A Novel Allelic Variant of OsAGPL2 Influences Rice Eating and Cooking Quality
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Determination of Starch Physicochemical Properties
2.3. Microscopic Analysis
2.4. Mapping of OsAGPL2
2.5. Haplotype Analysis
2.6. AGPase Enzyme Activity Assay and Sugar Component Determination
2.7. RNA Extraction and Gene Expression Analysis
3. Results
3.1. Phenotypic Differences of WT and h5
3.2. Significant Reduction in AC and GC in h5 Endosperm
3.3. Dispersed Spherical Starch Granules in h5 Endosperm
3.4. The Mutated Gene Is OsAGPL2
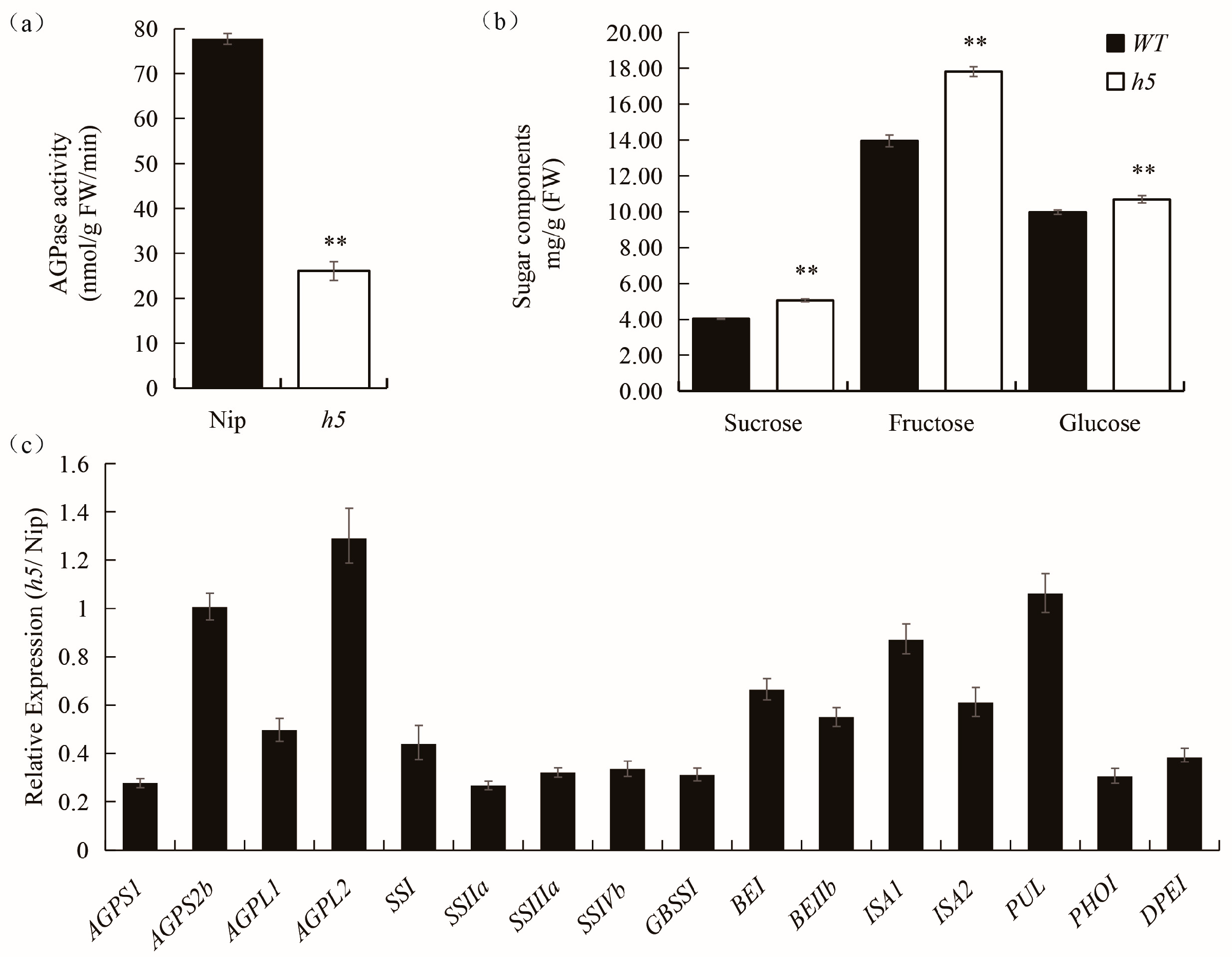
3.5. AGPase Activity Assay, Sugar Contents Analysis and Expression Analysis of Related Genes
3.6. OsAGPL2 Is Associated with AC and GC
4. Discussion
4.1. Impaired Starch Biosynthesis Pathway in the h5 Endosperm
4.2. Three SNPs in the 3′UTR and 5′UTR of OsAGPL2 as Key Superior Loci for Rice Quality Improvement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cai, Y.; Chen, Z.C.; Liu, J.J.; Yu, L.; Wang, Z.P.; Zhu, S.H.; Shi, W.; Pan, C.H.; Wu, Y.Y.; Li, Y.H.; et al. Genetic improvement of eating and cooking quality of rice cultivars in southern China. Plant Biotechnol. J. 2025, 23, 518–531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Q.; Yang, Y.; Chen, S.J.; Liu, X.J.; Zhu, J.H.; Zhou, L.H.; Lu, Y.; Li, Q.F.; Fan, X.L.; Tang, S.Z.; et al. A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J. Integr. Plant Biol. 2021, 63, 889–901. [Google Scholar] [CrossRef]
- Shao, Y.; Peng, Y.; Mao, B.G.; Lv, Q.M.; Yuan, D.Y.; Liu, X.L.; Zhao, B.G. Allelic variations of the Wx locus in cultivated rice and their use in the development of hybrid rice in China. PLoS ONE 2020, 15, e232279. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.; Zhou, L.H.; Feng, L.H.; Jiang, J.Y.; Huang, L.H.; Liu, Q.; Zhang, Y.D.; Zhang, C.Q.; Liu, Q.Q. Deciphering the role of Waxy gene mutations in enhancing rice grain quality. Foods 2024, 13, 1624. [Google Scholar] [CrossRef]
- Pan, L.X.; Sun, Z.Z.; Zhang, C.Q.; Li, B.; Yang, Q.Q.; Chen, F.; Fan, X.L.; Zhao, D.S.; Lv, Q.M.; Yuan, D.Y.; et al. Allelic diversification of the Wx and ALK loci in Indica restorer lines and their utilisation in hybrid rice breeding in china over the last 50 years. Int. J. Mol. Sci. 2022, 23, 5941. [Google Scholar] [CrossRef]
- Huang, L.C.; Gu, Z.W.; Chen, Z.Z.; Yu, J.W.; Chu, R.; Tan, H.Y.; Zhao, D.S.; Fan, X.L.; Zhang, C.Q.; Li, Q.F.; et al. Improving rice eating and cooking quality by coordinated expression of the major starch synthesis-related genes, SSII and Wx, in endosperm. Plant Mol. Biol. 2021, 106, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.Y.; Zeng, D.; Cheng, F.M.; Tian, Z.X.; Guo, L.B.; Su, Y.; Yan, M.X.; Jiang, H.; Dong, G.J.; Huang, Y.C.; et al. Alk, the key gene for gelatinization temperature, is a modifier gene for gel consistency in rice. J. Integr. Plant Biol. 2011, 53, 756–765. [Google Scholar]
- Yang, X.H.; Xia, X.Z.; Zeng, Y.; Nong, B.X.; Zhang, Z.Q.; Wu, Y.Y.; Xiong, F.Q.; Zhang, Y.X.; Liang, H.F.; Deng, G.F.; et al. Identification of candidate genes for gelatinization temperature, gel consistency and pericarp color by GWAS in rice based on SLAF-sequencing. PLoS ONE 2018, 13, e196690. [Google Scholar] [CrossRef]
- Zhang, A.P.; Gao, Y.; Li, Y.Y.; Ruan, B.P.; Yang, S.L.; Liu, C.L.; Zhang, B.; Jiang, H.Z.; Fang, G.N.; Ding, S.L.; et al. Genetic analysis for cooking and eating quality of super rice and fine mapping of a novel locus qGC10 for gel consistency. Front. Plant Sci. 2020, 11, 342. [Google Scholar] [CrossRef]
- Su, Y.; Rao, Y.C.; Hu, S.K.; Yang, Y.L.; Gao, Z.Y.; Zhang, G.H.; Liu, J.; Hu, J.; Yan, M.X.; Dong, G.J.; et al. Map-based cloning proves qGC-6, a major QTL for gel consistency of japonica/indica cross, responds by Waxy in rice (Oryza sativa L.). Theor. Appl. Genet. 2011, 123, 859–867. [Google Scholar] [CrossRef]
- Nie, S.; Chen, L.; Zheng, M.; Dong, J.; Ma, Y.; Zhou, L.; Wang, J.; Chen, J.; Hu, H.; Yang, T.; et al. GWAS and transcriptomic analysis identify OsRING315 as a new candidate gene controlling amylose content and gel consistency in rice. Rice 2024, 17, 38. [Google Scholar] [CrossRef]
- Tran, N.A.; Daygon, V.D.; Resurreccion, A.P.; Cuevas, R.P.; Corpuz, H.M.; Fitzgerald, M.A. A single nucleotide polymorphism in the Waxy gene explains a significant component of gel consistency. Theor. Appl. Genet. 2011, 123, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.C.; Tan, H.Y.; Zhang, C.Q.; Li, Q.F.; Liu, Q.Q. Starch biosynthesis in cereal endosperms: An updated review over the last decade. Plant Commun. 2021, 2, 100237. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; Wei, X.J.; Ren, Y.L.; Qiu, J.H.; Jiao, G.A.; Guo, X.P.; Tang, S.Q.; Wan, J.M.; Hu, P.S. OsBT1 encodes an ADP-glucose transporter involved in starch synthesis and compound granule formation in rice endosperm. Sci. Rep. 2017, 7, 40124. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhang, W.; Wang, Y.; Jin, J.; Xu, H.; Fu, Y.; Shan, Z.; Wang, X.; Teng, X.; Li, X.; et al. Rice like early STARVATION1 cooperates with FLOURY ENDOSPERM6 to modulate starch biosynthesis and endosperm development. Plant Cell 2024, 36, 1892–1912. [Google Scholar] [CrossRef]
- Xu, H.; Li, X.F.; Zhang, H.; Wang, L.C.; Zhu, Z.G.; Gao, J.P.; Li, C.S.; Zhu, Y. High temperature inhibits the accumulation of storage materials by inducing alternative splicing of OsbZIP58 during filling stage in rice. Plant Cell Environ. 2020, 43, 1879–1896. [Google Scholar] [CrossRef]
- Fu, F.F.; Xue, H.W. Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010, 154, 927–938. [Google Scholar] [CrossRef]
- Liu, Z.M.; Jiang, S.; Jiang, L.L.; Li, W.J.; Tang, Y.Q.; He, W.; Wang, M.L.; Xing, J.J.; Cui, Y.C.; Lin, Q.L.; et al. Transcription factor OsSGL is a regulator of starch synthesis and grain quality in rice. J. Exp. Bot. 2022, 73, 3417–3430. [Google Scholar] [CrossRef]
- Lee, S.K.; Hwang, S.K.; Han, M.H.; Eom, J.S.; Kang, H.G.; Han, Y.Y.; Choi, S.B.; Cho, M.H.; Bhoo, S.H.; An, G.H.; et al. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol. Biol. 2007, 65, 531–546. [Google Scholar] [CrossRef]
- Figueroa, C.M.; Asencion Diez, M.D.; Ballicora, M.A.; Iglesias, A.A. Structure, function, and evolution of plant ADP-glucose pyrophosphorylase. Plant Mol. Biol. 2022, 108, 307–323. [Google Scholar] [CrossRef]
- Zhang, D.P.; Wu, J.G.; Zhang, Y.J.; Shi, C.H. Phenotypic and candidate gene analysis of a new floury endosperm mutant (osagpl2-3) in Rice. Plant Mol. Biol. Report. 2012, 30, 1303–1312. [Google Scholar] [CrossRef]
- Tang, X.J.; Peng, C.; Zhang, J.; Cai, Y.; You, X.M.; Kong, F.; Yan, H.G.; Wang, G.X.; Wang, L.; Jin, J.; et al. ADP-glucose pyrophosphorylase large subunit 2 is essential for storage substance accumulation and subunit interactions in rice endosperm. Plant Sci. 2016, 249, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.J.; Jiao, G.A.; Lin, H.Y.; Sheng, Z.H.; Shao, G.N.; Xie, L.H.; Tang, S.Q.; Xu, Q.G.; Hu, A.P. Grain incomplete filling 2 regulates grain filling and starch synthesis during rice caryopsis development. J. Integr. Plant Biol. 2017, 59, 134–153. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, L.L.; Ren, Y.H.; Zhang, J.; Xu, A.H.; Wei, C.X. Gene mapping and mutation analysis of floury endosperm mutant M37 in rice. J. Yangzhou Univ. Agric. Life Sci. Ed. 2019, 40, 10–17. (In Chinese) [Google Scholar]
- Zhang, X.C.; Lu, F.F.; Lv, Y.S.; Luo, R.J.; Jiao, G.A.; Wu, Y.W.; Tang, S.Q.; Hu, P.S.; Wei, X.J. Identification and gene mapping-based clone of two chalkiness mutants in rice. Chin. J. Rice Sci. 2017, 31, 568–579. (In Chinese) [Google Scholar]
- Zhang, L.; You, R.; Chen, H.L.; Zhu, J.; Lin, L.S.; Wei, C.X. A new SNP in AGPL2, associated with floury endosperm in rice, is identified using a modified MutMap method. Agronomy 2023, 13, 1381. [Google Scholar] [CrossRef]
- Lei, J.; Teng, X.; Wang, Y.L.; Jiang, X.K.; Zhao, H.H.; Zheng, X.M.; Ren, Y.L.; Dong, H.H.; Wang, Y.L.; Duan, E.C.; et al. Plastidic pyruvate dehydrogenase complex E1 component subunit Alpha1 is involved in galactolipid biosynthesis required for amyloplast development in rice. Plant Biotechnol. J. 2022, 20, 437–453. [Google Scholar] [CrossRef]
- Huang, K.Y.; Lu, F.F.; Chen, P.F.; Jiao, G.A.; Lin, H.Y.; Zhang, J.; Zhao, S.L.; Cao, R.J.; Shao, G.N.; Sheng, Z.H.; et al. A large-scale gene regulatory network for rice endosperm starch biosynthesis and its application in genetic improvement of rice quality. Plant Biotechnol. J. 2025. online ahead of print. [Google Scholar] [CrossRef]
- Peng, C.; Wang, Y.H.; Liu, F.; Ren, Y.L.; Zhou, K.N.; Lv, J.; Zheng, M.; Zhao, S.L.; Zhang, L.; Wang, C.M.; et al. FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J. 2014, 77, 917–930. [Google Scholar] [CrossRef]
- Man, J.; Lin, L.; Wang, Z.; Wang, Y.; Liu, Q.; Wei, C. Different structures of heterogeneous starch granules from high-amylose rice. J. Agric. Food Chem. 2014, 62, 11254–11263. [Google Scholar] [CrossRef]
- Zhao, D.S.; Zhang, C.Q.; Li, Q.F.; Liu, Q.Q. Genetic control of grain appearance quality in rice. Biotechnol. Adv. 2022, 60, 108014. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, W.W.; Liu, S.J.; Tian, Y.L.; Liu, X.; Yan, H.G.; Cai, Y.; Teng, X.; Dong, H.; Chen, R.B.; et al. Rice FLOURY SHRUNKEN ENDOSPERM 5 encodes a putative plant organelle RNA recognition protein that is required for cis-splicing of mitochondrial nad4 intron1. Rice 2021, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Kharshiing, G.; Chrungoo, N.K. Wx alleles in rice: Relationship with apparent amylose content of starch and a possible role in rice domestication. J. Genet. 2021, 100, 65. [Google Scholar] [CrossRef]
- Liu, X.D.; Ding, Q.; Wang, W.S.; Pan, Y.L.; Tan, C.; Qiu, Y.B.; Chen, Y.; Li, H.j.; Li, Y.l.; Ye, N.; et al. Targeted deletion of the first intron of the Wxb allele via CRISPR/Cas9 significantly increases grain amylose content in rice. Rice 2022, 15, 1. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dan, Y.; Huang, F.; Xu, J.; He, Y.; Peng, R.; Li, C.; Song, J.; Hao, Y.; Tian, Z. A Novel Allelic Variant of OsAGPL2 Influences Rice Eating and Cooking Quality. Cells 2025, 14, 634. https://doi.org/10.3390/cells14090634
Dan Y, Huang F, Xu J, He Y, Peng R, Li C, Song J, Hao Y, Tian Z. A Novel Allelic Variant of OsAGPL2 Influences Rice Eating and Cooking Quality. Cells. 2025; 14(9):634. https://doi.org/10.3390/cells14090634
Chicago/Turabian StyleDan, Yuqing, Fudeng Huang, Junfeng Xu, Yong He, Ruixiao Peng, Chunshou Li, Jiayu Song, Yuanyuan Hao, and Zhihong Tian. 2025. "A Novel Allelic Variant of OsAGPL2 Influences Rice Eating and Cooking Quality" Cells 14, no. 9: 634. https://doi.org/10.3390/cells14090634
APA StyleDan, Y., Huang, F., Xu, J., He, Y., Peng, R., Li, C., Song, J., Hao, Y., & Tian, Z. (2025). A Novel Allelic Variant of OsAGPL2 Influences Rice Eating and Cooking Quality. Cells, 14(9), 634. https://doi.org/10.3390/cells14090634






