Flexible Players within the Sheaths: The Intrinsically Disordered Proteins of Myelin in Health and Disease
Abstract
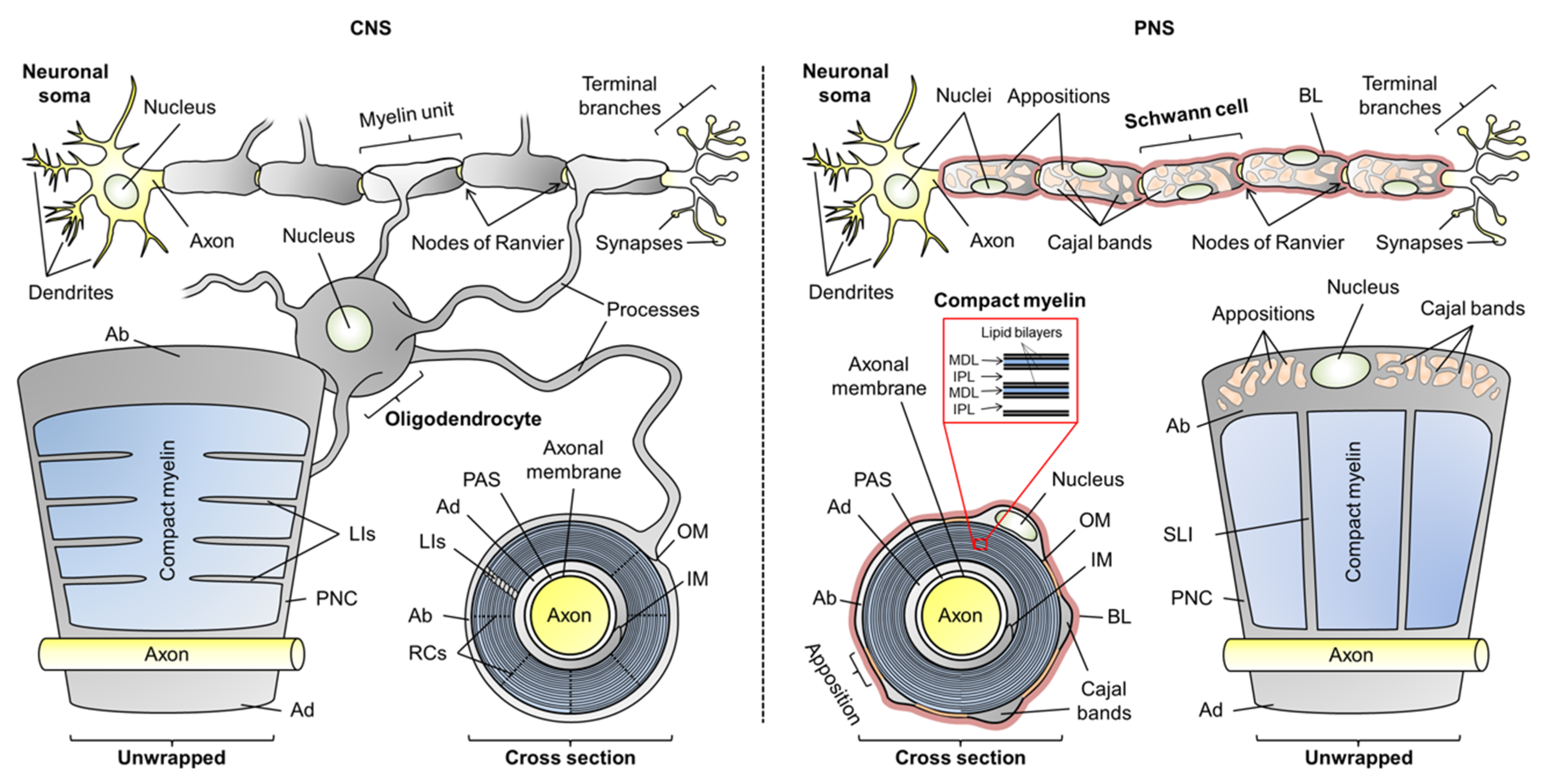
:1. Introduction
2. Intrinsically Disordered Proteins of Myelin
2.1. General Attributes of Myelin-Specific Proteins
2.2. Myelin Basic Protein
2.3. Myelin-Associated Oligodendrocytic Basic Protein
2.4. 2′,3′-Cyclic Nucleotide 3′-Phosphodiesterase
2.5. Juxtanodin
2.6. Myelin-Associated Glycoprotein
2.7. Periaxin
3. Selected Examples of IDPs in Demyelination
3.1. Basic Proteins and Multiple Sclerosis
3.2. Periaxin and Peripheral Neuropathies
4. Future Research Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, S.S.H.; Shultz, J.R.; Burish, M.J.; Harrison, K.H.; Hof, P.R.; Towns, L.C.; Wagers, M.W.; Wyatt, K.D. Shaping of white matter composition by biophysical scaling constraints. J. Neurosci. 2008, 28, 4047–4056. [Google Scholar] [CrossRef] [Green Version]
- Lundgaard, I.; Luzhynskaya, A.; Stockley, J.H.; Wang, Z.; Evans, K.A.; Swire, M.; Volbracht, K.; Gautier, H.O.B.; Franklin, R.J.M.; ffrench-Constant, C.; et al. Neuregulin and BDNF Induce a Switch to NMDA Receptor-Dependent Myelination by Oligodendrocytes. PLoS. Biol. 2013, 11, e1001743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salzer, J.L. Schwann Cell Myelination. Cold Spring Harbor Perspect. Biol. 2015, 7, a020529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nave, K. Myelination and the trophic support of long axons. Nat. Rev. Neurosci. 2010, 11, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Young, J.Z. The functioning of the giant nerve fibres of the squid. J. Exp. Biol. 1938, 15, 170–185. [Google Scholar]
- Simons, M.; Trotter, J. Wrapping it up: The cell biology of myelination. Curr. Opin. Neurobiol. 2007, 17, 533–540. [Google Scholar] [CrossRef]
- Zuchero, J.B.; Fu, M.; Sloan, S.A.; Ibrahim, A.; Olson, A.; Zaremba, A.; Dugas, J.C.; Wienbar, S.; Caprariello, A.V.; Kantor, C.; et al. CNS myelin wrapping is driven by actin disassembly. Dev. Cell 2015, 34, 152–167. [Google Scholar] [CrossRef] [Green Version]
- Hartline, D.K. What is myelin? Neuron Glia Biol. 2008, 4, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Fünfschilling, U.; Supplie, L.M.; Mahad, D.; Boretius, S.; Saab, A.S.; Edgar, J.; Brinkmann, B.G.; Kassmann, C.M.; Tzvetanova, I.D.; Möbius, W.; et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 2012, 485, 517–523. [Google Scholar] [CrossRef] [Green Version]
- Court, F.A.; Wrabetz, L.; Feltri, M.L. Basal lamina: Schwann cells wrap to the rhythm of space-time. Curr. Opin. Neurobiol. 2006, 16, 501–507. [Google Scholar] [CrossRef]
- Court, F.; Sherman, D.; Pratt, T.; Garry, E.; Ribchester, R.; Cottrell, D.; Fleetwood-Walker, S.; Brophy, P. Restricted growth of Schwann cells lacking Cajal bands slows conduction in myelinated nerves. Nature 2004, 431, 191–195. [Google Scholar] [CrossRef]
- Court, F.A.; Hewitt, J.E.; Davies, K.; Patton, B.L.; Uncini, A.; Wrabetz, L.; Feltri, M.L. A Laminin-2, Dystroglycan, Utrophin Axis Is Required for Compartmentalization and Elongation of Myelin Segments. J. Neurosci. 2009, 29, 3908–3919. [Google Scholar] [CrossRef] [Green Version]
- Sherman, D.L.; Wu, L.M.N.; Grove, M.; Gillespie, C.S.; Brophy, P.J. Drp2 and Periaxin Form Cajal Bands with Dystroglycan But Have Distinct Roles in Schwann Cell Growth. J. Neurosci. 2012, 32, 9419–9428. [Google Scholar] [CrossRef] [Green Version]
- Micu, I.; Plemel, J.R.; Caprariello, A.V.; Nave, K.; Stys, P.K. Axo-myelinic neurotransmission: A novel mode of cell signalling in the central nervous system. Nat. Rev. Neurosci. 2018, 19, 49–57. [Google Scholar] [CrossRef]
- Saher, G.; Brugger, B.; Lappe-Siefke, C.; Möbius, W.; Tozawa, R.; Wehr, M.; Wieland, F.; Ishibashi, S.; Nave, K. High cholesterol level is essential for myelin membrane growth. Nat. Neurosci. 2005, 8, 468–475. [Google Scholar] [CrossRef]
- Saher, G.; Quintes, S.; Nave, K. Cholesterol: A novel regulatory role in myelin formation. Neuroscientist 2011, 17, 79–93. [Google Scholar] [CrossRef]
- Aggarwal, S.; Yurlova, L.; Simons, M. Central nervous system myelin: Structure, synthesis and assembly. Trends Cell Biol. 2011, 21, 585–593. [Google Scholar] [CrossRef]
- Jo, E.; Boggs, J. Aggregation of Acidic Lipid Vesicles by Myelin Basic-Protein–Dependence on Potassium Concentration. Biochemistry 1995, 34, 13705–13716. [Google Scholar] [CrossRef]
- Luo, X.; Sharma, D.; Inouye, H.; Lee, D.; Avila, R.L.; Salmona, M.; Kirschner, D.A. Cytoplasmic domain of human myelin protein zero likely folded as beta-structure in compact myelin. Biophys. J. 2007, 92, 1585–1597. [Google Scholar] [CrossRef] [Green Version]
- Suresh, S.; Wang, C.; Nanekar, R.; Kursula, P.; Edwardson, J.M. Myelin basic protein and myelin protein 2 act synergistically to cause stacking of lipid bilayers. Biochemistry 2010, 49, 3456–3463. [Google Scholar] [CrossRef]
- Wang, C.; Neugebauer, U.; Bürck, J.; Myllykoski, M.; Baumgärtel, P.; Popp, J.; Kursula, P. Charge isomers of myelin basic protein: Structure and interactions with membranes, nucleotide analogues, and calmodulin. PLoS ONE 2011, 6, e19915. [Google Scholar] [CrossRef] [PubMed]
- Ruskamo, S.; Yadav, R.P.; Sharma, S.; Lehtimäki, M.; Laulumaa, S.; Aggarwal, S.; Simons, M.; Bürck, J.; Ulrich, A.S.; Juffer, A.H.; et al. Atomic resolution view into the structure-function relationships of the human myelin peripheral membrane protein P2. Acta Cryst. D 2014, 70, 165–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuusa, J.; Raasakka, A.; Ruskamo, S.; Kursula, P. Myelin-derived and putative molecular mimic peptides share structural properties in aqueous and membrane-like environments. Mult. Scler. Demyelinating Disord. 2017, 2, 4. [Google Scholar] [CrossRef] [Green Version]
- Raasakka, A.; Ruskamo, S.; Kowal, J.; Barker, R.; Baumann, A.; Martel, A.; Tuusa, J.; Myllykoski, M.; Bürck, J.; Ulrich, A.S.; et al. Membrane Association Landscape of Myelin Basic Protein Portrays Formation of the Myelin Major Dense Line. Sci. Rep. 2017, 7, 4974. [Google Scholar] [CrossRef]
- Schmitt, F.O.; Bear, R.S.; Clark, G.L. The Role of Lipoids in the X-Ray Diffraction Patterns of Nerve. Science 1935, 82, 44–45. [Google Scholar] [CrossRef]
- Schmitt, F.O.; Bear, R.S.; Palmer, K.J. X-ray diffraction studies on the structure of the nerve myelin sheath. J. Cell. Physiol. 1941, 18, 31–42. [Google Scholar] [CrossRef]
- Robertson, J.D. The Molecular Biology of Cell Membranes; Nachmansohn, D., Ed.; Molecular Biology. Elementary Processes of Nerve Conduction and Muscle Contraction; Academic Press: New York, NY, USA, 1960; pp. 87–151. [Google Scholar]
- Robertson, J.D. The Early Days of Electron-Microscopy of Nerve-Tissue and Membranes. Int. Rev. Cytol. 1987, 100, 129–201. [Google Scholar]
- Meller, K. Cryoelectron Microscopy of Vitrified Nerve Myelin. Cell Tissue Res. 1990, 262, 59–66. [Google Scholar] [CrossRef]
- Aggarwal, S.; Yurlova, L.; Snaidero, N.; Reetz, C.; Frey, S.; Zimmermann, J.; Paehler, G.; Janshoff, A.; Friedrichs, J.; Müller, D.J.; et al. A Size Barrier Limits Protein Diffusion at the Cell Surface to Generate Lipid-Rich Myelin-Membrane Sheets. Dev. Cell 2011, 21, 445–456. [Google Scholar] [CrossRef] [Green Version]
- Patzig, J.; Jahn, O.; Tenzer, S.; Wichert, S.P.; de Monasterio-Schrader, P.; Rosfa, S.; Kuharev, J.; Yan, K.; Bormuth, I.; Bremer, J.; et al. Quantitative and Integrative Proteome Analysis of Peripheral Nerve Myelin Identifies Novel Myelin Proteins and Candidate Neuropathy Loci. J. Neurosci. 2011, 31, 16369–16386. [Google Scholar] [CrossRef] [Green Version]
- de Monasterio-Schrader, P.; Jahn, O.; Tenzer, S.; Wichert, S.P.; Patzig, J.; Werner, H.B. Systematic approaches to central nervous system myelin. Cell. Mol. Life Sci. 2012, 69, 2879–2894. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Baek, R.; Kirschner, D.; Peterson, A.; Fujii, Y.; Nave, K.; Macklin, W.; Trapp, B. Evolution of a neuroprotective function of central nervous system myelin. J. Cell Biol. 2006, 172, 469–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campi, G.; Di Gioacchino, M.; Poccia, N.; Ricci, A.; Burghammer, M.; Bianconi, A. Intrinsic dynamical fluctuations of PNS myelin. arXiv 2017, arXiv:1705.09730. [Google Scholar]
- Shapiro, L.; Doyle, J.; Hensley, P.; Colman, D.; Hendrickson, W. Crystal structure of the extracellular domain from P-0, the major structural protein of peripheral nerve myelin. Neuron 1996, 17, 435–449. [Google Scholar] [CrossRef] [Green Version]
- Inouye, H.; Tsuruta, H.; Sedzik, J.; Uyemura, K.; Kirschner, D.A. Tetrameric Assembly of Full-Sequence Protein Zero Myelin Glycoprotein by Synchrotron X-Ray Scattering. Biophys. J. 1999, 76, 423–437. [Google Scholar] [CrossRef] [Green Version]
- Thompson, A.J.; Cronin, M.S.; Kirschner, D.A. Myelin protein zero exists as dimers and tetramers in native membranes of Xenopus laevis peripheral nerve. J. Neurosci. Res. 2002, 67, 766–771. [Google Scholar] [CrossRef]
- Favereaux, A.; Lagueny, A.; Vital, A.; Schmitter, J.; Chaignepain, S.; Ferrer, X.; Labatut-Cazabat, I.; Vital, C.; Petry, K. Serum IgG antibodies to P0 dimer and 35 kDa P0 related protein in neuropathy associated with monoclonal gammopathy. J. Neurol. Neurosurg. Psychiatry 2003, 74, 1262–1266. [Google Scholar] [CrossRef] [Green Version]
- Plotkowski, M.L.; Kim, S.; Phillips, M.L.; Partridge, A.W.; Deber, C.M.; Bowie, J.U. Transmembrane domain of myelin protein zero can form dimers: Possible implications for myelin construction. Biochemistry 2007, 46, 12164–12173. [Google Scholar] [CrossRef]
- Toyama, B.; Savas, J.; Park, S.; Harris, M.; Ingolia, N.; Yates, J., III; Hetzer, M. Identification of Long-Lived Proteins Reveals Exceptional Stability of Essential Cellular Structures. Cell 2013, 154, 971–982. [Google Scholar] [CrossRef] [Green Version]
- Boggs, J.M. Myelin basic protein: A multifunctional protein. Cell Mol. Life Sci. 2006, 63, 1945–1961. [Google Scholar] [CrossRef]
- Harauz, G.; Ladizhansky, V.; Boggs, J.M. Structural Polymorphism and Multifunctionality of Myelin Basic Protein. Biochemistry 2009, 48, 8094–8104. [Google Scholar] [CrossRef] [PubMed]
- Fulton, D.; Paez, P.M.; Campagnoni, A.T. The multiple roles of myelin protein genes during the development of the oligodendrocyte. ASN Neuro 2010, 2, e00027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raasakka, A.; Kursula, P. The myelin membrane-associated enzyme 2’,3’-cyclic nucleotide 3’-phosphodiesterase: On a highway to structure and function. Neurosci. Bull. 2014, 30, 956–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, H.; Myllykoski, M.; Ruskamo, S.; Wang, C.; Kursula, P. Myelin-specific proteins: A structurally diverse group of membrane-interacting molecules. Biofactors 2013, 39, 233–241. [Google Scholar] [CrossRef]
- Arroyo, E.; Scherer, S. On the molecular architecture of myelinated fibers. Histochem. Cell Biol. 2000, 113, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kursula, P. The current status of structural studies on proteins of the myelin sheath (review). Int. J. Mol. Med. 2001, 8, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Kursula, P. Structural properties of proteins specific to the myelin sheath. Amino Acids 2008, 34, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Orthmann-Murphy, J.L.; Abrams, C.K.; Scherer, S.S. Gap junctions couple astrocytes and oligodendrocytes. J. Mol. Neurosci. 2008, 35, 101–116. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Gomez, G.A.; Yap, A.S. Current perspectives on cadherin-cytoskeleton interactions and dynamics. Cell Health Cytoskelet. 2015, 7, 11–24. [Google Scholar]
- Luo, X.; Inouye, H.; Gross, A.A.R.; Hidalgo, M.M.; Sharma, D.; Lee, D.; Avila, R.L.; Salmona, M.; Kirschner, D.A. Cytoplasmic domain of zebrafish myelin protein zero: Adhesive role depends on beta-conformation. Biophys. J. 2007, 93, 3515–3528. [Google Scholar] [CrossRef] [Green Version]
- Raasakka, A.; Ruskamo, S.; Kowal, J.; Han, H.; Baumann, A.; Myllykoski, M.; Fasano, A.; Rossano, R.; Riccio, P.; Bürck, J.; et al. Molecular structure and function of myelin protein P0 in membrane stacking. Sci. Rep. 2019, 9, 642. [Google Scholar] [CrossRef] [Green Version]
- Raasakka, A.; Jones, N.; Hoffmann, S.V.; Kursula, P. Ionic strength and calcium regulate the membrane interactions of myelin basic protein and the cytoplasmic domain of myelin protein zero. Biochem. Biophys. Res. Commun. 2019, 511, 7–12. [Google Scholar] [CrossRef]
- Raasakka, A.; Ruskamo, S.; Barker, R.; Krokengen, O.C.; Vatne, G.H.; Kristiansen, C.K.; Hallin, E.I.; Skoda, M.W.A.; Bergmann, U.; Wacklin-Knecht, H.; et al. Neuropathy-related mutations alter the membrane binding properties of the human myelin protein P0 cytoplasmic tail. PLoS ONE 2019, 14, e0216833. [Google Scholar] [CrossRef] [Green Version]
- Myllykoski, M.; Baumgärtel, P.; Kursula, P. Conformations of peptides derived from myelin-specific proteins in membrane-mimetic conditions probed by synchrotron radiation CD spectroscopy. Amino Acids 2012, 42, 1467–1474. [Google Scholar] [CrossRef]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of Intrinsically Disordered Regions and Proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef] [PubMed]
- Sedzik, J.; Kirschner, D.A. Is myelin basic protein crystallizable? Neurochem Res. 1992, 17, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Pedraza, L. Nuclear transport of myelin basic protein. J. Neurosci. Res. 1997, 50, 258–264. [Google Scholar] [CrossRef]
- Moscarello, M.A. Myelin Basic Protein, the “Executive” Molecule of the Myelin Membrane; Juurlink, B.H.J., Devon, R.M., Doucette, J.R., Nazarali, A.J., Schreyer, D.J., Verge, V.M.K., Eds.; Cell Biology and Pathology of Myelin; Plenum Press: New York, NY, USA, 1997; pp. 13–22. [Google Scholar]
- Kim, J.; Mastronardi, F.; Wood, D.; Lubman, D.; Zand, R.; Moscarello, M. Multiple sclerosis–An important role for post-translational modifications of myelin basic protein in pathogenesis. Mol. Cell. Proteom. 2003, 2, 453–462. [Google Scholar] [CrossRef] [Green Version]
- Harauz, G.; Ishiyama, N.; Hill, C.; Bates, I.; Libich, D.; Fares, C. Myelin basic protein–diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron 2004, 35, 503–542. [Google Scholar] [CrossRef]
- Harauz, G.; Musse, A.A. A tale of two citrullines–Structural and functional aspects of myelin basic protein deimination in health and disease. Neurochem. Res. 2007, 32, 137–158. [Google Scholar] [CrossRef]
- Baron, W.; Hoekstra, D. On the biogenesis of myelin membranes: Sorting, trafficking and cell polarity. FEBS Lett. 2010, 584, 1760–1770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, C.; Bauer, N.M.; Schaefer, I.; White, R. Making myelin basic protein–from mRNA transport to localized translation. Front. Cell. Neurosci. 2013, 7, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vassall, K.A.; Bamm, V.V.; Harauz, G. MyelStones: The executive roles of myelin basic protein in myelin assembly and destabilization in multiple sclerosis. Biochem. J. 2015, 472, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Ridsdale, R.; Beniac, D.; Tompkins, T.; Moscarello, M.; Harauz, G. Three-dimensional structure of myelin basic protein II. Molecular modeling and considerations of predicted structures in multiple sclerosis. J. Biol. Chem. 1997, 272, 4269–4275. [Google Scholar] [CrossRef] [Green Version]
- Pribyl, T.M.; Campagnoni, C.W.; Kampf, K.; Kashima, T.; Handley, V.W.; McMahon, J.; Campagnoni, A.T. The Human Myelin Basic-Protein Gene is Included within a 179-Kilobase Transcription Unit–Expression in the Immune and Central Nervous Systems. Proc. Natl. Acad. Sci. USA 1993, 90, 10695–10699. [Google Scholar] [CrossRef] [Green Version]
- Campagnoni, A.T.; Pribyl, T.M.; Campagnoni, C.W.; Kampf, K.; Amurumarjee, S.; Landry, C.F.; Handley, V.W.; Newman, S.L.; Garbay, B.; Kitamura, K. Structure and Developmental Regulation of Golli-Mbp, a 105-Kilobase Gene that Encompasses the Myelin Basic-Protein Gene and is Expressed in Cells in the Oligodenrocyte Lineage in the Brain. J. Biol. Chem. 1993, 268, 4930–4938. [Google Scholar]
- Mendz, G.L.; Barden, J.A.; Martenson, R.E. Conformation of a Tetradecapeptide Epitope of Myelin Basic-Protein. Eur. J. Biochem. 1995, 231, 659–666. [Google Scholar] [CrossRef]
- Feng, J.; Hu, Y.K.; Xie, L.; Colwell, C.S.; Shao, X.M.; Sun, X.; Chen, B.; Tang, H.; Campagnoni, A.T. Golli protein negatively regulates store depletion-induced calcium influx in T cells. Immunity 2006, 24, 717–727. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.S.; Paez, P.M.; Spreuer, V.; Campagnoni, C.W.; Boggs, J.M.; Campagnoni, A.T.; Harauz, G. Classical 18.5-and 21.5-kDa isoforms of myelin basic protein inhibit calcium influx into oligodendroglial cells, in contrast to golli isoforms. J. Neurosci. Res. 2011, 89, 467–480. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhang, Y.; Li, D.; Feng, Y. Destabilization and mislocalization of myelin basic protein mRNAs in quaking dysmyelination lacking the QKI RNA-binding proteins. J. Neurosci. 2000, 20, 4944–4953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torvund-Jensen, J.; Steengaard, J.; Reimer, L.; Fihl, L.B.; Laursen, L.S. Transport and translation of MBP mRNA is regulated differently by distinct hnRNP proteins. J. Cell. Sci. 2014, 127, 1550–1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, A.; Campagnoni, C.; Kampf, K.; Feng, J.; Handley, V.; Schonmann, V.; Bongarzone, E.; Reyes, S.; Campagnoni, A. Identification of a protein that interacts with the Golli-Myelin basic protein and with nuclear-LIM interactor in the nervous system. J. Neurosci. Res. 2004, 75, 461–471. [Google Scholar] [CrossRef]
- Smith, G.S.T.; Seymour, L.V.; Boggs, J.M.; Harauz, G. The 21.5-kDa isoform of myelin basic protein has a non-traditional PY-nuclear-localization signal. Biochem. Biophys. Res. Commun. 2012, 422, 670–675. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.S.T.; Samborska, B.; Hawley, S.P.; Klaiman, J.M.; Gillis, T.E.; Jones, N.; Boggs, J.M.; Harauz, G. Nucleus-localized 21.5-kDa myelin basic protein promotes oligodendrocyte proliferation and enhances neurite outgrowth in coculture, unlike the plasma membrane-associated 18.5-kDa isoform. J. Neurosci. Res. 2013, 91, 349–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, G.S.T.; Homchaudhuri, L.; Boggs, J.M.; Harauz, G. Classic 18.5-and 21.5-kDa myelin basic protein isoforms associate with cytoskeletal and SH3-domain proteins in the immortalized N19-oligodendroglial cell line stimulated by phorbol ester and IGF-1. Neurochem. Res. 2012, 37, 1277–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, G.S.T.; De Avila, M.; Paez, P.M.; Spreuer, V.; Wills, M.K.B.; Jones, N.; Boggs, J.M.; Harauz, G. Proline substitutions and threonine pseudophosphorylation of the SH3 ligand of 18.5-kDa myelin basic protein decrease its affinity for the Fyn-SH3 domain and alter process development and protein localization in oligodendrocytes. J. Neurosci. Res. 2012, 90, 28–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Avila, M.; Vassall, K.A.; Smith, G.S.T.; Bamm, V.V.; Harauz, G. The proline-rich region of 18.5 kDa myelin basic protein binds to the SH3-domain of Fyn tyrosine kinase with the aid of an upstream segment to form a dynamic complex in vitro. Biosci. Rep. 2014, 34, 775–788. [Google Scholar] [CrossRef]
- Boggs, J.M.; Homchaudhuri, L.; Ranagaraj, G.; Liu, Y.; Smith, G.S.T.; Harauz, G. Interaction of myelin basic protein with cytoskeletal and signaling proteins in cultured primary oligodendrocytes and N19 oligodendroglial cells. BMC Res. Notes 2014, 7, 387. [Google Scholar] [CrossRef] [Green Version]
- Robb, N.D.; Chen, W.H. Myelin Basic Protein Interaction with Calmodulin and Gangliosides. J. Neurosci. Res. 1990, 25, 535–544. [Google Scholar]
- Harauz, G.; Ishiyama, N.; Bates, I. Analogous standard motifs in myelin basic protein and in MARCKS. Mol. Cell. Biochem. 2000, 209, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Libich, D.; Hill, C.; Haines, J.; Harauz, G. Myelin basic protein has multiple calmodulin-binding sites. Biochem. Biophys. Res. Commun. 2003, 308, 313–319. [Google Scholar] [CrossRef]
- Bamm, V.V.; De Avila, M.; Smith, G.S.T.; Ahmed, M.A.M.; Harauz, G. Structured Functional Domains of Myelin Basic Protein: Cross Talk between Actin Polymerization and Ca2+-Dependent Calmodulin Interaction. Biophys. J. 2011, 101, 1248–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavatorta, P.; Giovanelli, S.; Bobba, A.; Riccio, P.; Szabo, A.G.; Quagliariello, E. Myelin Basic-Protein Interaction with Zinc and Phosphate–Fluorescence Studies on the Water-Soluble Form of the Protein. Biophys. J. 1994, 66, 1174–1179. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.S.T.; Chen, L.; Bamm, V.V.; Dutcher, J.R.; Harauz, G. The interaction of zinc with membrane-associated 18.5 kDa myelin basic protein: An attenuated total reflectance-Fourier transform infrared spectroscopic study. Amino Acids 2010, 39, 739–750. [Google Scholar] [CrossRef]
- Baran, C.; Smith, G.S.T.; Bamm, V.V.; Harauz, G.; Lee, J.S. Divalent cations induce a compaction of intrinsically disordered myelin basic protein. Biochem. Biophys. Res. Commun. 2010, 391, 224–229. [Google Scholar] [CrossRef]
- Earl, C.; Chantry, A.; Mohammad, N.; Glynn, P. Zinc Ions Stabilize the Association of Basic-Protein with Brain Myelin Membranes. J. Neurochem. 1988, 51, 718–724. [Google Scholar] [CrossRef]
- Nawaz, S.; Kippert, A.; Saab, A.S.; Werner, H.B.; Lang, T.; Nave, K.; Simons, M. Phosphatidylinositol 4,5-Bisphosphate-Dependent Interaction of Myelin Basic Protein with the Plasma Membrane in Oligodendroglial Cells and Its Rapid Perturbation by Elevated Calcium. J. Neurosci. 2009, 29, 4794–4807. [Google Scholar] [CrossRef]
- Zhang, C.; Walker, A.K.; Zand, R.; Moscarello, M.A.; Yan, J.M.; Andrews, P.C. Myelin Basic Protein Undergoes a Broader Range of Modifications in Mammals than in Lower Vertebrates. J. Proteome Res. 2012, 11, 4791–4802. [Google Scholar] [CrossRef] [Green Version]
- Wood, D.D.; Moscarello, M.A. The Isolation, Characterization, and Lipid-Aggregating Properties of a Citrulline Containing Myelin Basic-Protein. J. Biol. Chem. 1989, 264, 5121–5127. [Google Scholar]
- Wood, D.; Bilbao, J.; OConnors, P.; Moscarello, M. Acute multiple sclerosis (Marburg type) is associated with developmentally immature myelin basic protein. Ann. Neurol. 1996, 40, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.S.; Chou, C.H.J.; Mazzei, G.J.; Dembure, P.; Kuo, J.F. Phospholipid-Sensitive Ca2+-Dependent Protein-Kinase Preferentially Phosphorylates Serine-115 of Bovine Myelin Basic-Protein. J. Neurochem. 1984, 43, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, A.; Nishiyama, K.; Nakanishi, H.; Uratsuji, Y.; Nomura, H.; Takeyama, Y.; Nishizuka, Y. Studies on the Phosphorylation of Myelin Basic-Protein by Protein-Kinase C and Adenosine 3’-5’-Monophosphate-Dependent Protein-Kinase. J. Biol. Chem. 1985, 260, 2492–2499. [Google Scholar]
- Schulz, P.; Cruz, T.F.; Moscarello, M.A. Endogenous Phosphorylation of Basic-Protein in Myelin of Varying Degrees of Compaction. Biochemistry 1988, 27, 7793–7799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erickson, A.K.; Payne, D.M.; Martino, P.A.; Rossomando, A.J.; Shabanowitz, J.; Weber, M.J.; Hunt, D.F.; Sturgill, T.W. Identification by Mass-Spectrometry of Threonine-97 in Bovine Myelin Basic-Protein as a Specific Phosphorylation Site for Mitogen-Activated Protein-Kinase. J. Biol. Chem. 1990, 265, 19728–19735. [Google Scholar] [PubMed]
- Ramwani, J.; Moscarello, M.A. Phosphorylation of Charge Isomers (Components) of Human Myelin Basic-Protein–Identification of Phosphorylated Sites. J. Neurochem. 1990, 55, 1703–1710. [Google Scholar] [CrossRef]
- Wang, Q.M.; Smith, J.B.; Harrison, M.L.; Geahlen, R.L. Identification of Tyrosine-67 in Bovine Brain Myelin Basic-Protein as a Specific Phosphorylation Site for Thymus-P56lck. Biochem. Biophys. Res. Commun. 1991, 178, 1393–1399. [Google Scholar] [CrossRef]
- Zand, R.; Li, M.; Jin, X.; Lubman, D. Determination of the sites of posttranslational modifications in the charge isomers of bovine myelin basic protein by capillary electrophoresis mass spectroscopy. Biochemistry 1998, 37, 2441–2449. [Google Scholar] [CrossRef]
- Wood, D.D.; Ackerley, C.A.; van den Brand, B.; Zhang, L.; Raijmakers, R.; Mastronardi, F.G.; Moscarello, M.A. Myelin localization of peptidylarginine deiminases 2 and 4: Comparison of PAD2 and PAD4 activities. Lab. Invest. 2008, 88, 354–364. [Google Scholar] [CrossRef] [Green Version]
- Beniac, D.R.; Wood, D.D.; Palaniyar, N.; Ottensmeyer, P.; Moscarello, M.A.; Harauz, G. Marburg’s variant of multiple sclerosis correlates with a less compact structure of myelin basic protein. Mol. Cell. Biol. Res. Commun. 1999, 1, 48–51. [Google Scholar] [CrossRef] [Green Version]
- McLaurin, J.; Ackerley, C.A.; Moscarello, M.A. Localization of Basic-Proteins in Human Myelin. J. Neurosci. Res. 1993, 35, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Boggs, J.M.; Rangaraj, G.; Hill, C.M.D.; Bates, I.R.; Heng, Y.M.; Harauz, G. Effect of arginine loss in myelin basic protein, as occurs in its deiminated charge isoform, on mediation of actin polymerization and actin binding to a lipid membrane in vitro. Biochemistry 2005, 44, 3524–3534. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, T.A.; Moscarello, M.A. Stimulation of Bovine Brain Phospholipase-C Activity by Myelin Basic-Protein Requires Arginyl Residues in Peptide Linkage. Arch. Biochem. Biophys. 1993, 302, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, H.; Martin, R.; Mariuzza, R. Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins. J. Mol. Biol. 2000, 304, 177–188. [Google Scholar] [CrossRef]
- Bates, I.; Feix, J.; Boggs, J.; Harauz, G. An immunodominant epitope of myelin basic protein is an amphipathic alpha-helix. J. Biol. Chem. 2004, 279, 5757–5764. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Doudevski, I.; Wood, D.; Moscarello, M.; Husted, C.; Genain, C.; Zasadzinski, J.; Israelachvili, J. Synergistic interactions of lipids and myelin basic protein. Proc. Natl. Acad. Sci. USA 2004, 101, 13466–13471. [Google Scholar] [CrossRef] [Green Version]
- Musse, A.A.; Gao, W.; Rangaraj, G.; Boggs, J.M.; Harauz, G. Myelin basic protein co-distributes with other PI(4,5)P-2-sequestering proteins in Triton X-100 detergent-resistant membrane microdomains. Neurosci. Lett. 2009, 450, 32–36. [Google Scholar] [CrossRef]
- Lee, D.W.; Banquy, X.; Kristiansen, K.; Kaufman, Y.; Boggs, J.M.; Israelachvili, J.N. Lipid domains control myelin basic protein adsorption and membrane interactions between model myelin lipid bilayers. Proc. Natl. Acad. Sci. USA 2014, 111, E768–E775. [Google Scholar] [CrossRef] [Green Version]
- Widder, K.; Träger, J.; Kerth, A.; Harauz, G.; Hinderberger, D. Interaction of Myelin Basic Protein with Myelin-like Lipid Monolayers at Air-Water Interface. Langmuir 2018, 34, 6095–6108. [Google Scholar] [CrossRef]
- Widder, K.; Harauz, G.; Hinderberger, D. Myelin basic protein (MBP) charge variants show different sphingomyelin-mediated interactions with myelin-like lipid monolayers. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183077. [Google Scholar] [CrossRef]
- Ishiyama, N.; Bates, I.; Hill, C.; Wood, D.; Matharu, P.; Viner, N.; Moscarello, M.; Harauz, G. The effects of deimination of myelin basic protein on structures formed by its interaction with phosphoinositide-containing lipid monolayers. J. Struct. Biol. 2001, 136, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Snaidero, N.; Paehler, G.; Frey, S.; Sanchez, P.; Zweckstetter, M.; Janshoff, A.; Schneider, A.; Weil, M.; Schaap, I.A.T.; et al. Myelin membrane assembly is driven by a phase transition of myelin basic proteins into a cohesive protein meshwork. PLoS Biol. 2013, 11, e1001577. [Google Scholar] [CrossRef] [Green Version]
- Beniac, D.; Luckevich, M.; Czarnota, G.; Tompkins, T.; Ridsdale, R.; Ottensmeyer, F.; Moscarello, M.; Harauz, G. Three-dimensional structure of myelin basic protein 1. Reconstruction via angular reconstitution of randomly oriented single particles. J. Biol. Chem. 1997, 272, 4261–4268. [Google Scholar] [CrossRef] [Green Version]
- Haas, H.; Oliveira, C.; Torriani, I.; Polverini, E.; Fasano, A.; Carlone, G.; Cavatorta, P.; Riccio, P. Small angle x-ray scattering from lipid-bound myelin basic protein in solution. Biophys. J. 2004, 86, 455–460. [Google Scholar] [CrossRef] [Green Version]
- Libich, D.S.; Harauz, G. Backbone dynamics of the 18.5 kDa isoform of myelin basic protein reveals transient alpha-helices and a calmodulin-binding site. Biophys. J. 2008, 94, 4847–4866. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, M.A.M.; De Avila, M.; Polverini, E.; Bessonov, K.; Bamm, V.V.; Harauz, G. Solution nuclear magnetic resonance structure and molecular dynamics simulations of a murine 18.5 kDa myelin basic protein segment (S72-S107) in association with dodecylphosphocholine micelles. Biochemistry 2012, 51, 7475–7487. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Peterson, J.; Gravel, M.; Braun, P.; Trapp, B. CNP overexpression induces aberrant oligodendrocyte membranes and inhibits MBP accumulation and myelin compaction. J. Neurosci. Res. 1997, 50, 238–247. [Google Scholar] [CrossRef]
- Snaidero, N.; Velte, C.; Myllykoski, M.; Raasakka, A.; Ignatev, A.; Werner, H.B.; Erwig, M.S.; Möbius, W.; Kursula, P.; Nave, K.; et al. Antagonistic functions of MBP and CNP establish cytosolic channels in CNS myelin. Cell Rep. 2017, 18, 314–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, Y.; Mizuno, R.; Nishimura, T.; Ogawa, Y.; Yoshikawa, H.; Fujimura, H.; Adachi, E.; Kishimoto, T.; Yanagihara, T.; Sakoda, S. Cloning and Expression of Myelin-Associated Oligodendrocytic Basic-Protein–a Novel Basic-Protein Constituting the Central-Nervous-System Myelin. J. Biol. Chem. 1994, 269, 31725–31730. [Google Scholar] [PubMed]
- Holz, A.; Schaeren-Wiemers, N.; Schaefer, C.; Pott, U.; Colello, R.; Schwab, M. Molecular and developmental characterization of novel cDNAs of the myelin-associated oligodendrocytic basic protein. J. Neurosci. 1996, 16, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Montague, P.; McCallion, A.; Davies, R.; Griffith, I. Myelin-Associated Oligodendrocytic Basic Protein: A Family of Abundant CNS Myelin Proteins in Search of a Function. Dev. Neurosci. 2006, 28, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Kosaras, B.; Kirschner, D.A. Radial Component of CNS Myelin–Junctional Subunit Structure and Supramolecular Assembly. J. Neurocytol. 1990, 19, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Yoshikawa, H.; Nagano, S.; Kondoh, G.; Sadahiro, S.; Gotow, T.; Yanagihara, T.; Sakoda, S. Myelin-associated oligodendrocytic basic protein is essential for normal arrangement of the radial component in central nervous system myelin. Eur. J. Neurosci. 1999, 11, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, H. Myelin-associated oligodendrocytic basic protein modulates the arrangement of radial growth of the axon and the radial component of myelin. Med. Electron Microsc. 2001, 34, 160–164. [Google Scholar] [CrossRef]
- Bourre, J.M.; Cloez, I.; Galliot, M.; Buisine, A.; Dumont, O.; Piciotti, M.; Prouillet, F.; Bourdon, R. Occurrence of manganese, copper and zinc in myelin. Alterations in the peripheral nervous system of dysmyelinating trembler mutant are at variance with brain mutants (quaking and shiverer). Neurochem. Int. 1987, 10, 281–286. [Google Scholar] [CrossRef]
- Stys, P.K.; Lehning, E.; Saubermann, A.J.; LoPachin, R.M. Intracellular Concentrations of Major Ions in Rat Myelinated Axons and Glia: Calculations Based on Electron Probe X-Ray Microanalyses. J. Neurochem. 1997, 68, 1920–1928. [Google Scholar] [CrossRef]
- Bonaventura, P.; Benedetti, G.; Albarède, F.; Miossec, P. Zinc and its role in immunity and inflammation. Autoimmun. Rev. 2015, 14, 277–285. [Google Scholar] [CrossRef]
- Choi, B.Y.; Jung, J.W.; Suh, S.W. The Emerging Role of Zinc in the Pathogenesis of Multiple Sclerosis. Int. J. Mol. Sci. 2017, 18, 2070. [Google Scholar] [CrossRef] [Green Version]
- Lundby, A.; Secher, A.; Lage, K.; Nordsborg, N.B.; Dmytriyev, A.; Lundby, C.; Olsen, J.V. Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat. Commun. 2012, 3, 876. [Google Scholar] [CrossRef] [Green Version]
- Cilia, E.; Pancsa, R.; Tompa, P.; Lenaerts, T.; Vranken, W.F. The DynaMine webserver: Predicting protein dynamics from sequence. Nucleic Acids Res. 2014, 42, W264–W270. [Google Scholar] [CrossRef] [Green Version]
- PONDR® Predictor of Natural Disordered Regions. Available online: http://www.pondr.com (accessed on 29 January 2020).
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schäfer, I.; Müller, C.; Luhmann, H.J.; White, R. MOBP levels are regulated by Fyn kinase and affect the morphological differentiation of oligodendrocytes. J. Cell. Sci. 2016, 129, 930–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapp, B.D.; Bernier, L.; Andrews, S.B.; Colman, D.R. Cellular and subcellular distribution of 2’,3’-cyclic nucleotide 3’-phosphodiesterase and its mRNA in the rat central nervous system. J. Neurochem. 1988, 51, 859–868. [Google Scholar] [CrossRef]
- Radtke, C.; Sasaki, M.; Lankford, K.L.; Gallo, V.; Kocsis, J.D. CNPase Expression in Olfactory Ensheathing Cells. J. Biomed. Biotechnol. 2011, 608496. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, R.; Minuk, J.; Cox, M.; Braun, P.; Gravel, M. CNP2 mRNA directs synthesis of both CNP1 and CNP2 polypeptides. J. Neurosci. Res. 1997, 50, 248–257. [Google Scholar] [CrossRef]
- McFerran, B.; Burgoyne, R. 2’,3’-Cyclic nucleotide 3’-phosphodiesterase is associated with mitochondria in diverse adrenal cell types. J. Cell. Sci. 1997, 110, 2979–2985. [Google Scholar] [PubMed]
- Lee, J.; O’Neill, R.; Park, M.; Gravel, M.; Braun, P. Mitochondrial localization of CNP2 is regulated by phosphorylation of the N-terminal targeting signal by PKC: Implications of a mitochondrial function for CNP2 in glial and non-glial cells. Mol. Cell. Neurosci. 2006, 31, 446–462. [Google Scholar] [CrossRef] [PubMed]
- Myllykoski, M.; Seidel, L.; Muruganandam, G.; Raasakka, A.; Torda, A.E.; Kursula, P. Structural and functional evolution of 2’,3’-cyclic nucleotide 3’-phosphodiesterase. Brain Res. 2016, 1641, 64–78. [Google Scholar] [CrossRef]
- Myllykoski, M.; Kursula, P. Expression, purification, and initial characterization of different domains of recombinant mouse 2’,3’-cyclic nucleotide 3’-phosphodiesterase, an enigmatic enzyme from the myelin sheath. BMC Res. Notes. 2010, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Myllykoski, M.; Itoh, K.; Kangas, S.; Heape, A.; Kang, S.; Lubeck, G.; Kursula, I.; Kursula, P. The N-terminal domain of the myelin enzyme 2’,3’-cyclic nucleotide 3’-phosphodiesterase: Direct molecular interaction with the calcium sensor calmodulin. J. Neurochem. 2012, 123, 515–524. [Google Scholar] [CrossRef]
- Stingo, S.; Masullo, M.; Polverini, E.; Laezza, C.; Ruggiero, I.; Arcone, R.; Ruozi, E.; Dal Piaz, F.; Malfitano, A.M.; D’Ursi, A.M.; et al. The N-terminal domain of 2’,3’-cyclic nucleotide 3’-phosphodiesterase harbors a GTP/ATP binding site. Chem. Biol. Drug Des. 2007, 70, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Drummond, G.I.; Iyer, N.T.; Keith, J. Hydrolysis of ribonucleoside 2′,3′-cyclic phosphates by a diesterase from brain. J. Biol. Chem. 1962, 237, 3535–3539. [Google Scholar]
- Kozlov, G.; Lee, J.; Elias, D.; Gravel, M.; Gutierrez, P.; Ekiel, I.; Braun, P.; Gehring, K. Structural evidence that brain cyclic nucleotide phosphodiesterase is a member of the 2H phosphodiesterase superfamily. J. Biol. Chem. 2003, 278, 46021–46028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, Y.; Tanaka, N.; Ichimiya, T.; Kurihara, T.; Nakamura, K. Crystal structure of the catalytic fragment of human brain 2’,3’-cyclic-nucleotide 3’-phosphodiesterase. J. Mol. Biol. 2005, 346, 789–800. [Google Scholar] [CrossRef]
- Myllykoski, M.; Raasakka, A.; Han, H.; Kursula, P. Myelin 2’,3’-Cyclic Nucleotide 3’-Phosphodiesterase: Active-Site Ligand Binding and Molecular Conformation. PLoS ONE 2012, 7, e32336. [Google Scholar] [CrossRef] [Green Version]
- Myllykoski, M.; Raasakka, A.; Lehtimäki, M.; Han, H.; Kursula, I.; Kursula, P. Crystallographic Analysis of the Reaction Cycle of 2’,3’-Cyclic Nucleotide 3’-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family. J. Mol. Biol. 2013, 425, 4307–4322. [Google Scholar] [CrossRef]
- Raasakka, A.; Myllykoski, M.; Laulumaa, S.; Lehtimäki, M.; Härtlein, M.; Moulin, M.; Kursula, I.; Kursula, P. Determinants of ligand binding and catalytic activity in the myelin enzyme 2’,3’-cyclic nucleotide 3’-phosphodiesterase. Sci. Rep. 2015, 5, 16520. [Google Scholar] [CrossRef]
- Lee, J.; Gravel, M.; Gao, E.; O’Neill, R.; Braun, P. Identification of essential residues in 2’,3’-cyclic nucleotide 3’-phosphodiesterase–Chemical modification and site-directed mutagenesis to investigate the role of cysteine and histidine residues in enzymatic activity. J. Biol. Chem. 2001, 276, 14804–14813. [Google Scholar] [CrossRef] [Green Version]
- Verrier, J.; Jackson, T.; Bansal, R.; Kochanek, P.M.; Jackson, E. Oligodendrocyte 2’,3’-Cyclic Nucleotide 3’-Phosphodiesterase Participates in Localized Adenosine Production: Possible Role in Traumatic Brain Injury. J. Neurotrauma 2012, 29, A168–A169. [Google Scholar]
- Verrier, J.D.; Jackson, T.C.; Bansal, R.; Kochanek, P.M.; Puccio, A.M.; Okonkwo, D.O.; Jackson, E.K. The brain in vivo expresses the 2’,3’-cAMP-adenosine pathway. J. Neurochem. 2012, 122, 115–125. [Google Scholar] [CrossRef]
- Verrier, J.D.; Jackson, T.C.; Gillespie, D.G.; Janesko-Feldman, K.; Bansal, R.; Goebbels, S.; Nave, K.; Kochanek, P.M.; Jackson, E.K. Role of CNPase in the oligodendrocytic extracellular 2’,3’-cAMP-adenosine pathway. Glia 2013, 61, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Gravel, M.; Robert, F.; Kottis, V.; Gallouzi, I.; Pelletier, J.; Braun, P.E. 2’,3’-Cyclic Nucleotide 3’-Phosphodiesterase: A Novel RNA-Binding Protein That Inhibits Protein Synthesis. J. Neurosci. Res. 2009, 87, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Bifulco, M.; Laezza, C.; Stingo, S.; Wolff, J. 2’,3’-Cyclic nucleotide 3’-phosphodiesterase: A membrane-bound, microtubule-associated protein and membrane anchor for tubulin. Proc. Natl. Acad. Sci. USA 2002, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Gravel, M.; Zhang, R.; Thibault, P.; Braun, P. Process outgrowth in oligodendrocytes is mediated by CNP, a novel microtubule assembly myelin protein. J. Cell Biol. 2005, 170, 661–673. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, D.A.; Braun, P.E. 2’,3’-Cyclic Nucleotide 3’-Phosphodiesterase Binds to Actin-Based Cytoskeletal Elements in an Isoprenylation-Independent Manner. J. Neurochem. 1996, 67, 943–951. [Google Scholar] [CrossRef]
- Azarashvili, T.; Krestinina, O.; Galvita, A.; Grachev, D.; Baburina, Y.; Stricker, R.; Evtodienko, Y.; Reiser, G. Ca2+-dependent permeability transition regulation in rat brain mitochondria by 2’,3’-cyclic nucleotides and 2’,3’-cyclic nucleotide 3’-phosphodiesterase. Am. J. Physiol. Cell Physiol. 2009, 296, C1428–C1439. [Google Scholar] [CrossRef] [Green Version]
- Baburina, Y.; Azarashvili, T.; Grachev, D.; Krestinina, O.; Galvita, A.; Stricker, R.; Reiser, G. Mitochondrial 2’,3’-cyclic nucleotide 3’-phosphodiesterase (CNP) interacts with mPTP modulators and functional complexes (I-V) coupled with release of apoptotic factors. Neurochem. Int. 2015, 90, 46–55. [Google Scholar] [CrossRef]
- Baburina, Y.; Odinokova, I.; Azarashvili, T.; Akatov, V.; Sotnikova, L.; Krestinina, O. Possible Involvement of 2’,3’-Cyclic Nucleotide-3’-Phosphodiesterase in the Protein Phosphorylation-Mediated Regulation of the Permeability Transition Pore. Int. J. Mol. Sci. 2018, 19, 3499. [Google Scholar] [CrossRef] [Green Version]
- Esposito, C.; Scrima, M.; Carotenuto, A.; Tedeschi, A.; Rovero, P.; D’Errico, G.; Malfitano, A.M.; Bifulco, M.; D’Ursi, A.M. Structures and micelle locations of the nonlipidated and lipidated C-terminal membrane anchor of 2’,3’-cyclic nucleotide-3’-phosphodiesterase. Biochemistry 2008, 47, 308–319. [Google Scholar] [CrossRef]
- Ruskamo, S.; Chukhlieb, M.; Vahokoski, J.; Bhargav, S.P.; Liang, F.; Kursula, I.; Kursula, P. Juxtanodin is an intrinsically disordered F-actin-binding protein. Sci. Rep. 2012, 2, 899. [Google Scholar] [CrossRef] [Green Version]
- Brockschnieder, D.; Sabanay, H.; Riethmacher, D.; Peles, E. Ermin, a myelinating oligodendrocyte-specific protein that regulates cell morphology. J. Neurosci. 2006, 26, 757–762. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Cao, Q.; Guo, A.; Chu, H.; Chan, Y.; Buschdorf, J.; Low, B.; Ling, E.; Liang, F. Juxtanodin: An oligodendroglial protein that promotes cellular arborization and 2’,3’-cyclic nucleotide-3’-phosphodiesterase trafficking. Proc. Natl. Acad. Sci. USA 2005, 102, 11527–11532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Jia, L.; Lu, B.; Liu, B.; Wang, W.; Wang, F.; Yang, G.; Bu, X.; Yao, L.; Zhang, B. Human Ermin (hErmin), a new oligodendrocyte-specific cytoskeletal protein related to epileptic seizure. Brain Res. 2011, 1367, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Xia, W.; Tang, J.; Tang, B.L.; Liang, F. Dephosphorylation-dependent Inhibitory Activity of Juxtanodin on Filamentous Actin Disassembly. J. Biol. Chem. 2010, 285, 28838–28849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, F.; Hwang, J.H.; Tang, N.W.; Hunziker, W. Juxtanodin in retinal pigment epithelial cells: Expression and biological activities in regulating cell morphology and actin cytoskeleton organization. J. Comp. Neurol. 2018, 526, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Fuxreiter, M.; Simon, I.; Friedrich, P.; Tompa, P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 2004, 338, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, F.; Gathje, H.; Kelm, S.; Dietz, F. Evolution of sialic acid-binding proteins: Molecular cloning and expression of fish siglec-4. Glycobiology 2004, 14, 959–968. [Google Scholar] [CrossRef] [Green Version]
- Jahn, O.; Tenzer, S.; Werner, H.B. Myelin Proteomics: Molecular Anatomy of an Insulating Sheath. Mol. Neurobiol. 2009, 40, 55–72. [Google Scholar] [CrossRef] [Green Version]
- Miescher, G.; Lutzelschwab, R.; Erne, B.; Ferracin, F.; Huber, S.; Steck, A. Reciprocal expression of myelin-associated glycoprotein splice variants in the adult human peripheral and central nervous systems. Mol. Brain Res. 1997, 52, 299–306. [Google Scholar] [CrossRef]
- Butt, A.; Ibrahim, M.; Gregson, N.; Berry, M. Differential expression of the L- and S-isoforms of myelin associated glycoprotein (MAG) in oligodendrocyte unit phenotypes in the adult rat anterior medullary velum. J. Neurocytol. 1998, 27, 271–280. [Google Scholar] [CrossRef]
- Keita, M.; Magy, L.; Heape, A.; Richard, L.; Piaser, M.; Vallat, J. Immunocytological studies of L-MAG expression regulation during myelination of embryonic brain cell cocultures. Dev. Neurosci. 2002, 24, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, H.; Sato, S.; Fujita, N.; Inuzuka, T.; Nakano, R.; Miyatake, T. Immunohistochemical Localization of Myelin-Associated Glycoprotein Isoforms during the Development in the Mouse-Brain. Brain Res. 1991, 563, 288–292. [Google Scholar] [CrossRef]
- Fujita, N.; Kemper, A.; Dupree, J.; Nakayasu, H.; Bartsch, U.; Schachner, M.; Maeda, N.; Suzuki, K.; Suzuki, K.; Popko, B. The cytoplasmic domain of the large myelin-associated glycoprotein isoform is needed for proper CNS but not peripheral nervous system myelination. J. Neurosci. 1998, 18, 1970–1978. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Flueck, B.; Kern, F.; Erne, B.; Steck, A.; Schaeren-Wiemers, N. Unraveling the differential expression of the two isoforms of myelin-associated glycoprotein in a mouse expressing GFP-tagged S-MAG specifically regulated and targeted into the different myelin compartments. Mol. Cell. Neurosci. 2006, 31, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Myllykoski, M.; Eichel, M.A.; Jung, R.B.; Kelm, S.; Werner, H.B.; Kursula, P. High-affinity heterotetramer formation between the large myelin-associated glycoprotein and the dynein light chain DYNLL1. J. Neurochem. 2018, 147, 764–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pronker, M.F.; Lemstra, S.; Snijder, J.; Heck, A.J.R.; Thies-Weesie, D.M.E.; Pasterkamp, R.J.; Janssen, B.J.C. Structural basis of myelin-associated glycoprotein adhesion and signalling. Nat. Commun. 2016, 7, 13584. [Google Scholar] [CrossRef] [PubMed]
- Quarles, R.H. Myelin-associated glycoprotein (MAG): Past, present and beyond. J. Neurochem. 2007, 100, 1431–1448. [Google Scholar] [CrossRef]
- Kursula, P.; Lehto, V.; Heape, A. The small myelin-associated glycoprotein binds to tubulin and microtubules. Mol. Brain Res. 2001, 87, 22–30. [Google Scholar] [CrossRef]
- Yin, X.; Crawford, T.; Griffin, J.; Tu, P.; Lee, V.; Li, C.; Roder, J.; Trapp, B. Myelin-associated glycoprotein is a myelin signal that modulates the caliber of myelinated axons. J. Neurosci. 1998, 18, 1953–1962. [Google Scholar] [CrossRef] [Green Version]
- Dashiell, S.; Tanner, S.; Pant, H.; Quarles, R. Myelin-associated glycoprotein modulates expression and phosphorylation of neuronal cytoskeletal elements and their associated kinases. J. Neurochem. 2002, 81, 1263–1272. [Google Scholar] [CrossRef] [Green Version]
- Kursula, P.; Meriläinen, G.; Lehto, V.; Heape, A. The small myelin-associated glycoprotein is a zinc-binding protein. J. Neurochem. 1999, 73, 2110–2118. [Google Scholar] [PubMed]
- Jaramillo, M.L.; Afar, D.E.H.; Almazan, G.; Bell, J.C. Identification of Tyrosine-620 as the Major Phosphorylation Site of Myelin-Associated Glycoprotein and its Implication in Interacting with Signaling Molecules. J. Biol. Chem. 1994, 269, 27240–27245. [Google Scholar] [PubMed]
- Kursula, P.; Tikkanen, G.; Lehto, V.; Nishikimi, M.; Heape, A. Calcium-dependent interaction between the large myelin-associated glycoprotein and S100 beta. J. Neurochem. 1999, 73, 1724–1732. [Google Scholar] [CrossRef] [PubMed]
- Barbar, E. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry 2008, 47, 503–508. [Google Scholar] [CrossRef]
- Clark, S.; Nyarko, A.; Loehr, F.; Karplus, P.A.; Barbar, E. The Anchored Flexibility Model in LC8 Motif Recognition: Insights from the Chica Complex. Biochemistry 2016, 55, 199–209. [Google Scholar] [CrossRef]
- Dytrych, L.; Sherman, D.; Gillespie, C.; Brophy, P. Two PDZ domain proteins encoded by the murine periaxin gene are the result of alternative intron retention and are differentially targeted in Schwann cells. J. Biol. Chem. 1998, 273, 5794–5800. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, M.B. Origin of Pdz (Dhr, Glgf) Domains. Trends Biochem. Sci. 1995, 20, 350. [Google Scholar] [CrossRef]
- Han, H.; Kursula, P. Periaxin and AHNAK Nucleoprotein 2 Form Intertwined Homodimers through Domain Swapping. J. Biol. Chem. 2014, 289, 14121–14131. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Zheng, J.J. PDZ domains and their binding partners: Structure, specificity, and modification. Cell Commun. Signal. 2010, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Shi, Y. L-periaxin interacts with S-periaxin through its PDZ domain. Neurosci. Lett. 2015, 609, 23–29. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.; Appel, R.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Walker, J., Ed.; The Proteomics Protocols Handbook; Humana Press: New Jersey, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Dempsey, B.R.; Rezvanpour, A.; Lee, T.; Barber, K.R.; Junop, M.S.; Shaw, G.S. Structure of an Asymmetric Ternary Protein Complex Provides Insight for Membrane Interaction. Structure 2012, 20, 1737–1745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozorowski, G.; Milton, S.; Luecke, H. Structure of a C-terminal AHNAK peptide in a 1:2:2 complex with S100A10 and an acetylated N-terminal peptide of annexin A2. Acta Cryst. D 2013, 69, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Zhang, L.; Xiao, H.; Yang, Y.; Shi, Y. Ezrin interacts with L-periaxin by the “head to head and tail to tail” mode and influences the location of L-periaxin in Schwann cell RSC96. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129520. [Google Scholar] [CrossRef] [PubMed]
- Sherman, D.; Brophy, P. A tripartite nuclear localization signal in the PDZ-domain protein L-periaxin. J. Biol. Chem. 2000, 275, 4537–4540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Zhang, L.; Yang, T. Nuclear Export of L-Periaxin, Mediated by Its Nuclear Export Signal in the PDZ Domain. PLoS ONE 2014, 9, e91953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherman, D.; Fabrizi, C.; Gillespie, C.; Brophy, P. Specific disruption of a Schwann cell dystrophin-related protein complex in a demyelinating neuropathy. Neuron 2001, 30, 677–687. [Google Scholar] [CrossRef] [Green Version]
- Brennan, K.M.; Bai, Y.; Pisciotta, C.; Wang, S.; Feely, S.M.E.; Hoegger, M.; Gutmann, L.; Moore, S.A.; Gonzalez, M.; Sherman, D.L.; et al. Absence of Dystrophin Related Protein-2 disrupts Cajal bands in a patient with Charcot-Marie-Tooth disease. Neuromusc. Disord. 2015, 25, 786–793. [Google Scholar] [CrossRef] [Green Version]
- Kyte, J.; Doolittle, R.F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Forbes, J.G.; Jin, A.J.; Ma, K.; Gutierrez-Cruz, G.; Tsai, W.L.; Wang, K. Titin PEVK segment: Charge-driven elasticity of the open and flexible polyampholyte. J. Muscle Res. Cell. Motil. 2005, 26, 291–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huttlin, E.L.; Jedrychowski, M.P.; Elias, J.E.; Goswami, T.; Rad, R.; Beausoleil, S.A.; Villen, J.; Haas, W.; Sowa, M.E.; Gygi, S.P. A Tissue-Specific Atlas of Mouse Protein Phosphorylation and Expression. Cell 2010, 143, 1174–1189. [Google Scholar] [CrossRef] [Green Version]
- Gerke, V.; Moss, S. Annexins: From structure to function. Physiol. Rev. 2002, 82, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, A.; Nakashima, K.; Yamagishi, K.; Hoshi, T.; Suzuki, A.; Baba, H. Localization of annexin II in the paranodal regions and Schmidt-Lanterman incisures in the peripheral nervous system. Glia 2007, 55, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 Proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef] [Green Version]
- Raasakka, A.; Linxweiler, H.; Brophy, P.J.; Sherman, D.L.; Kursula, P. Direct binding of the flexible C-terminal segment of periaxin to β4 integrin suggests a molecular basis for CMT4F. Front. Mol. Neurosci. 2019, 12, 84. [Google Scholar] [CrossRef]
- Kim, S.; Maynard, J.C.; Sasaki, Y.; Strickland, A.; Sherman, D.L.; Brophy, P.J.; Burlingame, A.L.; Milbrandt, J. Schwann Cell O-GlcNAc Glycosylation Is Required for Myelin Maintenance and Axon Integrity. J. Neurosci. 2016, 36, 9633–9646. [Google Scholar] [CrossRef]
- Einheber, S.; Milner, T.A.; Giancotti, F.; Salzer, J.L. Axonal Regulation of Schwann-Cell Integrin Expression Suggests a Role for Alpha-6-Beta-4 in Myelination. J. Cell Biol. 1993, 123, 1223–1236. [Google Scholar] [CrossRef] [Green Version]
- Masaki, T.; Matsumura, K.; Hirata, A.; Yamada, H.; Hase, A.; Arai, K.; Shimizu, T.; Yorifuji, H.; Motoyoshi, K.; Kamakura, K. Expression of dystroglycan and the laminin-alpha 2 chain in the rat peripheral nerve during development. Exp. Neurol. 2002, 174, 109–117. [Google Scholar] [CrossRef]
- Wang, M.M.; Zhang, X.; Lee, S.J.; Maripudi, S.; Keep, R.F.; Johnson, A.M.; Stamatovic, S.M.; Andjelkovic, A.V. Expression of periaxin (PRX) specifically in the human cerebrovascular system: PDZ domain-mediated strengthening of endothelial barrier function. Sci. Rep. 2018, 8, 10042. [Google Scholar] [CrossRef] [PubMed]
- Pancsa, R.; Schad, E.; Tantos, A.; Tompa, P. Emergent functions of proteins in non-stoichiometric supramolecular assemblies. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 970–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksenberg, J.; Seboun, E.; Hauser, S. Genetics of demyelinating diseases. Brain Pathol. 1996, 6, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, K.S. Mitochondrial Dysfunction in Demyelinating Diseases. Semin. Pediatr. Neurol. 2013, 20, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Stone, S. Unfolded protein response in myelin disorders. Neural Regen. Res. 2020, 15, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Torkildsen, O.; Brunborg, L.A.; Myhr, K.M.; Bø, L. The cuprizone model for demyelination. Acta Neurol. Scand. 2008, 117, 72–76. [Google Scholar] [CrossRef]
- Virtanen, J.O.; Jacobson, S. Viruses and Multiple Sclerosis. CNS Neurol. Disord. Drug Targets 2012, 11, 528–544. [Google Scholar] [CrossRef] [Green Version]
- Browne, P.; Chandraratna, D.; Angood, C.; Tremlett, H.; Baker, C.; Taylor, B.V.; Thompson, A.J. Atlas of Multiple Sclerosis 2013: A growing global problem with widespread inequity. Neurology 2014, 98, 1022–1024. [Google Scholar] [CrossRef] [Green Version]
- Namer, I.J.; Steibel, J.; Poulet, P.; Armspach, J.P.; Mohr, M.; Mauss, Y.; Chambron, J. Blood-Brain-Barrier Breakdown in Mbp-Specific T-Cell Induced Experimental Allergic Encephalomyelitis–a Quantitative Invivo Mri Study. Brain 1993, 116, 147–159. [Google Scholar] [CrossRef]
- Galea, I.; Bernardes-Silva, M.; Forse, P.A.; van Rooijen, N.; Liblau, R.S.; Perry, V.H. An antigen-specific pathway for CD8 T cells across the blood-brain barrier. J. Exp. Med. 2007, 204, 2023–2030. [Google Scholar] [CrossRef] [Green Version]
- Luo, C.; Jian, C.; Liao, Y.; Huang, Q.; Wu, Y.; Liu, X.; Zou, D.; Wu, Y. The role of microglia in multiple sclerosis. Neuropsychiatr. Dis. Treat. 2017, 13, 1661–1667. [Google Scholar] [CrossRef] [Green Version]
- Muraro, P.; Kalbus, M.; Afshar, G.; McFarland, H.; Martin, R. T cell response to 2’,3’-cyclic nucleotide 3’-phosphodiesterase (CNPase) in multiple sclerosis patients. J. Neuroimmunol. 2002, 130, 233–242. [Google Scholar] [CrossRef]
- Andersson, M.; Yu, M.; Söderström, M.; Weerth, S.; Baig, S.; Linington, C.; Solders, G.; Link, H. Multiple MAG peptides are recognized by circulating T and B lymphocytes in polyneuropathy and multiple sclerosis. Eur. J. Neurol. 2002, 9, 243–251. [Google Scholar] [CrossRef]
- Tejada-Simon, M.; Zang, Y.; Hong, J.; Rivera, V.; Zhang, J. Cross-reactivity with myelin basic protein and human herpesvirus-6 in multiple sclerosis. Ann. Neurol. 2003, 53, 189–197. [Google Scholar] [CrossRef]
- Musse, A.; Boggs, J.; Harauz, G. Deimination of membrane-bound myelin basic protein in multiple sclerosis exposes an immunodominant epitope. Proc. Natl. Acad. Sci. USA 2006, 103, 4422–4427. [Google Scholar] [CrossRef] [Green Version]
- Holz, A.; Bielekova, B.; Martin, R.; Oldstone, M. Myelin-associated oligodendrocytic basic protein: Identification of an encephalitogenic epitope and association with multiple sclerosis. J. Immunol. 2000, 164, 1103–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaushansky, N.; Eisenstein, M.; Zilkha-Falb, R.; Ben-Nun, A. The myelin-associated oligodendrocytic basic protein (MOBP) as a relevant primary target autoantigen in multiple sclerosis. Autoimmun. Rev. 2010, 9, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Yamamura, T.; Inobe, J.; Ohashi, T.; Takahashi, K.; Tabira, T. TCR repertoire to proteolipid protein (PLP) in multiple sclerosis (MS): Homologies between PLP-specific T cells and MS-associated T cells in TCR junctional sequences. Int. Immunol. 1996, 8, 123–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wucherpfennig, K.; Strominger, J. Molecular Mimicry in T-Cell-Mediated Autoimmunity–Viral Peptides Activate Human T-Cell Clones Specific for Myelin Basic-Protein. Cell 1995, 80, 695–705. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.; Lu, C.; Wang, X.; Xiao, B. Induction of experimental autoimmune encephalomyelitis in Lewis rats by a viral peptide with limited homology to myelin basic protein. Exp. Neurol. 2007, 206, 231–239. [Google Scholar] [CrossRef]
- Wucherpfennig, K.W.; Sette, A.; Southwood, S.; Oseroff, C.; Matsui, M.; Strominger, J.L.; Hafler, D.A. Structural Requirements for Binding of an Immunodominant Myelin Basic-Protein Peptide to Dr2 Isotypes and for its Recognition by Human T-Cell Clones. J. Exp. Med. 1994, 179, 279–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, K.G.; Catz, I.; Steinman, L. Fine Specificity of the Antibody-Response to Myelin Basic-Protein in the Central-Nervous-System in Multiple-Sclerosis–the Minimal B-Cell Epitope and a Model of its Features. Proc. Natl. Acad. Sci. USA 1995, 92, 11061–11065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, K.; Pyrdol, J.; Gauthier, L.; Wiley, D.; Wucherpfennig, K. Crystal structure of HLA-DR2 (DRA*0101, DRB1*1501) complexed with a peptide from human myelin basic protein. J. Exp. Med. 1998, 188, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Huang, Y.P.; Lue, J.; Quandt, J.A.; Martin, R.; Mariuzza, R.A. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005, 24, 2968–2979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfister, H.W.; Einhaupl, K.M.; Wick, M.; Fatehmoghadam, A.; Huber, M.; Schielke, E.; Goebel, F.D.; Matuschke, A.; Heinrich, B.; Bogner, J.R.; et al. Myelin Basic-Protein in the Cerebrospinal-Fluid of Patients Infected with Hiv. J. Neurol. 1989, 236, 288–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, L.; Inman, R. Mechanisms of disease: Molecular mimicry and autoimmunity. N. Engl. J. Med. 1999, 341, 2068–2074. [Google Scholar] [CrossRef]
- D’Souza, C.; Wood, D.; She, Y.; Moscarello, M. Autocatalytic cleavage of myelin basic protein: An alternative to molecular mimicry. Biochemistry 2005, 44, 12905–12913. [Google Scholar] [CrossRef]
- Pritzker, L.; Joshi, S.; Gowan, J.; Harauz, G.; Moscarello, M. Deimination of myelin basic protein. 1. Effect of deimination of arginyl residues of myelin basic protein on its structure and susceptibility to digestion by cathepsin D. Biochemistry 2000, 39, 5374–5381. [Google Scholar] [CrossRef]
- Medveczky, P.; Antal, J.; Patthy, A.; Kekesi, K.; Juhasz, G.; Szilagyi, L.; Graf, L. Myelin basic protein, an autoantigen in multiple sclerosis, is selectively processed by human trypsin 4. FEBS Lett. 2006, 580, 545–552. [Google Scholar] [CrossRef] [Green Version]
- D’Souza, C.A.; Moscarello, M.A. Differences in susceptibility of MBP charge isomers to digestion by stromelysin-1 (MMP-3) and release of an immunodominant epitope. Neurochem. Res. 2006, 31, 1045–1054. [Google Scholar] [CrossRef]
- Williams, K.R.; Williams, N.D.; Konigsberg, W.; Yu, R.K. Acidic Lipids Enhance Cathepsin-D Cleavage of the Myelin Basic-Protein. J. Neurosci. Res. 1986, 15, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Shaharabani, R.; Ram-On, M.; Avinery, R.; Aharoni, R.; Arnon, R.; Talmon, Y.; Beck, R. Structural transition in myelin membrane as initiator of multiple sclerosis. J. Am. Chem. Soc. 2016, 138, 12159–12165. [Google Scholar] [CrossRef] [PubMed]
- Shaharabani, R.; Ram-On, M.; Talmon, Y.; Beck, R. Pathological transitions in myelin membranes driven by environmental and multiple sclerosis conditions. Proc. Natl. Acad. Sci. USA 2018, 115, 11156–11161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frankenhaeuser, B. The effect of calcium on the myelinated nerve fibre. J. Physiol. 1957, 137, 245–260. [Google Scholar] [CrossRef]
- Haak, L.L.; Grimaldi, M.; Russell, J.T. Mitochondria in myelinating cells: Calcium signaling in oligodendrocyte precursor cells. Cell Calcium 2000, 28, 297–306. [Google Scholar] [CrossRef]
- Cheli, V.T.; Santiago González, D.A.; Namgyal Lama, T.; Spreuer, V.; Handley, V.; Murphy, G.G.; Paez, P.M. Conditional Deletion of the L-Type Calcium Channel Cav1.2 in Oligodendrocyte Progenitor Cells Affects Postnatal Myelination in Mice. J. Neurosci. 2016, 36, 10853–10869. [Google Scholar] [CrossRef] [Green Version]
- Krasnow, A.M.; Ford, M.C.; Valdivia, L.E.; Wilson, S.W.; Attwell, D. Regulation of developing myelin sheath elongation by oligodendrocyte calcium transients in vivo. Nat. Neurosci. 2018, 21, 24–28. [Google Scholar] [CrossRef]
- Miller, R.H. Calcium control of myelin sheath growth. Nat. Neurosci. 2018, 21, 2–3. [Google Scholar] [CrossRef]
- Friess, M.; Hammann, J.; Unichenko, P.; Luhmann, H.J.; White, R.; Kirischuk, S. Intracellular ion signaling influences myelin basic protein synthesis in oligodendrocyte precursor cells. Cell Calcium 2016, 60, 322–330. [Google Scholar] [CrossRef]
- Paez, P.M.; Spreuer, V.; Handley, V.; Feng, J.M.; Campagnoni, C.; Campagnoni, A.T. Increased expression of golli myelin basic proteins enhances calcium influx into oligodendroglial cells. J. Neurosci. 2007, 27, 12690–12699. [Google Scholar] [CrossRef] [Green Version]
- Tsang, D.; Tsang, Y.S.; Ho, W.K.; Wong, R.N. Myelin Basic Protein Is a Zinc-Binding Protein in Brain: Possible Role in Myelin Compaction. Neurochem. Res. 1997, 22, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Riccio, P.; Giovannelli, S.; Bobba, A.; Romito, E.; Fasano, A.; Bleve-Zacheo, T.; Favilla, R.; Quagliariello, E.; Cavatorta, P. Specificity of zinc binding to myelin basic protein. Neurochem. Res. 1995, 20, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Weil, M.; Möbius, W.; Winkler, A.; Ruhwedel, T.; Wrzos, C.; Romanelli, E.; Bennett, J.L.; Enz, L.; Goebels, N.; Nave, K.; et al. Loss of Myelin Basic Protein Function Triggers Myelin Breakdown in Models of Demyelinating Diseases. Cell Rep. 2016, 16, 314–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramchandren, S. Charcot-Marie-Tooth Disease and Other Genetic Polyneuropathies. Contin. (Minneap. Minn.) 2017, 23, 1360–1377. [Google Scholar] [CrossRef]
- Roda, R.H.; McCray, B.A.; Klein, C.J.; Hoke, A. Novel hemizygous nonsense mutation in DRP2 is associated with inherited neuropathy. Neurol. Genet. 2018, 4, e220. [Google Scholar] [CrossRef] [Green Version]
- Datta, S.; Kataria, S.; Govindarajan, R. A Case Report on Charcot-Marie-Tooth Disease with a Novel Periaxin Gene Mutation. Cureus 2019, 11, e5111. [Google Scholar] [CrossRef] [Green Version]
- Takashima, H.; Boerkoel, C.; De Jonghe, P.; Ceuterick, C.; Martin, J.; Voit, T.; Schroder, J.; Williams, A.; Brophy, P.; Timmerman, V.; et al. Periaxin mutations cause a broad spectrum of demyelinating neuropathies. Ann. Neurol. 2002, 51, 709–715. [Google Scholar] [CrossRef]
- Otagiri, T.; Sugai, K.; Kijima, K.; Arai, H.; Sawaishi, Y.; Shimohata, M.; Hayasaka, K. Periaxin mutation in Japanese patients with Charcot-Marie-Tooth disease. J. Hum. Genet. 2006, 51, 625–628. [Google Scholar] [CrossRef] [Green Version]
- Guilbot, A.; Williams, A.; Ravise, N.; Verny, C.; Brice, A.; Sherman, D.; Brophy, P.; LeGuern, E.; Delague, V.; Bareil, C.; et al. A mutation in periaxin is responsible for CMT4F, an autosomal recessive form of Charcot-Marie-Tooth disease. Hum. Mol. Genet. 2001, 10, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Marchesi, C.; Milani, M.; Morbin, M.; Cesani, M.; Lauria, G.; Scaioli, V.; Piccolo, G.; Fabrizi, G.M.; Cavallaro, T.; Taroni, F.; et al. Four novel cases of periaxin-related neuropathy and review of the literature. Neurology 2010, 75, 1830–1838. [Google Scholar] [CrossRef]
- Nouioua, S.; Hamadouche, T.; Funalot, B.; Bernard, R.; Bellatache, N.; Bouderba, R.; Grid, D.; Assami, S.; Benhassine, T.; Levy, N.; et al. Novel mutations in the PRX and the MTMR2 genes are responsible for unusual Charcot-Marie-Tooth disease phenotypes. Neuromusc. Disord. 2011, 21, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Boerkoel, C.; Takashima, H.; Stankiewicz, P.; Garcia, C.; Leber, S.; Rhee-Morris, L.; Lupski, J. Periaxin mutations cause recessive Dejerine-Sottas neuropathy. Am. J. Hum. Genet. 2001, 68, 325–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.J.; Hyun, Y.S.; Nam, S.H.; Koo, H.; Bin Hong, Y.; Chung, K.W.; Choi, B. Novel Compound Heterozygous Nonsense PRX Mutations in a Korean Dejerine-Sottas Neuropathy Family. J. Clin. Neurol. 2015, 11, 92–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabzinska, D.; Drac, H.; Sherman, D.; Kostera-Pruszczyk, A.; Brophy, P.; Kochanski, A.; Hausmanowa-Petrusewicz, I. Charcot-Marie-Tooth type 4F disease caused by S399fsx410 mutation in the PRX gene. Neurology 2006, 66, 745–747. [Google Scholar] [CrossRef]
- Schabhüttl, M.; Wieland, T.; Senderek, J.; Baets, J.; Timmerman, V.; De Jonghe, P.; Reilly, M.M.; Stieglbauer, K.; Laich, E.; Windhager, R.; et al. Whole-exome sequencing in patients with inherited neuropathies: Outcome and challenges. J. Neurol. 2014, 261, 970–982. [Google Scholar] [CrossRef]
- Tokunaga, S.; Hashiguchi, A.; Yoshimura, A.; Maeda, K.; Suzuki, T.; Haruki, H.; Nakamura, T.; Okamoto, Y.; Takashima, H. Late-onset Charcot-Marie-Tooth disease 4F caused by periaxin gene mutation. Neurogenetics 2012, 13, 359–365. [Google Scholar] [CrossRef]
- Auer-Grumbach, M.; Fischer, C.; Papic, L.; John, E.; Plecko, B.; Bittner, R.E.; Bernert, G.; Pieber, T.R.; Miltenberger, G.; Schwarz, R.; et al. Two novel mutations in the GDAP and PRX genes in early onset Charcot-Marie-Tooth syndrome. Neuropediatrics 2008, 39, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Nagase, T.; Kikuno, R.; Nakayama, M.; Hirosawa, M.; Ohara, O. Prediction of the coding sequences of unidentified human genes. XVIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000, 7, 273–281. [Google Scholar] [CrossRef]
- Kijima, K.; Numakura, C.; Shirahata, E.; Sawaishi, Y.; Shimohata, M.; Igarashi, S.; Tanaka, T.; Hayasaka, K. Periaxin mutation causes early-onset but slow-progressive Charcot-Marie-Tooth disease. J. Hum. Genet. 2004, 49, 376–379. [Google Scholar] [CrossRef]
- Parman, Y.; Battaloglu, E.; Baris, I.; Bilir, B.; Poyraz, M.; Bissar-Tadmouri, N.; Williams, A.; Ammar, N.; Nelis, E.; Timmerman, V.; et al. Clinicopathological and genetic study of early-onset demyelinating neuropathy. Brain 2004, 127, 2540–2550. [Google Scholar] [CrossRef] [Green Version]
- Sherman, D.L.; Brophy, P.J. A murine model of Charcot-Marie-Tooth disease 4F reveals a role for the C-terminus of periaxin in the formation and stabilization of Cajal bands. Wellcome Open Res. 2018, 3, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renouil, M.; Stojkovic, T.; Jacquemont, M.L.; Lauret, K.; Boue, P.; Fourmaintraux, A.; Randrianaivo, H.; Tallot, M.; Mignard, D.; Roelens, P.; et al. Charcot-Marie-Tooth disease associated with periaxin mutations (CMT4F): Clinical, electrophysiological and genetic analysis of 24 patients. Rev. Neurol. 2013, 169, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Barankova, L.; Siskova, D.; Hühne, K.; Vyhnalkova, E.; Sakmaryova, I.; Bojar, M.; Rautenstrauss, B.; Seeman, P. A 71-nucleotide deletion in the periaxin gene in a Romani patient with early-onset slowly progressive demyelinating CMT. Eur. J. Neurol. 2008, 15, 548–551. [Google Scholar] [CrossRef]
- Konrat, R. NMR contributions to structural dynamics studies of intrinsically disordered proteins. J. Magn. Reson. 2014, 241, 74–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallace, B.A. The role of circular dichroism spectroscopy in the era of integrative structural biology. Curr. Opin. Struct. Biol. 2019, 58, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Kikhney, A.G.; Svergun, D.I. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015, 589, 2570–2577. [Google Scholar] [CrossRef] [Green Version]
- Schuler, B.; Muller-Spath, S.; Soranno, A.; Nettels, D. Application of confocal single-molecule FRET to intrinsically disordered proteins. Methods Mol. Biol. 2012, 896, 21–45. [Google Scholar]
- Baul, U.; Chakraborty, D.; Mugnai, M.L.; Straub, J.E.; Thirumalai, D. Sequence Effects on Size, Shape, and Structural Heterogeneity in Intrinsically Disordered Proteins. J. Phys. Chem. B 2019, 123, 3462–3474. [Google Scholar] [CrossRef]
- Ghosh, I.; Considine, N.; Maunus, E.; Sun, L.; Zhang, A.; Buswell, J.; Evans, T.C., Jr.; Xu, M. Site-Specific Protein Labeling by Intein-Mediated Protein Ligation. Methods Mol. Biol. 2011, 705, 87–107. [Google Scholar]
- Stevens, A.J.; Sekar, G.; Shah, N.H.; Mostafavi, A.Z.; Cowburn, D.; Muir, T.W. A promiscuous split intein with expanded protein engineering applications. Proc. Natl. Acad. Sci. USA 2017, 114, 8538–8543. [Google Scholar] [CrossRef] [Green Version]
- Binder, H.; Arnold, K.; Ulrich, A.S.; Zschornig, O. Interaction of Zn2+ with phospholipid membranes. Biophys. Chem. 2001, 90, 57–74. [Google Scholar] [CrossRef]
- Gillooly, D.; Simonsen, A.; Stenmark, H. Cellular functions of phosphatidylinositol 3-phosphate and FYVE domain proteins. Biochem. J. 2001, 355, 249–258. [Google Scholar] [CrossRef] [PubMed]









| Mutation 1 | Neuropathy | (Potential) Molecular Impact | Reference(s) |
|---|---|---|---|
| R82fs | DSS | Tail loss; loss of interactions | [258] |
| L132fs | CMT4F | Tail loss; loss of interactions | [259] |
| R196stop | CMT4F | [260] | |
| S259fs | CMT4F | Loss of hydrophobic, PEVK-rich, AHNAK2 homology and acidic regions; loss of interactions | [261] |
| R364stop | CMT4F | Loss of PEVK-rich, AHNAK2 homology and acidic regions; loss of interactions | [262] |
| R368stop | DSS | Loss of PEVK-rich, AHNAK2 homology and acidic regions; loss of interactions | [263] |
| R392stop | DSS | Loss of PEVK-rich, AHNAK2 homology and acidic regions; loss of interactions | [264] |
| S399fs | CMT4F | Loss of PEVK-rich, AHNAK2 homology and acidic regions; loss of interactions | [265] |
| A406T | DSS | [263] | |
| E495Q | DSS | [263] | |
| V525A | CMT4F | [260,266] | |
| Q547stop | CMT4F | Loss of PEVK-rich (partial), AHNAK2 homology and acidic regions; loss of interactions | [261] |
| D651N | CMT4F | [267] | |
| R679stop | DSS | Loss of PEVK-rich (partial), AHNAK2 homology and acidic regions; loss of interactions | [264] |
| E682stop | CMT4F | Loss of PEVK-rich (partial), AHNAK2 homology and acidic regions; loss of interactions | [261] |
| A700fs | CMT4F | [268] | |
| C715stop | DSS | Loss of PEVK-rich (partial), AHNAK2 homology and acidic regions; loss of interactions | [258] |
| V763fs | DSS | Loss of PEVK-rich (partial), AHNAK2 homology and acidic regions; loss of interactions | [263] |
| K808fs | CMT4F | Loss of AHNAK2 homology and acidic regions; loss of interactions | [261] |
| V882A | DSS | [263,269] | |
| I921M | DSS | [263] | |
| S929fs | DSS | Loss of AHNAK2 homology and acidic regions; loss of interactions | [263] |
| K935E | DSS | [263] | |
| K935stop | DSS | Loss of acidic domain; loss of integrin interaction | [263] |
| K1062N | CMT4F | (Loss of predicted AnxA2/S100A10 interaction?) | [257] |
| R1070stop | CMT4F | Loss of acidic domain; loss of integrin interaction | [208,259,267,270,271,272] |
| P1083R | DSS | [265] | |
| E1085fs | CMT4F | Loss of acidic domain; loss of integrin interaction | [273] |
| K1095fs | CMT4F | Loss of acidic domain; loss of integrin interaction | [274] |
| G1132R | DSS | [263] | |
| E1259K | DSS | [263] | |
| R1335Q 2 | CMT | [266] | |
| E1359del | DSS | [263] | |
| R1411C | DSS | [263] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raasakka, A.; Kursula, P. Flexible Players within the Sheaths: The Intrinsically Disordered Proteins of Myelin in Health and Disease. Cells 2020, 9, 470. https://doi.org/10.3390/cells9020470
Raasakka A, Kursula P. Flexible Players within the Sheaths: The Intrinsically Disordered Proteins of Myelin in Health and Disease. Cells. 2020; 9(2):470. https://doi.org/10.3390/cells9020470
Chicago/Turabian StyleRaasakka, Arne, and Petri Kursula. 2020. "Flexible Players within the Sheaths: The Intrinsically Disordered Proteins of Myelin in Health and Disease" Cells 9, no. 2: 470. https://doi.org/10.3390/cells9020470
APA StyleRaasakka, A., & Kursula, P. (2020). Flexible Players within the Sheaths: The Intrinsically Disordered Proteins of Myelin in Health and Disease. Cells, 9(2), 470. https://doi.org/10.3390/cells9020470






