Systematic Comparison of FBS and Medium Variation Effect on Key Cellular Processes Using Morphological Profiling
Abstract
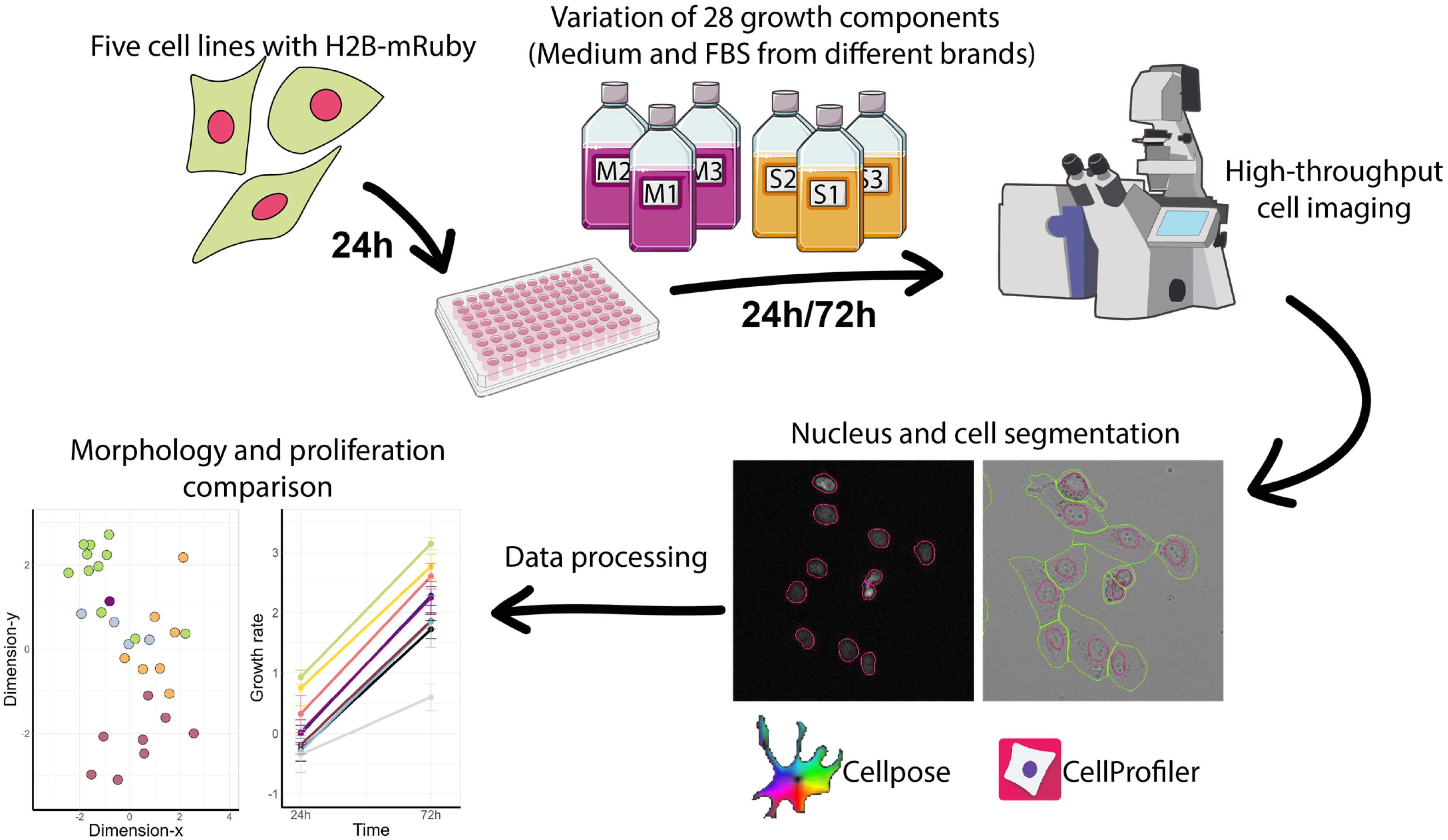
1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. Cell Imaging
2.3. Morphological Profiling and Cell Proliferation Analysis
2.4. ERK1/2 Activity Measurement
2.5. Mitochondria and Lysosome Staining
2.6. Drug Treatment and Cell Viability Measurements
2.7. Data and Statistics Analysis
3. Results
3.1. Cell Proliferation
3.2. Morphological Changes
3.3. ERK1/2 Activity and Response to EGF Stimulation
3.4. Drug Sensitivity
3.5. Mitochondrial Potential and Lysosomes Accumulation
3.6. SH-SY5Y Cell Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FBS | fetal bovine serum |
| DMEM | Dulbecco’s Modified Eagle Medium |
| EGF | epidermal growth factor |
| ERK1/2 | mitogen-activated protein kinase 1 (MAPK 3 and MAPK1), Extracellular signal-regulated kinases 1/2 |
References
- Weiskirchen, S.; Schroder, S.K.; Buhl, E.M.; Weiskirchen, R. A Beginner’s Guide to Cell Culture: Practical Advice for Preventing Needless Problems. Cells 2023, 12, 682. [Google Scholar] [CrossRef]
- Yao, T.; Asayama, Y. Animal-cell culture media: History, characteristics, and current issues. Reprod. Med. Biol. 2017, 16, 99–117. [Google Scholar] [CrossRef]
- Baker, M. 1500 scientists lift the lid on reproducibility. Nature 2016, 533, 452–454. [Google Scholar] [CrossRef]
- Makin, T.R.; Orban de Xivry, J.J. Ten common statistical mistakes to watch out for when writing or reviewing a manuscript. Elife 2019, 8, e48175. [Google Scholar] [CrossRef]
- Horbach, S.; Halffman, W. The ghosts of HeLa: How cell line misidentification contaminates the scientific literature. PLoS ONE 2017, 12, e0186281. [Google Scholar] [CrossRef]
- Callaway, E. Contamination hits cell work. Nature 2014, 511, 518. [Google Scholar] [CrossRef]
- Yu, M.; Selvaraj, S.K.; Liang-Chu, M.M.; Aghajani, S.; Busse, M.; Yuan, J.; Lee, G.; Peale, F.; Klijn, C.; Bourgon, R.; et al. A resource for cell line authentication, annotation and quality control. Nature 2015, 520, 307–311. [Google Scholar] [CrossRef]
- Liu, S.; Yang, W.; Li, Y.; Sun, C. Fetal bovine serum, an important factor affecting the reproducibility of cell experiments. Sci. Rep. 2023, 13, 1942. [Google Scholar] [CrossRef]
- van der Valk, J. Fetal bovine serum-a cell culture dilemma. Science 2022, 375, 143–144. [Google Scholar] [CrossRef]
- Hirsch, E.; Bornemissza, Z.; Nagy, Z.K.; Marosi, G.J.; Farkas, A. Quantitative and qualitative analysis of cell culture media powders for mammalian cells by Raman microscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2024, 310, 123906. [Google Scholar] [CrossRef]
- Lee, D.Y.; Yun, S.H.; Lee, S.Y.; Lee, J.; Mariano, E., Jr.; Joo, S.T.; Choi, I.; Choi, J.S.; Kim, G.D.; Lee, J.; et al. Analysis of commercial fetal bovine serum (FBS) and its substitutes in the development of cultured meat. Food Res. Int. 2023, 174, 113617. [Google Scholar] [CrossRef]
- van der Valk, J.; Gstraunthaler, G. Fetal Bovine Serum (FBS)—A pain in the dish? Altern. Lab Anim. 2017, 45, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Bryan, N.; Andrews, K.D.; Loughran, M.J.; Rhodes, N.P.; Hunt, J.A. Elucidating the contribution of the elemental composition of fetal calf serum to antigenic expression of primary human umbilical-vein endothelial cells in vitro. Biosci. Rep. 2011, 31, 199–210. [Google Scholar] [CrossRef]
- Sasaki, H.; Enomoto, J.; Ikeda, Y.; Honda, H.; Fukuda, J.; Kato, R. Comparisons of cell culture medium using distribution of morphological features in microdevice. J. Biosci. Bioeng. 2016, 121, 117–123. [Google Scholar] [CrossRef]
- Fang, C.Y.; Wu, C.C.; Fang, C.L.; Chen, W.Y.; Chen, C.L. Long-term growth comparison studies of FBS and FBS alternatives in six head and neck cell lines. PLoS ONE 2017, 12, e0178960. [Google Scholar] [CrossRef]
- Pandolfino, M.C.; Saiagh, S.; Knol, A.C.; Dreno, B. Comparison of three culture media for the establishment of melanoma cell lines. Cytotechnology 2010, 62, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.R.; Qu, L.H.; Ma, L.M. Differential impacts of charcoal-stripped fetal bovine serum on c-Myc among distinct subtypes of breast cancer cell lines. Biochem. Biophys. Res. Commun. 2020, 526, 267–272. [Google Scholar] [CrossRef]
- Yang, N.; Sin, D.D.; Dorscheid, D.R. Various factors affect lipopolysaccharide sensitization in cell cultures. Biotechniques 2020, 69, 126–132. [Google Scholar] [CrossRef]
- Soutar, M.P.M.; Kempthorne, L.; Annuario, E.; Luft, C.; Wray, S.; Ketteler, R.; Ludtmann, M.H.R.; Plun-Favreau, H. FBS/BSA media concentration determines CCCP’s ability to depolarize mitochondria and activate PINK1-PRKN mitophagy. Autophagy 2019, 15, 2002–2011. [Google Scholar] [CrossRef]
- Khasawneh, R.R.; Al Sharie, A.H.; Abu-El Rub, E.; Serhan, A.O.; Obeidat, H.N. Addressing the impact of different fetal bovine serum percentages on mesenchymal stem cells biological performance. Mol. Biol. Rep. 2019, 46, 4437–4441. [Google Scholar] [CrossRef]
- Vicogne, D.; Houdou, M.; Garat, A.; Climer, L.; Lupashin, V.; Morelle, W.; Foulquier, F. Fetal bovine serum impacts the observed N-glycosylation defects in TMEM165 KO HEK cells. J. Inherit. Metab. Dis. 2020, 43, 357–366. [Google Scholar] [CrossRef]
- Larsson, P.; Engqvist, H.; Biermann, J.; Werner Ronnerman, E.; Forssell-Aronsson, E.; Kovacs, A.; Karlsson, P.; Helou, K.; Parris, T.Z. Optimization of cell viability assays to improve replicability and reproducibility of cancer drug sensitivity screens. Sci. Rep. 2020, 10, 5798. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.Y.; Chang, G.C.; Hsu, S.L.; Huang, Y.R.; Chiu, L.Y.; Sheu, G.T. Effect of folic acid and vitamin B12 on pemetrexed antifolate chemotherapy in nutrient lung cancer cells. Biomed. Res. Int. 2013, 2013, 389046. [Google Scholar] [CrossRef] [PubMed]
- Lund, P.; Pilgaard, L.; Duroux, M.; Fink, T.; Zachar, V. Effect of growth media and serum replacements on the proliferation and differentiation of adipose-derived stem cells. Cytotherapy 2009, 11, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Lee, A.; Kim, K. The effect of serum types on Chondrogenic differentiation of adipose-derived stem cells. Biomater. Res. 2018, 22, 6. [Google Scholar] [CrossRef]
- Golikov, M.V.; Valuev-Elliston, V.T.; Smirnova, O.A.; Ivanov, A.V. Physiological Media in Studies of Cell Metabolism. Mol. Biol. 2022, 56, 629–637. [Google Scholar] [CrossRef]
- Golikov, M.V.; Karpenko, I.L.; Lipatova, A.V.; Ivanova, O.N.; Fedyakina, I.T.; Larichev, V.F.; Zakirova, N.F.; Leonova, O.G.; Popenko, V.I.; Bartosch, B.; et al. Cultivation of Cells in a Physiological Plasmax Medium Increases Mitochondrial Respiratory Capacity and Reduces Replication Levels of RNA Viruses. Antioxidants 2021, 11, 97. [Google Scholar] [CrossRef]
- Okano, S.; Hurley, D.J.; Vandenplas, M.L.; Moore, J.N. Effect of fetal bovine serum and heat-inactivated fetal bovine serum on microbial cell wall-induced expression of procoagulant activity by equine and canine mononuclear cells in vitro. Am. J. Vet. Res. 2006, 67, 1020–1024. [Google Scholar] [CrossRef]
- Shibayama, K.; Nagasawa, M.; Ando, T.; Minami, M.; Wachino, J.; Suzuki, S.; Arakawa, Y. Usefulness of adult bovine serum for Helicobacter pylori culture media. J. Clin. Microbiol. 2006, 44, 4255–4257. [Google Scholar] [CrossRef]
- Sovadinova, I.; Kuroda, K.; Palermo, E.F. Unexpected Enhancement of Antimicrobial Polymer Activity against Staphylococcus aureus in the Presence of Fetal Bovine Serum. Molecules 2021, 26, 4512. [Google Scholar] [CrossRef]
- Rossiter, N.J.; Huggler, K.S.; Adelmann, C.H.; Keys, H.R.; Soens, R.W.; Sabatini, D.M.; Cantor, J.R. CRISPR screens in physiologic medium reveal conditionally essential genes in human cells. Cell Metab. 2021, 33, 1248–1263.e9. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, E.; Segura-Cabrera, A.; Pacini, C.; Picco, G.; Behan, F.M.; Jaaks, P.; Coker, E.A.; van der Meer, D.; Barthorpe, A.; Lightfoot, H.; et al. Drug mechanism-of-action discovery through the integration of pharmacological and CRISPR screens. Mol. Syst. Biol. 2020, 16, e9405. [Google Scholar] [CrossRef] [PubMed]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Goncalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef] [PubMed]
- Seashore-Ludlow, B.; Rees, M.G.; Cheah, J.H.; Cokol, M.; Price, E.V.; Coletti, M.E.; Jones, V.; Bodycombe, N.E.; Soule, C.K.; Gould, J.; et al. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer Discov. 2015, 5, 1210–1223. [Google Scholar] [CrossRef]
- Chandrasekaran, S.N.; Cimini, B.A.; Goodale, A.; Miller, L.; Kost-Alimova, M.; Jamali, N.; Doench, J.G.; Fritchman, B.; Skepner, A.; Melanson, M.; et al. Three million images and morphological profiles of cells treated with matched chemical and genetic perturbations. Nat. Methods 2024, 21, 1114–1121. [Google Scholar] [CrossRef]
- Mills, C.E.; Subramanian, K.; Hafner, M.; Niepel, M.; Gerosa, L.; Chung, M.; Victor, C.; Gaudio, B.; Yapp, C.; Nirmal, A.J.; et al. Multiplexed and reproducible high content screening of live and fixed cells using Dye Drop. Nat. Commun. 2022, 13, 6918. [Google Scholar] [CrossRef]
- Dravid, A.; Raos, B.; Svirskis, D.; O’Carroll, S.J. Optimised techniques for high-throughput screening of differentiated SH-SY5Y cells and application for neurite outgrowth assays. Sci. Rep. 2021, 11, 23935. [Google Scholar] [CrossRef]
- Regot, S.; Hughey, J.J.; Bajar, B.T.; Carrasco, S.; Covert, M.W. High-sensitivity measurements of multiple kinase activities in live single cells. Cell 2014, 157, 1724–1734. [Google Scholar] [CrossRef]
- Lebedev, T.D.; Khabusheva, E.R.; Mareeva, S.R.; Ivanenko, K.A.; Morozov, A.V.; Spirin, P.V.; Rubtsov, P.M.; Snezhkina, A.V.; Kudryavtseva, A.V.; Sorokin, M.I.; et al. Identification of cell type-specific correlations between ERK activity and cell viability upon treatment with ERK1/2 inhibitors. J. Biol. Chem. 2022, 298, 102226. [Google Scholar] [CrossRef]
- Mikheeva, A.M.; Bogomolov, M.A.; Gasca, V.A.; Sementsov, M.V.; Spirin, P.V.; Prassolov, V.S.; Lebedev, T.D. Improving the power of drug toxicity measurements by quantitative nuclei imaging. Cell Death Discov. 2024, 10, 181. [Google Scholar] [CrossRef]
- Stringer, C.; Pachitariu, M. Cellpose3: One-click image restoration for improved cellular segmentation. Nat. Methods 2025. [Google Scholar] [CrossRef] [PubMed]
- Stirling, D.R.; Swain-Bowden, M.J.; Lucas, A.M.; Carpenter, A.E.; Cimini, B.A.; Goodman, A. CellProfiler 4: Improvements in speed, utility and usability. BMC Bioinform. 2021, 22, 433. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Shipley, M.M.; Mangold, C.A.; Szpara, M.L. Differentiation of the SH-SY5Y Human Neuroblastoma Cell Line. J. Vis. Exp. 2016, 53193. [Google Scholar] [CrossRef]
- Lebedev, T.D.; Vagapova, E.R.; Prassolov, V.S. The Different Impact of ERK Inhibition on Neuroblastoma, Astrocytoma, and Rhabdomyosarcoma Cell Differentiation. Acta Naturae 2021, 13, 69–77. [Google Scholar] [CrossRef]
- Ramezani, M.; Weisbart, E.; Bauman, J.; Singh, A.; Yong, J.; Lozada, M.; Way, G.P.; Kavari, S.L.; Diaz, C.; Leardini, E.; et al. A genome-wide atlas of human cell morphology. Nat. Methods 2025. [Google Scholar] [CrossRef]
- Silva-Carvalho, A.E.; Neves, F.A.R.; Saldanha-Araujo, F. The immunosuppressive mechanisms of mesenchymal stem cells are differentially regulated by platelet poor plasma and fetal bovine serum supplemented media. Int. Immunopharmacol. 2020, 79, 106172. [Google Scholar] [CrossRef]
- Clark, K.C.; Kol, A.; Shahbenderian, S.; Granick, J.L.; Walker, N.J.; Borjesson, D.L. Canine and Equine Mesenchymal Stem Cells Grown in Serum Free Media Have Altered Immunophenotype. Stem Cell Rev. Rep. 2016, 12, 245–256. [Google Scholar] [CrossRef]
- Gross, S.M.; Rotwein, P. Quantification of growth factor signaling and pathway cross talk by live-cell imaging. Am. J. Physiol. Cell Physiol. 2017, 312, C328–C340. [Google Scholar] [CrossRef]
- Nishikawa, M.; Kimura, H.; Yanagawa, N.; Hamon, M.; Hauser, P.; Zhao, L.; Jo, O.D.; Yanagawa, N. An optimal serum-free defined condition for in vitro culture of kidney organoids. Biochem. Biophys. Res. Commun. 2018, 501, 996–1002. [Google Scholar] [CrossRef]
- Alves, S.; Castro, L.; Fernandes, M.S.; Francisco, R.; Castro, P.; Priault, M.; Chaves, S.R.; Moyer, M.P.; Oliveira, C.; Seruca, R.; et al. Colorectal cancer-related mutant KRAS alleles function as positive regulators of autophagy. Oncotarget 2015, 6, 30787–30802. [Google Scholar] [CrossRef] [PubMed]
- Janoueix-Lerosey, I.; Lequin, D.; Brugieres, L.; Ribeiro, A.; de Pontual, L.; Combaret, V.; Raynal, V.; Puisieux, A.; Schleiermacher, G.; Pierron, G.; et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 2008, 455, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, K.; Sequist, L.V.; Arcila, M.E.; Lovly, C.M.; Chen, X.; Rudin, C.M.; Moran, T.; Camidge, D.R.; Vnencak-Jones, C.L.; Berry, L.; et al. Characteristics of lung cancers harboring NRAS mutations. Clin. Cancer Res. 2013, 19, 2584–2591. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Lei, T.; Guo, P.; Yu, J.; Xu, Q.; Luo, Y.; Ke, R.; Huang, D. Mechanisms shaping the role of ERK1/2 in cellular senescence (Review). Mol. Med. Rep. 2019, 19, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Heckenbach, I.; Mkrtchyan, G.V.; Ezra, M.B.; Bakula, D.; Madsen, J.S.; Nielsen, M.H.; Oro, D.; Osborne, B.; Covarrubias, A.J.; Idda, M.L.; et al. Nuclear morphology is a deep learning biomarker of cellular senescence. Nat. Aging 2022, 2, 742–755. [Google Scholar] [CrossRef]
- Mohamed, H.E.; Asker, M.E.; Kotb, N.S.; El Habab, A.M. Human platelet lysate efficiency, stability, and optimal heparin concentration required in culture of mammalian cells. Blood Res. 2020, 55, 35–43. [Google Scholar] [CrossRef]
- Baker, M. Reproducibility: Respect your cells! Nature 2016, 537, 433–435. [Google Scholar] [CrossRef]
- Shin, J.; Rhim, J.; Kwon, Y.; Choi, S.Y.; Shin, S.; Ha, C.W.; Lee, C. Comparative analysis of differentially secreted proteins in serum-free and serum-containing media by using BONCAT and pulsed SILAC. Sci. Rep. 2019, 9, 3096. [Google Scholar] [CrossRef]
- Nonnis, S.; Maffioli, E.; Zanotti, L.; Santagata, F.; Negri, A.; Viola, A.; Elliman, S.; Tedeschi, G. Effect of fetal bovine serum in culture media on MS analysis of mesenchymal stromal cells secretome. EuPA Open Proteom. 2016, 10, 28–30. [Google Scholar] [CrossRef]
- Giannasi, C.; Niada, S.; Della Morte, E.; Casati, S.R.; De Palma, C.; Brini, A.T. Serum starvation affects mitochondrial metabolism of adipose-derived stem/stromal cells. Cytotherapy 2023, 25, 704–711. [Google Scholar] [CrossRef]
- Vultaggio-Poma, V.; Scussel Bergamin, L.; Falzoni, S.; Tarantini, M.; Giuliani, A.L.; Sandona, D.; Polverino De Laureto, P.; Di Virgilio, F. Fetal bovine serum contains biologically available ATP. Purinergic Signal 2024, 20, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Kim, S.J.; Langley, R.R.; Fidler, I.J. Modulation of the cancer cell transcriptome by culture media formulations and cell density. Int. J. Oncol. 2015, 46, 2067–2075. [Google Scholar] [CrossRef] [PubMed]
- Santiappillai, N.T.; Abuhammad, S.; Slater, A.; Kirby, L.; McArthur, G.A.; Sheppard, K.E.; Smith, L.K. CDK4/6 Inhibition Reprograms Mitochondrial Metabolism in BRAF(V600) Melanoma via a p53 Dependent Pathway. Cancers 2021, 13, 524. [Google Scholar] [CrossRef] [PubMed]
- Foy, R.; Lew, K.X.; Saurin, A.T. The search for CDK4/6 inhibitor biomarkers has been hampered by inappropriate proliferation assays. NPJ Breast Cancer 2024, 10, 19. [Google Scholar] [CrossRef]
- Guan, M.; Zhang, Y.; Huang, Q.; He, L.; Fang, Q.; Zhang, J.; Gao, S.; Fang, J.; Ma, Y.; Su, K.; et al. Fetal bovine serum inhibits neomycin-induced apoptosis of hair cell-like HEI-OC-1 cells by maintaining mitochondrial function. Am. J. Transl. Res. 2019, 11, 1343–1358. [Google Scholar]
- Torres-Quesada, O.; Doerrier, C.; Strich, S.; Gnaiger, E.; Stefan, E. Physiological Cell Culture Media Tune Mitochondrial Bioenergetics and Drug Sensitivity in Cancer Cell Models. Cancers 2022, 14, 3917. [Google Scholar] [CrossRef]




| Code | Serum | Medium |
|---|---|---|
| S1/M1 | 10% FBS #10270106 (Gibco) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
| M3 | 10% FBS #10270106 (Gibco) | DMEM #DMEM-HA/RPMI-1640 #RMPI-XA (Capricorn Scientific) |
| M4 | 10% FBS #10270106 (Gibco) | DMEM #SH30022.01/RPMI-1640 #SH30027.01 (Cytiva) |
| M5 | 10% FBS #10270106 (Gibco) | DMEM #AL066/RPMI-1640 #AL028 (HiMedia) |
| M6 | 10% FBS #10270106 (Gibco) | DMEM #C410E/RPMI-1640 #C310∏ (PanEco ltd) |
| M7 | 10% FBS #10270106 (Gibco) | DMEM G4511-500ML/RPMI-1640 #G4531-500ML (Servicebio) |
| M8 | 10% FBS #10270106 (Gibco) | DMEM #D0822/RPMI-1640 #R8758 (Sigma Aldrich) |
| S2 | 10% FBS #500SA (Dia-M) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
| S3 | 10% FBS #FB-1200 (Biosera) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
| S4 | 10% FBS #FBS-12B (Capricorn Scientific) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
| S5 | 10% FBS heat-inactivated #FBS-HI-12A (Capricorn Scientific) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
| S6 | 10% FBS #FBS-16A (Capricorn Scientific) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
| S7 | 10% FBS #35-015-CV (Corning) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
| S8 | 10% FBS #SH30088.03 (Cytiva) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
| S9 | 10% FBS #RM10432 (HiMedia) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
| S10 | 10% FBS heat-inactivated #RM9955 (HiMedia) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
| S11 | 10% FBS #F800822 (Globe Kang) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
| S12 | 10% Serum replacement #TCL280 (HiMedia) | DMEM #61965026/RPMI-1640 #21870076 (Gibco) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebedev, T.; Mikheeva, A.; Gasca, V.; Spirin, P.; Prassolov, V. Systematic Comparison of FBS and Medium Variation Effect on Key Cellular Processes Using Morphological Profiling. Cells 2025, 14, 336. https://doi.org/10.3390/cells14050336
Lebedev T, Mikheeva A, Gasca V, Spirin P, Prassolov V. Systematic Comparison of FBS and Medium Variation Effect on Key Cellular Processes Using Morphological Profiling. Cells. 2025; 14(5):336. https://doi.org/10.3390/cells14050336
Chicago/Turabian StyleLebedev, Timofey, Alesya Mikheeva, Valentina Gasca, Pavel Spirin, and Vladimir Prassolov. 2025. "Systematic Comparison of FBS and Medium Variation Effect on Key Cellular Processes Using Morphological Profiling" Cells 14, no. 5: 336. https://doi.org/10.3390/cells14050336
APA StyleLebedev, T., Mikheeva, A., Gasca, V., Spirin, P., & Prassolov, V. (2025). Systematic Comparison of FBS and Medium Variation Effect on Key Cellular Processes Using Morphological Profiling. Cells, 14(5), 336. https://doi.org/10.3390/cells14050336







