.; Cunningham, K.W.; Okamoto, K.; Shaw, J.M. Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J. Cell Biol. 2004, 167, 87–98. [Google Scholar] [CrossRef]
- Vlahou, G.; Eliáš, M.; von Kleist-Retzow, J.C.; Wiesner, R.J.; Rivero, F. The Ras related GTPase Miro is not required for mitochondrial transport in Dictyostelium discoideum. Eur. J. Cell Biol. 2011, 90, 342–355. [Google Scholar] [CrossRef]
- Shen, Y.; Ng, L.F.; Low, N.P.; Hagen, T.; Gruber, J.; Inoue, T.C. Elegans miro-1 Mutation Reduces the amount of Mitochondria and Extends Life Span. PLoS ONE 2016, 11, e0153233. [Google Scholar] [CrossRef]
- Baltrusaitis, E.E.; Ravitch, E.E.; Fenton, A.R.; Perez, T.A.; Holzbaur, E.L.F.; Dominguez, R. Interaction between the mitochondrial adaptor MIRO and the motor adaptor TRAK. J. Biol. Chem. 2023, 299, 105441. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef]
- Cox, A.D.; Der, C.J. Ras history: The saga continues. Small GTPases 2010, 1, 2–27. [Google Scholar] [CrossRef]
- Aspenström, P. Fast-cycling Rho GTPases. Small GTPases 2020, 11, 248–255. [Google Scholar] [CrossRef]
- Peters, D.T.; Kay, L.; Eswaran, J.; Lakey, J.H.; Soundararajan, M. Human Miro Proteins Act as NTP Hydrolases through a Novel, Non-Canonical Catalytic Mechanism. Int. J. Mol. Sci. 2018, 19, 3839. [Google Scholar] [CrossRef]
- Lee, S.; Lee, K.S.; Huh, S.; Liu, S.; Lee, D.Y.; Hong, S.H.; Yu, K.; Lu, B. Polo Kinase Phosphorylates Miro to Control ER-Mitochondria Contact Sites and Mitochondrial Ca(2+) Homeostasis in Neural Stem Cell Development. Dev. Cell 2016, 37, 174–189. [Google Scholar] [CrossRef]
- Suzuki, M.; Danilchanka, O.; Mekalanos, J.J. Vibrio cholerae T3SS effector VopE modulates mitochondrial dynamics and innate immune signaling by targeting Miro GTPases. Cell Host Microbe 2014, 16, 581–591. [Google Scholar] [CrossRef]
- Smith, K.P.; Focia, P.J.; Chakravarthy, S.; Landahl, E.C.; Klosowiak, J.L.; Rice, S.E.; Freymann, D.M. Insight into human Miro1/2 domain organization based on the structure of its N-terminal GTPase. J. Struct. Biol. 2020, 212, 107656. [Google Scholar] [CrossRef] [PubMed]
- Koshiba, T.; Holman, H.A.; Kubara, K.; Yasukawa, K.; Kawabata, S.; Okamoto, K.; MacFarlane, J.; Shaw, J.M. Structure-function analysis of the yeast mitochondrial Rho GTPase, Gem1p: Implications for mitochondrial inheritance. J. Biol. Chem. 2011, 286, 354–362. [Google Scholar] [CrossRef]
- Saotome, M.; Safiulina, D.; Szabadkai, G.; Das, S.; Fransson, Å.; Aspenström, P.; Rizzuto, R.; Hajnóczky, G. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc. Natl. Acad. Sci. USA 2008, 105, 20728–20733. [Google Scholar] [CrossRef] [PubMed]
- Macaskill, A.F.; Rinholm, J.E.; Twelvetrees, A.E.; Arancibia-Carcamo, I.L.; Muir, J.; Fransson, Å.; Aspenström, P.; Attwell, D.; Kittler, J.T. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron 2009, 61, 541–555. [Google Scholar] [CrossRef]
- Jaffe, A.B.; Hall, A. Rho GTPases: Biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005, 21, 247–269. [Google Scholar] [CrossRef] [PubMed]
- Glater, E.E.; Megeath, L.J.; Stowers, R.S.; Schwarz, T.L. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 2006, 173, 545–557. [Google Scholar] [CrossRef]
- Russo, G.J.; Louie, K.; Wellington, A.; Macleod, G.T.; Hu, F.; Panchumarthi, S.; Zinsmaier, K.E. Drosophila Miro is required for both anterograde and retrograde axonal mitochondrial transport. J. Neurosci. 2009, 29, 5443–5455. [Google Scholar] [CrossRef]
- Kornmann, B.; Osman, C.; Walter, P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. USA 2011, 108, 14151–14156. [Google Scholar] [CrossRef]
- Niescier, R.F.; Hong, K.; Park, D.; Min, K.T. MCU Interacts with Miro1 to Modulate Mitochondrial Functions in Neurons. J. Neurosci. 2018, 38, 4666–4677. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; López-Doménech, G.; Halff, E.F.; Covill-Cooke, C.; Ivankovic, D.; Melandri, D.; Arancibia-Cárcamo, I.L.; Burden, J.J.; Lowe, A.R.; Kittler, J.T. Miro clusters regulate ER-mitochondria contact sites and link cristae organization to the mitochondrial transport machinery. Nat. Commun. 2019, 10, 4399. [Google Scholar] [CrossRef] [PubMed]
- Green, A.; Hossain, T.; Eckmann, D.M. Mitochondrial dynamics involves molecular and mechanical events in motility, fusion and fission. Front. Cell Dev. Biol. 2022, 10, 1010232. [Google Scholar] [CrossRef] [PubMed]
- Misko, A.; Jiang, S.; Wegorzewska, I.; Milbrandt, J.; Baloh, R.H. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J. Neurosci. 2010, 30, 4232–4240. [Google Scholar] [CrossRef] [PubMed]
- Züchner, S.; Mersiyanova, I.V.; Muglia, M.; Bissar-Tadmouri, N.; Rochelle, J.; Dadali, E.L.; Zappia, M.; Nelis, E.; Patitucci, A.; Senderek, J.; et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 2004, 36, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Fatiga, F.F.; Wang, L.J.; Hsu, T.; Capuno, J.I.; Fu, C.Y. Miro1 functions as an inhibitory regulator of MFN at elevated mitochondrial Ca2+ levels. J. Cell Biochem. 2021, 122, 1848–1862. [Google Scholar] [CrossRef] [PubMed]
- López-Doménech, G.; Covill-Cooke, C.; Ivankovic, D.; Halff, E.F.; Sheehan, D.F.; Norkett, R.; Birsa, N.; Kittler, J.T. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J. 2018, 37, 321–336. [Google Scholar] [CrossRef] [PubMed]
- Brustovetsky, T.; Khanna, R.; Brustovetsky, N. Involvement of CRMP2 in Regulation of Mitochondrial Morphology and Motility in Huntington’s Disease. Cells 2021, 10, 3172. [Google Scholar] [CrossRef] [PubMed]
- Covill-Cooke, C.; Toncheva, V.S.; Kittler, J.T. Regulation of peroxisomal trafficking and distribution. Cell. Mol. Life Sci. 2021, 78, 1929–1941. [Google Scholar] [CrossRef]
- Covill-Cooke, C.; Toncheva, V.S.; Drew, J.; Birsa, N.; López-Doménech, G.; Kittler, J.T. Peroxisomal fission is modulated by the mitochondrial Rho-GTPases, Miro1 and Miro2. EMBO Rep. 2020, 21, e49865. [Google Scholar] [CrossRef]
- Melwani, P.K.; Pandey, B.N. Tunneling nanotubes: The intercellular conduits contributing to cancer pathogenesis and its therapy. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 189028. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, T.; Mukherjee, S.; Pattnaik, B.; Kumar, M.; Singh, S.; Kumar, M.; Rehman, R.; Tiwari, B.K.; Jha, K.A.; Barhanpurkar, A.P.; et al. Miro1 regulates intercellular mitochondrial transport & enhances mesenchymal stem cell rescue efficacy. EMBO J. 2014, 33, 994–1010. [Google Scholar] [CrossRef]
- Cleary, J.M.; Hancock, W.O. Molecular mechanisms underlying microtubule growth dynamics. Curr. Biol. 2021, 31, R560–R573. [Google Scholar] [CrossRef] [PubMed]
- Mills, K.M.; Brocardo, M.G.; Henderson, B.R. APC binds the Miro/Milton motor complex to stimulate transport of mitochondria to the plasma membrane. Mol. Biol. Cell 2016, 27, 466–482. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Svitkina, T.M. Adenomatous Polyposis Coli (APC) in cell migration. Eur. J. Cell Biol. 2022, 101, 151228. [Google Scholar] [CrossRef] [PubMed]
- Brocardo, M.; Lei, Y.; Tighe, A.; Taylor, S.S.; Mok, M.T.; Henderson, B.R. Mitochondrial targeting of adenomatous polyposis coli protein is stimulated by truncating cancer mutations: Regulation of Bcl-2 and implications for cell survival. J. Biol. Chem. 2008, 283, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Weiner, A.T.; Seebold, D.Y.; Michael, N.L.; Guignet, M.; Feng, C.; Follick, B.; Yusko, B.A.; Wasilko, N.P.; Torres-Gutierrez, P.; Rolls, M.M. Identification of Proteins Required for Precise Positioning of Apc2 in Dendrites. G3 2018, 8, 1841–1853. [Google Scholar] [CrossRef] [PubMed]
- Balmik, A.A.; Chinnathambi, S. Inter-relationship of Histone Deacetylase-6 with Tau-cytoskeletal organization and remodeling. Eur. J. Cell Biol. 2022, 101, 151202. [Google Scholar] [CrossRef] [PubMed]
- Kalinski, A.L.; Kar, A.N.; Craver, J.; Tosolini, A.P.; Sleigh, J.N.; Lee, S.J.; Hawthorne, A.; Brito-Vargas, P.; Miller-Randolph, S.; Passino, R.; et al. Deacetylation of Miro1 by HDAC6 blocks mitochondrial transport and mediates axon growth inhibition. J. Cell Biol. 2019, 218, 1871–1890. [Google Scholar] [CrossRef]
- Morris, R.L.; Hollenbeck, P.J. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J. Cell Biol. 1995, 131, 1315–1326. [Google Scholar] [CrossRef]
- Pathak, D.; Sepp, K.J.; Hollenbeck, P.J. Evidence that myosin activity opposes microtubule-based axonal transport of mitochondria. J. Neurosci. 2010, 30, 8984–8992. [Google Scholar] [CrossRef]
- Quintero, O.A.; DiVito, M.M.; Adikes, R.C.; Kortan, M.B.; Case, L.B.; Lier, A.J.; Panaretos, N.S.; Slater, S.Q.; Rengarajan, M.; Feliu, M.; et al. Human Myo19 is a novel myosin that associates with mitochondria. Curr. Biol. 2009, 19, 2008–2013. [Google Scholar] [CrossRef]
- Morris, R.L.; Hollenbeck, P.J. The regulation of bidirectional mitochondrial transport is coordinated with axonal outgrowth. J. Cell Sci. 1993, 104, 917–927. [Google Scholar] [CrossRef]
- Dresbach, T.; Qualmann, B.; Kessels, M.M.; Garner, C.C.; Gundelfinger, E.D. The presynaptic cytomatrix of brain synapses. Cell. Mol. Life Sci. 2001, 58, 94–116. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, J.; Yu, M.; Xie, Y.; Huang, Y.; Wolff, D.W.; Abel, P.W.; Tu, Y. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene 2013, 32, 4814–4824. [Google Scholar] [CrossRef]
- Cunniff, B.; McKenzie, A.J.; Heintz, N.H.; Howe, A.K. AMPK activity regulates trafficking of mitochondria to the leading edge during cell migration and matrix invasion. Mol. Biol. Cell 2016, 27, 2662–2674. [Google Scholar] [CrossRef]
- Pollard, T.D.; Cooper, J.A. Actin, a central player in cell shape and movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef]
- Schuler, M.H.; Lewandowska, A.; Caprio, G.D.; Skillern, W.; Upadhyayula, S.; Kirchhausen, T.; Shaw, J.M.; Cunniff, B. Miro1-mediated mitochondrial positioning shapes intracellular energy gradients required for cell migration. Mol. Biol. Cell 2017, 28, 2159–2169. [Google Scholar] [CrossRef]
- Cangkrama, M.; Liu, H.; Whipman, J.; Zubair, M.; Matsushita, M.; Di Filippo, M.; Kopf, M.; Innocenti, M.; Werner, S.A. Protumorigenic mDia2-MIRO1 Axis Controls Mitochondrial Positioning and Function in Cancer-Associated Fibroblasts. Cancer Res. 2022, 82, 3701–3717. [Google Scholar] [CrossRef]
- Traut, T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 1994, 140, 1–22. [Google Scholar] [CrossRef]
- Zsurka, G.; Kunz, W.S. Mitochondrial involvement in neurodegenerative diseases. IUBMB Life 2013, 65, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, V.; Palermo, G.; Del Prete, E.; Mancuso, M.; Ceravolo, R. Understanding the Multiple Role of Mitochondria in Parkinson’s Disease and Related Disorders: Lesson from Genetics and Protein-Interaction Network. Front. Cell Dev. Biol. 2021, 9, 636506. [Google Scholar] [CrossRef]
- Horvath, R.; Medina, J.; Reilly, M.M.; Shy, M.E.; Zuchner, S. Peripheral neuropathy in mitochondrial disease. Handb. Clin. Neurol. 2023, 194, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Bano, D.; Ehninger, D.; Bagetta, G. Decoding metabolic signatures in Alzheimer’s disease: A mitochondrial perspective. Cell Death Discov. 2023, 9, 432. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.W.; Guo, M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson’s disease. Curr. Opin. Neurobiol. 2007, 17, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Weihofen, A.; Thomas, K.J.; Ostaszewski, B.L.; Cookson, M.R.; Selkoe, D.J. Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry 2009, 48, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Winter, D.; Ashrafi, G.; Schlehe, J.; Wong, Y.L.; Selkoe, D.; Rice, S.; Steen, J.; LaVoie, M.J.; Schwarz, T.L. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 2011, 147, 893–906. [Google Scholar] [CrossRef]
- Safiulina, D.; Kuum, M.; Choubey, V.; Gogichaishvili, N.; Liiv, J.; Hickey, M.A.; Cagalinec, M.; Mandel, M.; Zeb, A.; Liiv, M.; et al. Miro proteins prime mitochondria for Parkin translocation and mitophagy. EMBO J. 2019, 38, e99384. [Google Scholar] [CrossRef] [PubMed]
- Shlevkov, E.; Kramer, T.; Schapansky, J.; LaVoie, M.J.; Schwarz, T.L. Miro phosphorylation sites regulate Parkin recruitment and mitochondrial motility. Proc. Natl. Acad. Sci. USA 2016, 113, E6097–E6106. [Google Scholar] [CrossRef]
- Kazlauskaite, A.; Martínez-Torres, R.J.; Wilkie, S.; Kumar, A.; Peltier, J.; Gonzalez, A.; Johnson, C.; Zhang, J.; Hope, A.G.; Peggie, M.; et al. Binding to serine 65-phosphorylated ubiquitin primes Parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep. 2015, 16, 939–954. [Google Scholar] [CrossRef]
- López-Doménech, G.; Howden, J.H.; Covill-Cooke, C.; Morfill, C.; Patel, J.V.; Bürli, R.; Crowther, D.; Birsa, N.; Brandon, N.J.; Kittler, J.T. Loss of neuronal Miro1 disrupts mitophagy and induces hyperactivation of the integrated stress response. EMBO J. 2021, 40, e100715. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.H.; Shaltouki, A.; Gonzalez, A.E.; Bettencourt da Cruz, A.; Burbulla, L.F.; St Lawrence, E.; Schüle, B.; Krainc, D.; Palmer, T.D.; Wang, X. Functional Impairment in Miro Degradation and Mitophagy Is a Shared Feature in Familial and Sporadic Parkinson’s Disease. Cell Stem Cell 2016, 19, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.H.; Li, L.; Vanhauwaert, R.; Nguyen, K.T.; Davis, M.D.; Bu, G.; Wszolek, Z.K.; Wang, X. Miro1 Marks Parkinson’s Disease Subset and Miro1 Reducer Rescues Neuron Loss in Parkinson’s Models. Cell Metab. 2019, 30, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, D.; Berenguer-Escuder, C.; Bellet, M.E.; Scheibner, D.; Bohler, J.; Massart, F.; Rapaport, D.; Skupin, A.; Fouquier d’Hérouël, A.; Sharma, M.; et al. Mutations in RHOT1 Disrupt Endoplasmic Reticulum-Mitochondria Contact Sites Interfering with Calcium Homeostasis and Mitochondrial Dynamics in Parkinson’s Disease. Antioxid. Redox Signal. 2019, 31, 1213–1234. [Google Scholar] [CrossRef] [PubMed]
- Iijima-Ando, K.; Sekiya, M.; Maruko-Otake, A.; Ohtake, Y.; Suzuki, E.; Lu, B.; Iijima, K.M. Loss of axonal mitochondria promotes tau-mediated neurodegeneration and Alzheimer’s disease-related tau phosphorylation via PAR-1. PLoS Genet. 2012, 8, e1002918. [Google Scholar] [CrossRef] [PubMed]
- Panchal, K.; Tiwari, A.K. Miro, a Rho GTPase genetically interacts with Alzheimer’s disease-associated genes (Tau, Abeta(42) and Appl) in Drosophila melanogaster. Biol. Open 2020, 9, bio049569. [Google Scholar] [CrossRef]
- Kunst, C.B.; Mezey, E.; Brownstein, M.J.; Patterson, D. Mutations in SOD1 associated with amyotrophic lateral sclerosis cause novel protein interactions. Nat. Genet. 1997, 15, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Moller, A.; Bauer, C.S.; Cohen, R.N.; Webster, C.P.; De Vos, K.J. Amyotrophic lateral sclerosis-associated mutant SOD1 inhibits anterograde axonal transport of mitochondria by reducing Miro1 levels. Hum. Mol. Genet. 2017, 26, 4668–4679. [Google Scholar] [CrossRef]
- Cagalinec, M.; Safiulina, D.; Liiv, M.; Liiv, J.; Choubey, V.; Wareski, P.; Veksler, V.; Kaasik, A. Principles of the mitochondrial fusion and fission cycle in neurons. J. Cell Sci. 2013, 126, 2187–2197. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aspenström, P. Miro GTPases at the Crossroads of Cytoskeletal Dynamics and Mitochondrial Trafficking. Cells 2024, 13, 647. https://doi.org/10.3390/cells13070647
Aspenström P. Miro GTPases at the Crossroads of Cytoskeletal Dynamics and Mitochondrial Trafficking. Cells. 2024; 13(7):647. https://doi.org/10.3390/cells13070647
Chicago/Turabian StyleAspenström, Pontus. 2024. "Miro GTPases at the Crossroads of Cytoskeletal Dynamics and Mitochondrial Trafficking" Cells 13, no. 7: 647. https://doi.org/10.3390/cells13070647





