Transcriptional Basis for Differential Thermosensitivity of Seedlings of Various Tomato Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth and Treatment
2.2. RNA Extraction and NGS Analysis
2.3. Quantitative RT-PCR
2.4. Read Alignment and Analysis of MACE Sequencing
2.5. Principal Component Analysis (PCA)
2.6. Classification of the Genes
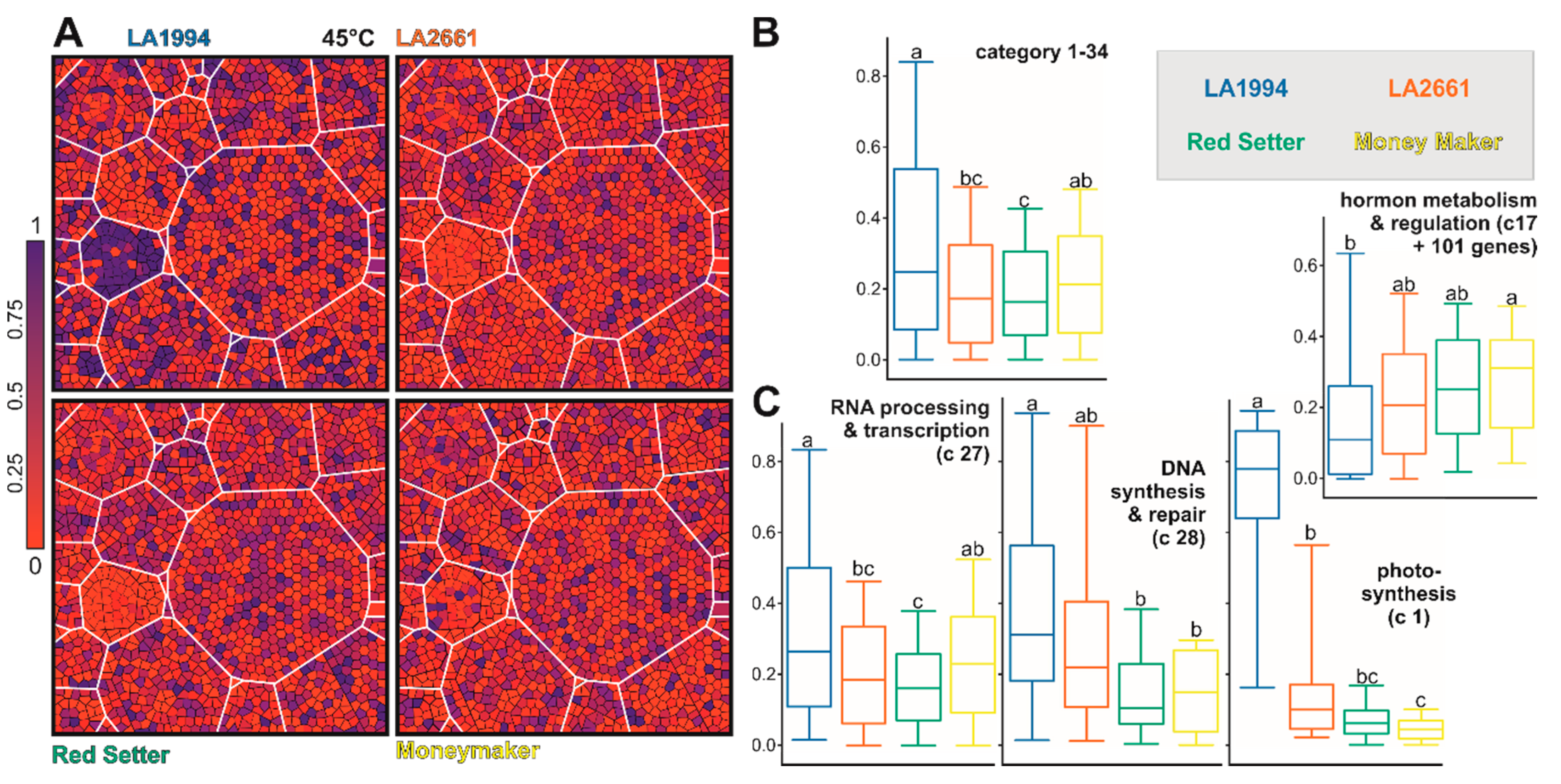
2.7. Functional Assignment and Voronoi Treemap Representation
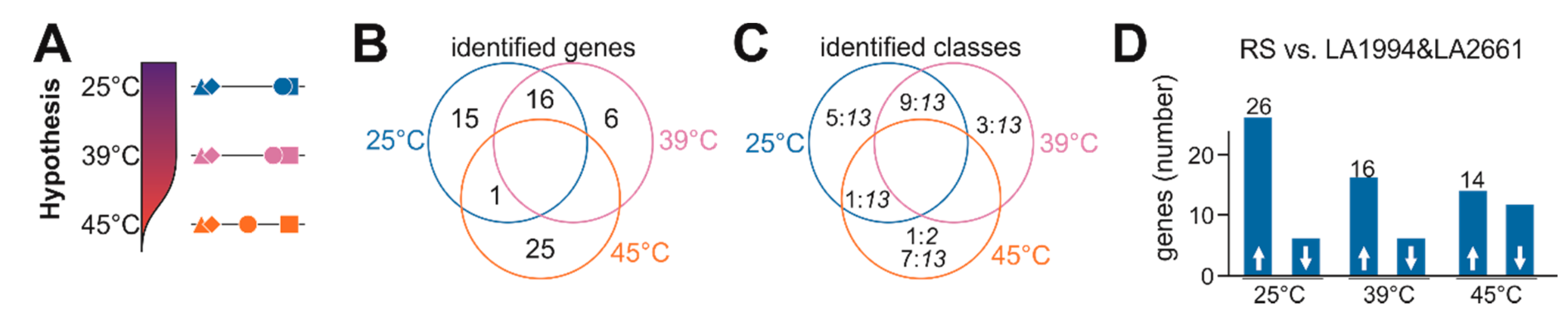
2.8. Hypothesis-Driven Gene Selection
- Genes were selected in case a significant change in transcript abundance (p < 0.05) was observed for Red Setter at 25 °C, after 39 °C or after 45 °C HS treatment when compared to solely LA1994 and LA2661 at the according condition. Further, the change of transcript abundance between Red Setter and each of the other two genotypes, LA1994 and LA2661, had to be either positive or negative for both.
- Genes selected in (i) were further considered in case a difference of transcript abundance in Moneymaker (p < 0.05) at 25 °C or after 39 °C treatment was observed when compared to solely LA1994 and LA2661 at the according condition. Further, the change of transcript abundance between Moneymaker, LA1994 and LA2661 had to be positive or negative for both, and similar to the change found for Red Setter when compared to LA1994 and LA2661. These genes were selected as genes contributing to the difference between the tolerant and sensitive lines at 25 °C and 39 °C.
- Genes selected in (i) and fulfilling the following rule:log2(TPM45 °C − TPM25 °C)Red Setter > 0 and log2(TPM45 °C-TPM25 °C)Red Setter − log2(TPM45 °C − TPM25 °C)Moneymaker > 1orlog2(TPM45 °C − TPM25 °C)Red Setter < 0 and log2(TPM45 °C − TPM25 °C)Moneymaker − log2(TPM45 °C − TPM25 °C)Red Setter > 1were selected as genes contributing to the difference between tolerant and sensitive lines at 45 °C.
3. Results
3.1. Differences of Genotypes in Seedling Thermotolerance
3.2. Common Transcriptome Responses of Tomato Genotypes to Elevated Temperatures
3.3. Variation in the Transcriptome Profile of Tomato Genotypes at Different Temperatures
3.4. Specific Transcripts Involved in Cultivar-Specific Heat Stress Responses
4. Discussion
4.1. The Global Response of Tomato Seedlings to Elevated Temperatures and Its Variability
4.2. Genotype-Specific Transcriptome Responses to Elevated Temperatures
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bokszczanin, K.L.; Fragkostefanakis, S. Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front. Plant Sci. 2013, 4, 315. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Salaria, N.; Thakur, K.; Kukreja, S.; Gautam, S.; Goutam, U. Functional genomic approaches to improve crop plant heat stress tolerance [version 1; peer review: 2 approved]. F1000Research 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Lawas, L.M.F.; Zuther, E.; Jagadish, S.K.; Hincha, D.K. Molecular mechanisms of combined heat and drought stress resilience in cereals. Curr. Opin. Plant Biol. 2018, 45, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Driedonks, N.; Xu, J.; Peters, J.L.; Park, S.; Rieu, I. Multi-level interactions between heat shock factors, heat shock proteins, and the redox system regulate acclimation to heat. Front. Plant Sci. 2015, 6, 999. [Google Scholar] [CrossRef] [Green Version]
- Guy, C.; Kaplan, F.; Kopka, J.; Selbig, J.; Hincha, D.K. Metabolomics of temperature stress. Physiol. Plant. 2008, 132, 220–235. [Google Scholar] [CrossRef]
- Dobra, J.; Motyka, V.; Dobrev, P.; Malbeck, J.; Prasil, I.T.; Haisel, D.; Gaudinova, A.; Havlova, M.; Gubis, J.; Vankova, R. Comparison of hormonal responses to heat, drought and combined stress in tobacco plants with elevated proline content. J. Plant Physiol. 2010, 167, 1360–1370. [Google Scholar] [CrossRef]
- Talanova, V.V.; Akimova, T.V.; Titov, A.F. Effect of Whole Plant and Local Heating on the ABA Content in Cucumber Seedling Leaves and Roots and on Their Heat Tolerance. Russ. J. Plant Physiol. 2003, 50, 90–94. [Google Scholar] [CrossRef]
- Dobrá, J.; Černý, M.; Štorchová, H.; Dobrev, P.; Skalák, J.; Jedelský, P.L.; Lukšanová, H.; Gaudinová, A.; Pešek, B.; Malbecka, J.; et al. The impact of heat stress targeting on the hormonal andtranscriptomic response in Arabidopsis. Plant Sci. 2015, 231, 52–61. [Google Scholar] [CrossRef]
- Ozga, J.A.; Kaur, H.; Savada, R.P.; Reinecke, D.M. Hormonal regulation of reproductive growth under normal and heat-stress conditions in legume and other model crop species. J. Exp. Bot. 2017, 68, 1885–1894. [Google Scholar] [CrossRef]
- Yeh, C.H.; Kaplinsky, N.J.; Hu, C.; Charng, Y.Y. Some like it hot, some like it warm: Phenotyping to explore thermotolerance diversity. Plant Sci. 2012, 195, 10–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Queitsch, C.; Hong, S.W.; Vierling, E.; Lindquist, S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 2000, 12, 479–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharf, K.D.; Nover, L. Heat-shock-induced alterations of ribosomal protein phosphorylation in plant cell cultures. Cell 1982, 30, 427–437. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Mesihovic, A.; Simm, S.; Paupière, M.J.; Hu, Y.; Paul, P.; Mishra, S.K.; Tschiersch, B.; Theres, K.; Bovy, A.; et al. HsfA2 Controls the Activity of Developmentally and Stress-Regulated Heat Stress Protection Mechanisms in Tomato Male Reproductive Tissues. Plant Physiol. 2016, 170, 2461–2477. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient Gene Editing in Tomato in the First Generation Using the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated9 System. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Wang, S.; Huang, Z.; Zhang, S.; Liao, Q.; Zhang, C.; Lin, T.; Qin, M.; Peng, M.; Yang, C.; et al. Rewiring of the Fruit Metabolome in Tomato Breeding. Cell 2018, 172, 249–261. [Google Scholar] [CrossRef] [Green Version]
- Keller, M.; Consortium, S.; Simm, S. The coupling of transcriptome and proteome adaptation during development and heat stress response of tomato pollen. BMC Genom. 2018, 19, 447. [Google Scholar] [CrossRef]
- Liu, H.-C.; Charng, Y.-Y. Common and Distinct Functions of Arabidopsis Class A1 and A2 Heat Shock Factors in Diverse Abiotic Stress Responses and Development. Plant Physiol. 2013, 163, 276–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharf, K.-D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Vierling, E. The Roles of Heat Shock Proteins in Plants. Annu. Rev. Plant Biol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Müller, F.; Xu, J.; Kristensen, L.; Wolters-Arts, M.; De Groot, P.F.M.; Jansma, S.Y.; Mariani, C.; Park, S.; Rieu, I. High-temperature-induced defects in tomato (Solanum lycopersicum) anther and pollen development are associated with reduced expression of B-class floral patterning genes. PLoS ONE 2016, 11, e0167614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Firon, N.; Shaked, R.; Peet, M.M.; Pharr, D.M.; Zamski, E.; Rosenfeld, K.; Althan, L.; Pressman, E. Pollen grains of heat tolerant tomato cultivars retain higher carbohydrate concentration under heat stress conditions. Sci. Hortic. 2006, 109, 212–217. [Google Scholar] [CrossRef]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.-D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Mesihovic, A.; Jiménez-Gómez, J.M.; Röth, S.; Gebhardt, P.; Bublak, D.; Bovy, A.; Scharf, K.D.; Schleiff, E.; Fragkostefanakis, S. Natural variation in HsfA2 pre-mRNA splicing is associated with changes in thermotolerance during tomato domestication. New Phytol. 2020, 225, 1297–1310. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [Green Version]
- Bashir, K.; Matsui, A.; Rasheed, S.; Seki, M. Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress. F1000Research 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with imageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fragkostefanakis, S.; Simm, S.; Paul, P.; Bublak, D.; Scharf, K.D.; Schleiff, E. Chaperone network composition in Solanum lycopersicum explored by transcriptome profiling and microarray meta-analysis. Plant Cell Environ. 2015, 38, 693–709. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)-from genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Rescheneder, P.; Von Haeseler, A. NextGenMap: Fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 2013, 29, 2790–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Zhou, R.; Yu, X.; Kjær, K.H.; Rosenqvist, E.; Ottosen, C.O.; Wu, Z. Screening and validation of tomato genotypes under heat stress using Fv/Fm to reveal the physiological mechanism of heat tolerance. Environ. Exp. Bot. 2015, 118, 1–11. [Google Scholar] [CrossRef]
- Xu, J.; Driedonks, N.; Rutten, M.J.M.; Vriezen, W.H.; de Boer, G.-J.; Rieu, I. Mapping quantitative trait loci for heat tolerance of reproductive traits in tomato (Solanum lycopersicum). Mol. Breed. 2017, 37, 58. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Simm, S.; El-Shershaby, A.; Hu, Y.; Bublak, D.; Mesihovic, A.; Darm, K.; Mishra, S.K.; Tschiersch, B.; Theres, K.; et al. The repressor and co-activator HsfB1 regulates the major heat stress transcription factors in tomato. Plant Cell Environ. 2019, 42, 874–890. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K.-D. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef] [Green Version]
- Minoia, S.; Cellini, F.; Bendahmane, A.; D’Onofrio, O.; Petrozza, A.; Carriero, F.; Piron, F.; Mosca, G.; Sozio, G. A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Notes 2010, 3, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roden, J.C.; King, B.W.; Trout, D.; Mortazavi, A.; Wold, B.J.; Hart, C.E. Mining gene expression data by interpreting principal components. BMC Bioinform. 2006, 7, 194. [Google Scholar] [CrossRef] [Green Version]
- Brown, B.C.; Bray, N.L.; Pachter, L. Expression reflects population structure. PLoS Genet. 2018, 14, e1007841. [Google Scholar] [CrossRef] [Green Version]
- Lenz, M.; Muller, F.J.; Zenke, M.; Schuppert, A. Principal components analysis and the reported low intrinsic dimensionality of gene expression microarray data. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schramm, F.; Ganguli, A.; Kiehlmann, E.; Englich, G.; Walch, D.; Von Koskull-Döring, P. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 2006, 60, 759–772. [Google Scholar] [CrossRef]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Hartl, F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.M.; Kim, D.W.; Woo, M.S.; Jeong, H.S.; Son, Y.S.; Akhter, S.; Choi, G.J.; Bahk, J.D. Functional characterization of ArabidopsisHsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions. Plant Cell Environ. 2014, 37, 1202–1222. [Google Scholar] [CrossRef]
- Kolmos, E.; Chow, B.Y.; Pruneda-Paz, J.L.; Kay, S.A. HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc. Natl. Acad. Sci. USA 2014, 111, 16172–16177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Guzmán, M.; Apostolova, N.; Bellés, J.M.; Barrero, J.M.; Piqueras, P.; Ponce, M.R.; Micol, J.L.; Serrano, R.; Rodríguez, P.L. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 2002, 14, 1833–1846. [Google Scholar] [CrossRef] [Green Version]
- Larkindale, J.; Hall, J.D.; Knight, M.R.; Vierling, E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005, 138, 882–897. [Google Scholar] [CrossRef] [Green Version]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Varaud, E.; Brioudes, F.; Szécsi, J.; Leroux, J.; Brown, S.; Perrot-Rechenmann, C.; Bendahmane, M. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 2011, 23, 973–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadakuduti, S.S.; Pollard, M.; Kosma, D.K.; Allen, C.; Ohlrogge, J.B.; Barry, C.S. Pleiotropic phenotypes of the sticky peel mutant provide new insight into the role of CUTIN DEFICIENT2 in epidermal cell function in tomato. Plant Physiol. 2012, 159, 945–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.; Wang, S.; Kim, J.; Yan, J.; Yan, X.; Pang, Q.; Hua, J. Nuclear Pore Complex Components Have Temperature-Influenced Roles in Plant Growth and Immunity. Plant. Cell Environ. 2020, 43, 1452–1466. [Google Scholar] [CrossRef] [PubMed]
- Andolfo, G.; Ferriello, F.; Tardella, L.; Ferrarini, A.; Sigillo, L.; Frusciante, L.; Ercolano, M.R. Tomato genome-wide transcriptional responses to Fusarium wilt and Tomato Mosaic Virus. PLoS ONE 2014, 9, e94963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Q.; Lin, J.; Liu, J.Z.; Wang, X.; Lim, W.; Oh, M.; Park, J.; Rajashekar, C.B.; Whitham, S.A.; Cheng, N.H.; et al. Ectopic expression of Arabidopsis glutaredoxin AtGRXS17 enhances thermotolerance in tomato. Plant Biotechnol. J. 2012, 10, 945–955. [Google Scholar] [CrossRef]
- Wu, Q.; Hu, Y.; Sprague, S.A.; Kakeshpour, T.; Park, J.; Nakata, P.A.; Cheng, N.; Hirschi, K.D.; White, F.F.; Park, S. Expression of a monothiol glutaredoxin, AtGRXS17, in tomato (Solanum lycopersicum) enhances drought tolerance. Biochem. Biophys. Res. Commun. 2017, 491, 1034–1039. [Google Scholar] [CrossRef]
- Kampranis, S.C.; Damianova, R.; Atallah, M.; Toby, G.; Kondi, G.; Tsichlis, P.N.; Makris, A.M. A novel plant glutathione S-transferase/peroxidase suppresses Bax lethality in yeast. J. Biol. Chem. 2000, 275, 29207–29216. [Google Scholar] [CrossRef] [Green Version]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Fragkostefanakis, S.; Schleiff, E.; Simm, S. Transcriptional Basis for Differential Thermosensitivity of Seedlings of Various Tomato Genotypes. Genes 2020, 11, 655. https://doi.org/10.3390/genes11060655
Hu Y, Fragkostefanakis S, Schleiff E, Simm S. Transcriptional Basis for Differential Thermosensitivity of Seedlings of Various Tomato Genotypes. Genes. 2020; 11(6):655. https://doi.org/10.3390/genes11060655
Chicago/Turabian StyleHu, Yangjie, Sotirios Fragkostefanakis, Enrico Schleiff, and Stefan Simm. 2020. "Transcriptional Basis for Differential Thermosensitivity of Seedlings of Various Tomato Genotypes" Genes 11, no. 6: 655. https://doi.org/10.3390/genes11060655
APA StyleHu, Y., Fragkostefanakis, S., Schleiff, E., & Simm, S. (2020). Transcriptional Basis for Differential Thermosensitivity of Seedlings of Various Tomato Genotypes. Genes, 11(6), 655. https://doi.org/10.3390/genes11060655




