Short Tandem Repeats (STRs) as Biomarkers for the Quantitative Follow-Up of Chimerism after Stem Cell Transplantation: Methodological Considerations and Clinical Application
Abstract
1. Introduction
2. Patients and Methods
2.1. Patients and Samples
2.2. Genotyping
2.3. Data Analysis
3. Results
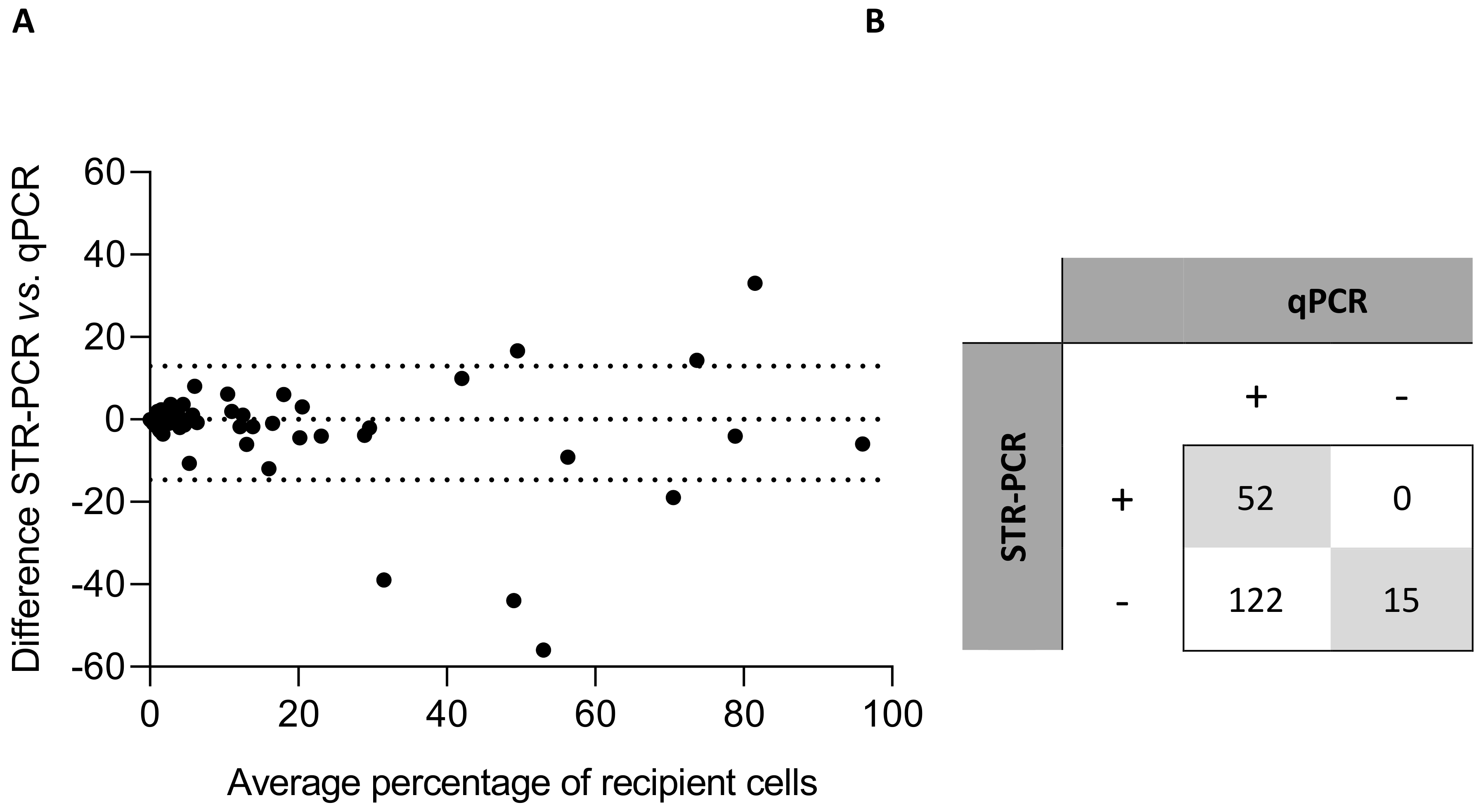
3.1. Correlation Analysis
3.2. Analysis of Artificial Mixtures
3.3. Informative Loci
3.4. Implementation of qPCR for Follow-Up of Patients in Complete Chimerism
3.5. Detection of Impending Relapse
4. Discussion
4.1. Comparison between STR-PCR and qPCR: Technical Aspects
- (1)
- The center of origin can provide baseline STR genotyping (when the same commercial kit is used).
- (2)
- Donor is usually available for DNA sample extraction and recipient is available for buccal swab DNA extraction. A post-transplant buccal swab is not suitable for recipient DNA sampling for qPCR purposes, since donor leukocytes may contaminate epithelial DNA and therefore, MC would be detected due to the high sensitivity of the technique. This is not an issue for STR-PCR, because it is easy to compare donor and recipient buccal swab samples and neglect those peaks that correspond to donor alleles, so that recipient peaks can be readily identified.
- (3)
- If only donor or recipient is available for DNA extraction, post-transplant samples could even be searched for recipient-derived peaks, based on the low probability (even in the case of transplantation from sibling donors, Figure 3) that donor and recipient share identical STR genotyping (3.3 x 10−12 for two randomly selected Caucasian individuals, as per user instructions of the manufacturer of a standard STR-PCR kit).
4.2. Comparison between STR-PCR and qPCR: Clinical Aspects
4.3. Methodological Algorithm for Quantitative Chimerism Follow-Up
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Copelan, E.A. Hematopoietic stem-cell transplantation. N. Engl. J. Med. 2006, 354, 1813–1826. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.; Hamerton, J.; Barnes, D.; Loutit, J. Cytological identification of radiation-chimaeras. Nature 1956, 177, 452–454. [Google Scholar] [CrossRef]
- Alcoceba, M.; Martínez-Laperche, C.; Buces, E.; Corral, R.; Buño, I. Seguimiento genético del trasplante hematopoyético alogénico. In Genética Hematológica. Manual Práctico; Sociedad Española de Hematología y Hemoterapia (SEHH): Madrid, Spain, 2016; ISBN 978-84-608-1494-8. [Google Scholar]
- Dubovsky, J.; Daxberger, H.; Fritsch, G.; Printz, D.; Peters, C.; Matthes, S.; Gadner, H.; Lion, T.; Muller-Bérat, N. Kinetics of chimerism during the early post-transplant period in pediatric patients with malignant and non-malignant hematologic disorders: Implications for timely detection of engraftment, graft failure and rejection. Leukemia 1999, 13, 2060–2069. [Google Scholar] [CrossRef]
- Breuer, S.; Preuner, S.; Fritsch, G.; Daxberger, H.; Koenig, M.; Poetschger, U.; Lawitschka, A.; Peters, C.; Mann, G.; Lion, T.; et al. Early recipient chimerism testing in the T- and NK-cell lineages for risk assessment of graft rejection in pediatric patients undergoing allogeneic stem cell transplantation. Leukemia 2012, 26, 509–519. [Google Scholar] [CrossRef]
- Baron, F.; Little, M.-T.; Storb, R. Kinetics of engraftment following allogeneic hematopoietic cell transplantation with reduced-intensity or nonmyeloablative conditioning. Blood Rev. 2005, 19, 153–164. [Google Scholar] [CrossRef]
- Balon, J.; Hałaburda, K.; Bieniaszewska, M.; Reichert, M.; Bieniaszewski, L.; Piekarska, A.; Pawłowski, R.; Hellmann, A. Early complete donor hematopoietic chimerism in peripheral blood indicates the risk of extensive graft-versus-host disease. Bone Marrow Transplant. 2005, 35, 1083–1088. [Google Scholar] [CrossRef]
- Jiang, Y.; Wan, L.; Qin, Y.; Wang, X.; Yan, S.; Xie, K.; Wang, C. Chimerism status is correlated to acute graft-versus-host disease after allogeneic stem cell transplantation. Int. J. Hematol. 2014, 99, 323–328. [Google Scholar] [CrossRef]
- Martínez-Laperche, C.; Noriega, V.; Kwon, M.; Balsalobre, P.; González-Rivera, M.; Serrano, D.; Anguita, J.; Gayoso, J.; Díez-Martín, J.L.; Buño, I. Achievement of early complete donor chimerism in CD25+-activated leukocytes is a strong predictor of the development of graft-versus-host-disease after stem cell transplantation. Exp. Hematol. 2015, 43, 4–13. [Google Scholar] [CrossRef]
- Bornhäuser, M.; Oelschlaegel, U.; Platzbecker, U.; Bug, G.; Lutterbeck, K.; Kiehl, M.G.; Schetelig, J.; Kiani, A.; Illmer, T.; Schaich, M.; et al. Monitoring of donor chimerism in sorted CD34+ peripheral blood cells allows the sensitive detection of imminent relapse after allogeneic stem cell transplantation. Haematologica 2009, 94, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Bader, P.; Niethammer, D.; Willasch, A.; Kreyenberg, H.; Klingebiel, T. How and when should we monitor chimerism after allogeneic stem cell transplantation? Bone Marrow Transplant. 2005, 35, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Sairafi, D.; Remberger, M.; Uhlin, M.; Ljungman, P.; Ringdén, O.; Mattsson, J. Leukemia lineage-specific chimerism analysis and molecular monitoring improve outcome of donor lymphocyte infusions. Biol. Blood Marrow Transplant. 2010, 16, 1728–1737. [Google Scholar] [CrossRef] [PubMed]
- Horn, B.; Wahlstrom, J.T.; Melton, A.; Liou, A.; Ouachee-Chardin, M.; Sunkersett, G.; Willert, J.; Hwang, J.; Expose-Spencer, J.; Cowan, M.C.; et al. Early mixed chimerism-based preemptive immunotherapy in children undergoing allogeneic hematopoietic stem cell transplantation for acute leukemia. Pediatr. Blood Cancer 2017, 64. [Google Scholar] [CrossRef] [PubMed]
- Lion, T.; Watzinger, F.; Preuner, S.; Kreyenberg, H.; Tilanus, M.; de Weger, R.; van Loon, J.; de Vries, L.; Cavé, H.; Acquaviva, C.; et al. The EuroChimerism concept for a standardized approach to chimerism analysis after allogeneic stem cell transplantation. Leukemia 2012, 26, 1821–1828. [Google Scholar] [CrossRef]
- Clark, J.R.; Scott, S.D.; Jack, A.L.; Lee, H.; Mason, J.; Carter, G.I.; Pearce, L.; Jackson, T.; Clouston, H.; Sproul, A.; et al. Monitoring of chimerism following allogeneic haematopoietic stem cell transplantation (HSCT): Technical recommendations for the use of short tandem repeat (STR) based techniques, on behalf of the United Kingdom National External Quality Assessment Service for Leucocyte Immunophenotyping Chimerism Working Group. Br. J. Haematol. 2015, 168, 26–37. [Google Scholar] [CrossRef]
- Thiede, C.; Florek, M.; Bornhäuser, M.; Ritter, M.; Mohr, B.; Brendel, C.; Ehninger, G.; Neubauer, A. Rapid quantification of mixed chimerism using multiplex amplification of short tandem repeat markers and fluorescence detection. Bone Marrow Transplant. 1999, 23, 1055–1060. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Buño, I.; Nava, P.; Simón, A.; González-Rivera, M.; Jiménez, J.L.; Balsalobre, P.; Serrano, D.; Carrión, R.; Gómez-Pineda, A.; Díez-Martín, J.L. A comparison of fluorescent in situ hybridization and multiplex short tandem repeat polymerase chain reaction for quantifying chimerism after stem cell transplantation. Haematologica 2005, 90, 1373–1379. [Google Scholar]
- Alizadeh, M.; Bernard, M.; Danic, B.; Dauriac, C.; Birebent, B.; Lapart, C.; Lamy, T.; Le Prisé, P.-Y.; Beauplet, A.; Bories, D.; et al. Quantitative assessment of hematopoietic chimerism after bone marrow transplantation by real-time quantitative polymerase chain reaction. Blood 2002, 99, 4618–4625. [Google Scholar] [CrossRef]
- Maas, F.; Schaap, N.; Kolen, S.; Zoetbrood, A.; Buño, I.; Dolstra, H.; de Witte, T.; Schattenberg, A.; van de Wiel-van Kemenade, E. Quantification of donor and recipient hemopoietic cells by real-time PCR of single nucleotide polymorphisms. Leukemia 2003, 17, 621–629. [Google Scholar] [CrossRef]
- Jiménez-Velasco, A.; Barrios, M.; Román-Gómez, J.; Navarro, G.; Buño, I.; Castillejo, J.A.; Rodríguez, A.I.; García-Gemar, G.; Torres, A.; Heiniger, A.I. Reliable quantification of hematopoietic chimerism after allogeneic transplantation for acute leukemia using amplification by real-time PCR of null alleles and insertion/deletion polymorphisms. Leukemia 2005, 19, 336–343. [Google Scholar] [CrossRef]
- Kim, S.Y.; Jeong, M.H.; Park, N.; Ra, E.; Park, H.; Seo, S.H.; Kim, J.Y.; Seong, M.-W.; Park, S.S. Chimerism monitoring after allogeneic hematopoietic stem cell transplantation using quantitative real-time PCR of biallelic insertion/deletion polymorphisms. J. Mol. Diagn. JMD 2014, 16, 679–688. [Google Scholar] [CrossRef]
- Masmas, T.N.; Madsen, H.O.; Petersen, S.L.; Ryder, L.P.; Svejgaard, A.; Alizadeh, M.; Vindeløv, L.L. Evaluation and automation of hematopoietic chimerism analysis based on real-time quantitative polymerase chain reaction. Biol. Blood Marrow Transplant. 2005, 11, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Bach, C.; Tomova, E.; Goldmann, K.; Weisbach, V.; Roesler, W.; Mackensen, A.; Winkler, J.; Spriewald, B.M. Monitoring of hematopoietic chimerism by real-time quantitative PCR of micro insertions/deletions in samples with low DNA quantities. Transfus. Med. Hemother. 2015, 42, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Vicente, D.C.; Laranjeira, A.B.A.; Miranda, E.C.M.; Yunes, J.A.; de Souza, C.A. Chimerism interpretation with a highly sensitive quantitative PCR method: 6 months median latency before chimerism drop below 0.1. Bone Marrow Transplant. 2016, 51, 874–875. [Google Scholar] [CrossRef] [PubMed]
- Jacque, N.; Nguyen, S.; Golmard, J.-L.; Uzunov, M.; Garnier, A.; Leblond, V.; Vernant, J.-P.; Bories, D.; Dhédin, N. Chimerism analysis in peripheral blood using indel quantitative real-time PCR is a useful tool to predict post-transplant relapse in acute leukemia. Bone Marrow Transplant. 2015, 50, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Tyler, J.; Kumer, L.; Fisher, C.; Casey, H.; Shike, H. Personalized Chimerism Test that Uses Selection of Short Tandem Repeat or Quantitative PCR Depending on Patient’s Chimerism Status. J. Mol. Diagn. JMD 2019, 21, 483–490. [Google Scholar] [CrossRef]
- Kwon, M.; Martínez-Laperche, C.; Balsalobre, P.; Serrano, D.; Anguita, J.; Gayoso, J.; Díez-Martín, J.L.; Buño, I. Early peripheral blood and T-cell chimerism dynamics after umbilical cord blood transplantation supported with haploidentical cells. Bone Marrow Transplant. 2014, 49, 212–218. [Google Scholar] [CrossRef][Green Version]
- Frankfurt, O.; Zitzner, J.R.; Tambur, A.R. Real-time qPCR for chimerism assessment in allogeneic hematopoietic stem cell transplants from unrelated adult and double umbilical cord blood. Hum. Immunol. 2015, 76, 155–160. [Google Scholar] [CrossRef]
- Sellmann, L.; Rabe, K.; Bünting, I.; Dammann, E.; Göhring, G.; Ganser, A.; Stadler, M.; Weissinger, E.M.; Hambach, L. Diagnostic value of highly-sensitive chimerism analysis after allogeneic stem cell transplantation. Bone Marrow Transplant. 2018, 53, 1457–1465. [Google Scholar] [CrossRef]
- Willasch, A.M.; Kreyenberg, H.; Shayegi, N.; Rettinger, E.; Meyer, V.; Zabel, M.; Lang, P.; Kremens, B.; Meisel, R.; Strahm, B.; et al. Monitoring of hematopoietic chimerism after transplantation for pediatric myelodysplastic syndrome: Real-time or conventional short tandem repeat PCR in peripheral blood or bone marrow? Biol. Blood Marrow Transplant. 2014, 20, 1918–1925. [Google Scholar] [CrossRef]
- Antin, J.H.; Childs, R.; Filipovich, A.H.; Giralt, S.; Mackinnon, S.; Spitzer, T.; Weisdorf, D. Establishment of complete and mixed donor chimerism after allogeneic lymphohematopoietic transplantation: Recommendations from a workshop at the 2001 Tandem Meetings of the International Bone Marrow Transplant Registry and the American Society of Blood and Marrow Transplantation. Biol. Blood Marrow Transplant. 2001, 7, 473–485. [Google Scholar] [CrossRef]
- Mackinnon, S.; Barnett, L.; Bourhis, J.H.; Black, P.; Heller, G.; O’Reilly, R.J. Myeloid and lymphoid chimerism after T-cell-depleted bone marrow transplantation: Evaluation of conditioning regimens using the polymerase chain reaction to amplify human minisatellite regions of genomic DNA. Blood 1992, 80, 3235–3241. [Google Scholar] [CrossRef] [PubMed]
- Childs, R.; Clave, E.; Contentin, N.; Jayasekera, D.; Hensel, N.; Leitman, S.; Read, E.J.; Carter, C.; Bahceci, E.; Young, N.S.; et al. Engraftment kinetics after nonmyeloablative allogeneic peripheral blood stem cell transplantation: Full donor T-cell chimerism precedes alloimmune responses. Blood 1999, 94, 3234–3241. [Google Scholar] [CrossRef]
- Lion, T.; Daxberger, H.; Dubovsky, J.; Filipcik, P.; Fritsch, G.; Printz, D.; Peters, C.; Matthes-Martin, S.; Lawitschka, A.; Gadner, H. Analysis of chimerism within specific leukocyte subsets for detection of residual or recurrent leukemia in pediatric patients after allogeneic stem cell transplantation. Leukemia 2001, 15, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Mattsson, J.; Uzunel, M.; Remberger, M.; Ringdén, O. T cell mixed chimerism is significantly correlated to a decreased risk of acute graft-versus-host disease after allogeneic stem cell transplantation. Transplantation 2001, 71, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Mattsson, J.; Uzunel, M.; Tammik, L.; Aschan, J.; Ringdén, O. Leukemia lineage-specific chimerism analysis is a sensitive predictor of relapse in patients with acute myeloid leukemia and myelodysplastic syndrome after allogeneic stem cell transplantation. Leukemia 2001, 15, 1976–1985. [Google Scholar] [CrossRef]
- Mohty, M.; Avinens, O.; Faucher, C.; Viens, P.; Blaise, D.; Eliaou, J.-F. Predictive factors and impact of full donor T-cell chimerism after reduced intensity conditioning allogeneic stem cell transplantation. Haematologica 2007, 92, 1004–1006. [Google Scholar] [CrossRef]
- Haugaard, A.K.; Madsen, H.O.; Marquart, H.V.; Rosthøj, S.; Masmas, T.N.; Heilmann, C.; Müller, K.G.; Ifversen, M. Highly sensitive chimerism detection in blood is associated with increased risk of relapse after allogeneic hematopoietic cell transplantation in childhood leukemia. Pediatr. Transplant. 2019, 23, e13549. [Google Scholar] [CrossRef]
- Lamba, R.; Abella, E.; Kukuruga, D.; Klein, J.; Savasan, S.; Abidi, M.H.; Mohamed, A.; Peres, E. Mixed hematopoietic chimerism at day 90 following allogenic myeloablative stem cell transplantation is a predictor of relapse and survival. Leukemia 2004, 18, 1681–1686. [Google Scholar] [CrossRef]
- Terwey, T.H.; Hemmati, P.G.; Nagy, M.; Pfeifer, H.; Gökbuget, N.; Brüggemann, M.; Le Duc, T.M.; le Coutre, P.; Dörken, B.; Arnold, R. Comparison of chimerism and minimal residual disease monitoring for relapse prediction after allogeneic stem cell transplantation for adult acute lymphoblastic leukemia. Biol. Blood Marrow Transplant. 2014, 20, 1522–1529. [Google Scholar] [CrossRef]
- Stumph, J.; Vnencak-Jones, C.L.; Koyama, T.; Frangoul, H. Comparison of peripheral blood and bone marrow samples for detection of post transplant mixed chimerism. Bone Marrow Transplant. 2008, 41, 589–590. [Google Scholar] [CrossRef][Green Version]
- Bach, C.; Steffen, M.; Roesler, W.; Winkler, J.; Mackensen, A.; Stachel, K.-D.; Metzler, M.; Spriewald, B.M. Systematic comparison of donor chimerism in peripheral blood and bone marrow after hematopoietic stem cell transplantation. Blood Cancer J. 2017, 7, e566. [Google Scholar] [CrossRef] [PubMed]
- Thiede, C. Diagnostic chimerism analysis after allogeneic stem cell transplantation: New methods and markers. Am. J. Pharm. Genom.-Relat. Res. Drug Dev. Clin. Pract. 2004, 4, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Saito, B.; Fukuda, T.; Yokoyama, H.; Kurosawa, S.; Takahashi, T.; Fuji, S.; Takahashi, N.; Tajima, K.; Kim, S.-W.; Mori, S.-I.; et al. Impact of T cell chimerism on clinical outcome in 117 patients who underwent allogeneic stem cell transplantation with a busulfan-containing reduced-intensity conditioning regimen. Biol. Blood Marrow Transplant. 2008, 14, 1148–1155. [Google Scholar] [CrossRef] [PubMed]
- Maris, M.B.; Sandmaier, B.M.; Storer, B.E.; Maloney, D.G.; Shizuru, J.A.; Agura, E.; Kliem, C.; Pulsipher, M.; Maziarz, R.T.; McSweeney, P.A.; et al. Unrelated donor granulocyte colony-stimulating factor-mobilized peripheral blood mononuclear cell transplantation after nonmyeloablative conditioning: The effect of postgrafting mycophenolate mofetil dosing. Biol. Blood Marrow Transplant. 2006, 12, 454–465. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Suárez-González, J.; Martínez-Laperche, C.; Kwon, M.; Balsalobre, P.; Carbonell, D.; Chicano, M.; Rodríguez-Macías, G.; Serrano, D.; Gayoso, J.; Díez-Martín, J.L.; et al. Donor Cell-Derived Hematologic Neoplasms after Hematopoietic Stem Cell Transplantation: A Systematic Review. Biol. Blood Marrow Transplant. 2018, 24, 1505–1513. [Google Scholar] [CrossRef] [PubMed]
- Suárez-González, J.; Martínez-Laperche, C.; Martínez, N.; Rodríguez-Macías, G.; Kwon, M.; Balsalobre, P.; Carbonell, D.; Chicano, M.; Serrano, D.; Triviño, J.C.; et al. Whole-exome sequencing reveals acquisition of mutations leading to the onset of donor cell leukemia after hematopoietic transplantation: A model of leukemogenesis. Leukemia 2018, 32, 1822–1826. [Google Scholar] [CrossRef]
- Suárez-González, J.; Triviño, J.C.; Bautista, G.; García-Marco, J.A.; Figuera, Á.; Balas, A.; Vicario, J.L.; Ortuño, F.J.; Teruel, R.; Álamo, J.M.; et al. Exome sequencing reveals heterogeneous clonal dynamics in donor cell myeloid neoplasms after stem cell transplantation. Haematologica 2020. [Google Scholar] [CrossRef]




| Total–n | 57 |
|---|---|
| Sex Female—n (%) | 20 (35) |
| Age, years—median (range) | 46 (6–66) |
| Diagnosis—n (%) | |
| - AML | 42 (74) |
| - ALL | 10 (18) |
| - MDS/MPN | 3 (5) |
| - NHL | 2 (3) |
| Time to switch, months—median (range) | 13 (1–48) |
| qPCR performed in PB—n (%) | 57 (100) |
| Number of PB samples—mean (range) | 6.12 (1–16) |
| Recipient DNA 0.1–1% in PB—n (%) | 4 (7) |
| qPCR performed in BM—n (%) | 46 (81) |
| Number of BM samples—mean (range) | 2.14 (0–9) |
| Recipient DNA 0.1–1% in BM—n (%) - One sample - More than one sample | 27 (59) 11 (24) 16 (35) |
| Molecular MRD marker—n (%) | |
| - None | 13 (23) |
| - WT1 | 23 (40) |
| - NPM1 | 12 (21) |
| - Other | 10 (18) |
| Positive molecular MRD marker—n (%) | 2 (5) |
| Relapse—n (%) | 1 (2) |
| Follow-up—median (range) | 32 (9–67) |
| Patient ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Diagnosis | AML | MDS (RAEB2) | CLL | MF | AML | AML | ALL | AML |
| Relapse (days after HSCT) | 1815 | 90 | 1703 | 253 | 479 | 322 | 218 | 363 |
| Date of prior sample (days before relapse) | 363 | 21 | 72 | 7 | 85 | 112 | 45 | 31 |
| Type of prior sample | PB | PB | PB | PB | PB | PB | PB | BM |
| Result qPCR in prior sample | CC | MC | MC | MC | MC | MC | MC | CC |
| Quantification qPCR in prior sample (% recipient) | 0.017 | 0.2 | 1.5 | 1 | 0.15 | 0.159 | 0.1 | 0.003 |
| MM | WT1 | None | None | None | NPM1 | WT1 | None | WT1 |
| Result MM at relapse | Pos | - | - | - | Pos | Pos | - | Neg |
| Result MM in prior sample | Neg | - | - | - | Neg | Neg | - | Neg |
| STR-PCR | qPCR | |
|---|---|---|
| DNA needed | 0.2–1 ng | 2250 ng |
| Total turnaround time (10 samples) | 2.5 working days | 1 working day |
| Hands-on time (10 samples) | 1.5 h | 2.5 h |
| Analysis time (10 samples) | 0.5 h | 0.5 h |
| Cost per sample (€) | 35€ | 150€ |
| Devices needed | Screening and follow-up:
| Screening:
|
| Objective | Patients | Leukocyte Lineage | Technique | Chronogram |
|---|---|---|---|---|
| Engraftment | All | PB T-cells (B-cells, NK-cells) | STR-PCR | From day 15, every other week until CC (More frequently if needed) |
| BM | STR-PCR | Day +30 | ||
| GVHD | All | T-cells (Activated leukocytes) | STR-PCR | Every other week until CC |
| Follow up after CC | “Non leukemic” diseases 1 | PB and BM | STR-PCR | Days 90, 180, 365 |
| Follow up after CC—detection of relapse | “Medullary” diseases 2 with MRD marker | PB | STR-PCR | Monthly during first year, every 3 months for second year |
| BM | STR-PCR | Every 3 months for first year | ||
| “Medullary” diseases2 without MRD marker | PB | qPCR | Monthly during first year, every 3 months for second year | |
| BM | qPCR | Every 3 months for first year | ||
| “Medullary” diseases2 CD34+ | CD34+ from BM | STR-PCR | Every 3 months for first year |
| Monday | Tuesday | Wednesday | Thursday | Friday |
|---|---|---|---|---|
| Collection of samples Immunomagnetic separation DNA extraction | Collection of samples Immunomagnetic separation DNA extraction | PCR | Capillary electrophoresis Data Analysis Report | Transplant Committee meeting |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarro-Bailón, A.; Carbonell, D.; Escudero, A.; Chicano, M.; Muñiz, P.; Suárez-González, J.; Bailén, R.; Oarbeascoa, G.; Kwon, M.; Díez-Martín, J.L.; et al. Short Tandem Repeats (STRs) as Biomarkers for the Quantitative Follow-Up of Chimerism after Stem Cell Transplantation: Methodological Considerations and Clinical Application. Genes 2020, 11, 993. https://doi.org/10.3390/genes11090993
Navarro-Bailón A, Carbonell D, Escudero A, Chicano M, Muñiz P, Suárez-González J, Bailén R, Oarbeascoa G, Kwon M, Díez-Martín JL, et al. Short Tandem Repeats (STRs) as Biomarkers for the Quantitative Follow-Up of Chimerism after Stem Cell Transplantation: Methodological Considerations and Clinical Application. Genes. 2020; 11(9):993. https://doi.org/10.3390/genes11090993
Chicago/Turabian StyleNavarro-Bailón, Almudena, Diego Carbonell, Asunción Escudero, María Chicano, Paula Muñiz, Julia Suárez-González, Rebeca Bailén, Gillen Oarbeascoa, Mi Kwon, José Luis Díez-Martín, and et al. 2020. "Short Tandem Repeats (STRs) as Biomarkers for the Quantitative Follow-Up of Chimerism after Stem Cell Transplantation: Methodological Considerations and Clinical Application" Genes 11, no. 9: 993. https://doi.org/10.3390/genes11090993
APA StyleNavarro-Bailón, A., Carbonell, D., Escudero, A., Chicano, M., Muñiz, P., Suárez-González, J., Bailén, R., Oarbeascoa, G., Kwon, M., Díez-Martín, J. L., Martínez-Laperche, C., & Buño, I. (2020). Short Tandem Repeats (STRs) as Biomarkers for the Quantitative Follow-Up of Chimerism after Stem Cell Transplantation: Methodological Considerations and Clinical Application. Genes, 11(9), 993. https://doi.org/10.3390/genes11090993






