Bioinformatics Analysis Revealed Novel 3?UTR Variants Associated with Intellectual Disability
Abstract
:1. Introduction
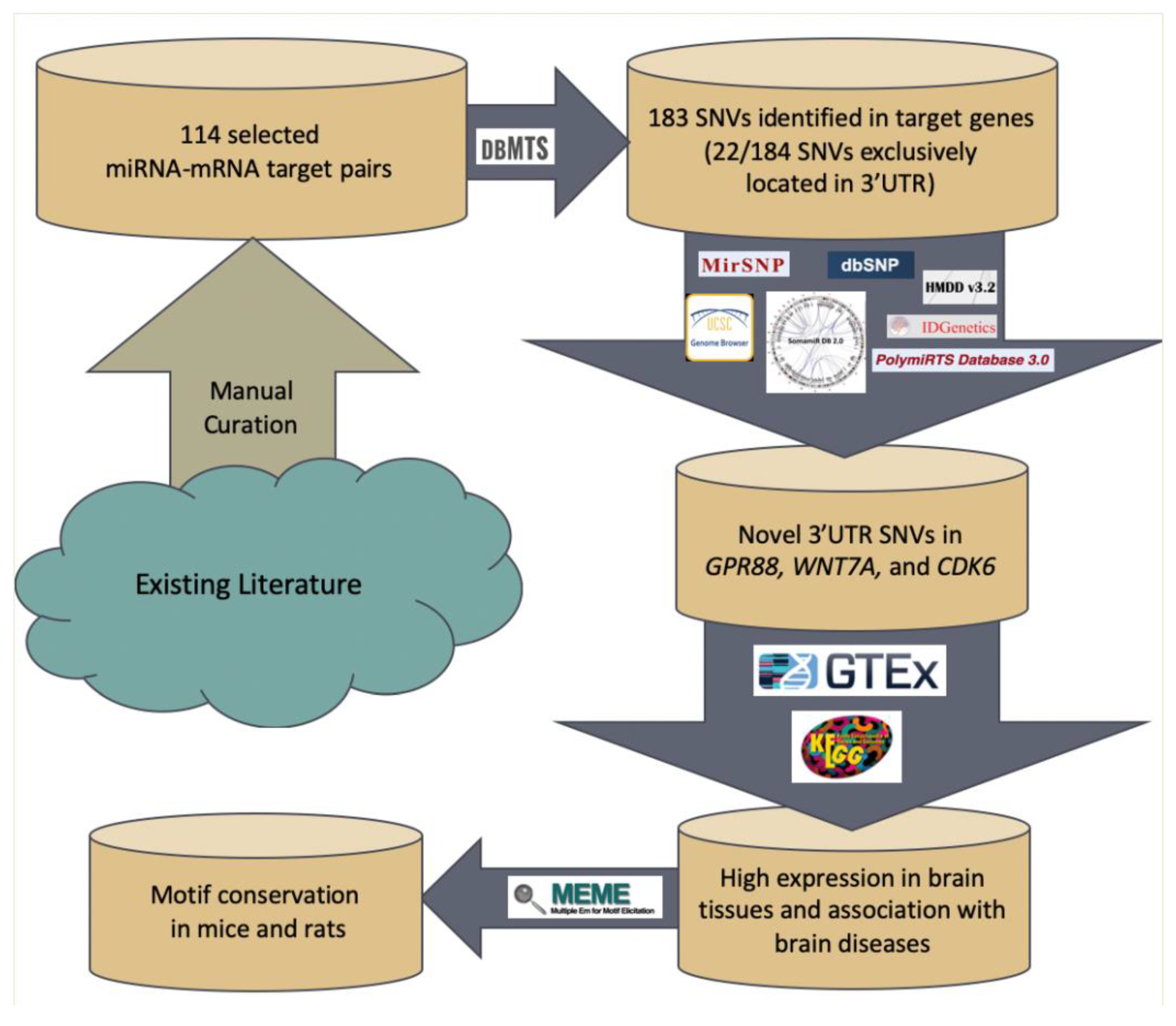
2. Materials and Methods
2.1. Input Data Curation from Literature
2.2. miRNA-Gene Pair Selection
2.3. dbMTS Database Variant Identification
2.4. Functional Annotation of Candidate ID Gene Associated with Novel 3′UTR SNVs
2.5. Expression Data and Association with Brain Diseases in Genes Exclusively Expressed in 3′UTR Region
2.6. Pathway and Brain Diseases Association with of Genes Exclusively Expressed in 3′UTR Region
2.7. Conservation Across Other Species
3. Results
3.1. miRNA-Gene Pair Selection Screen
3.2. Analysis of Result from dbMTS
3.3. Novel SNV Identification
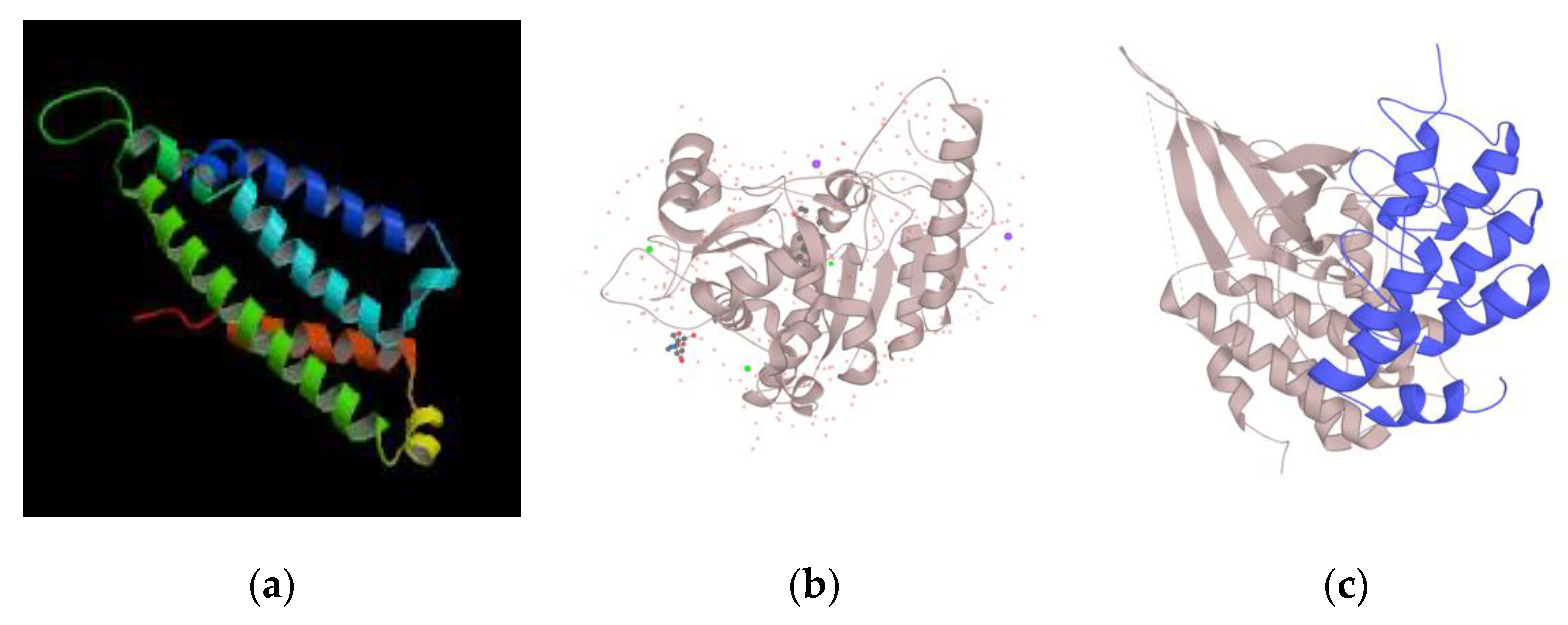
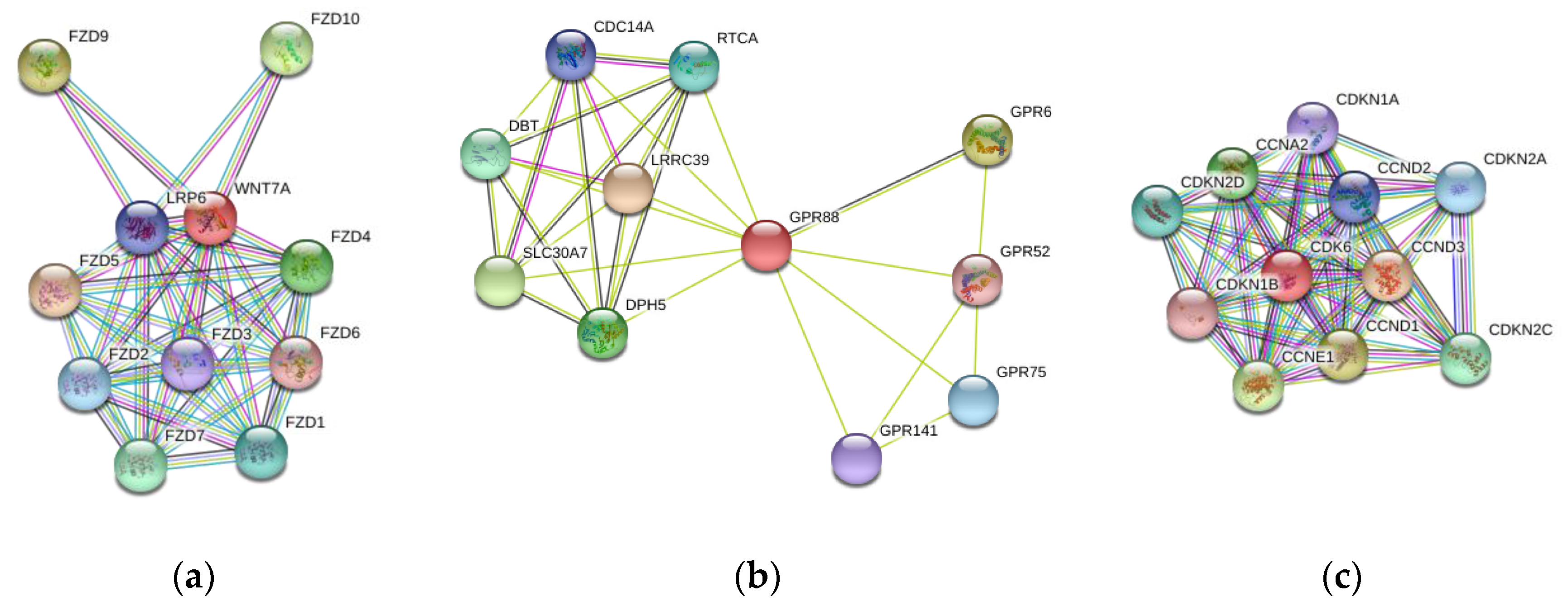
3.4. Functional Annotation of Candidate ID Gene Associated with Novel 3′UTR SNVs
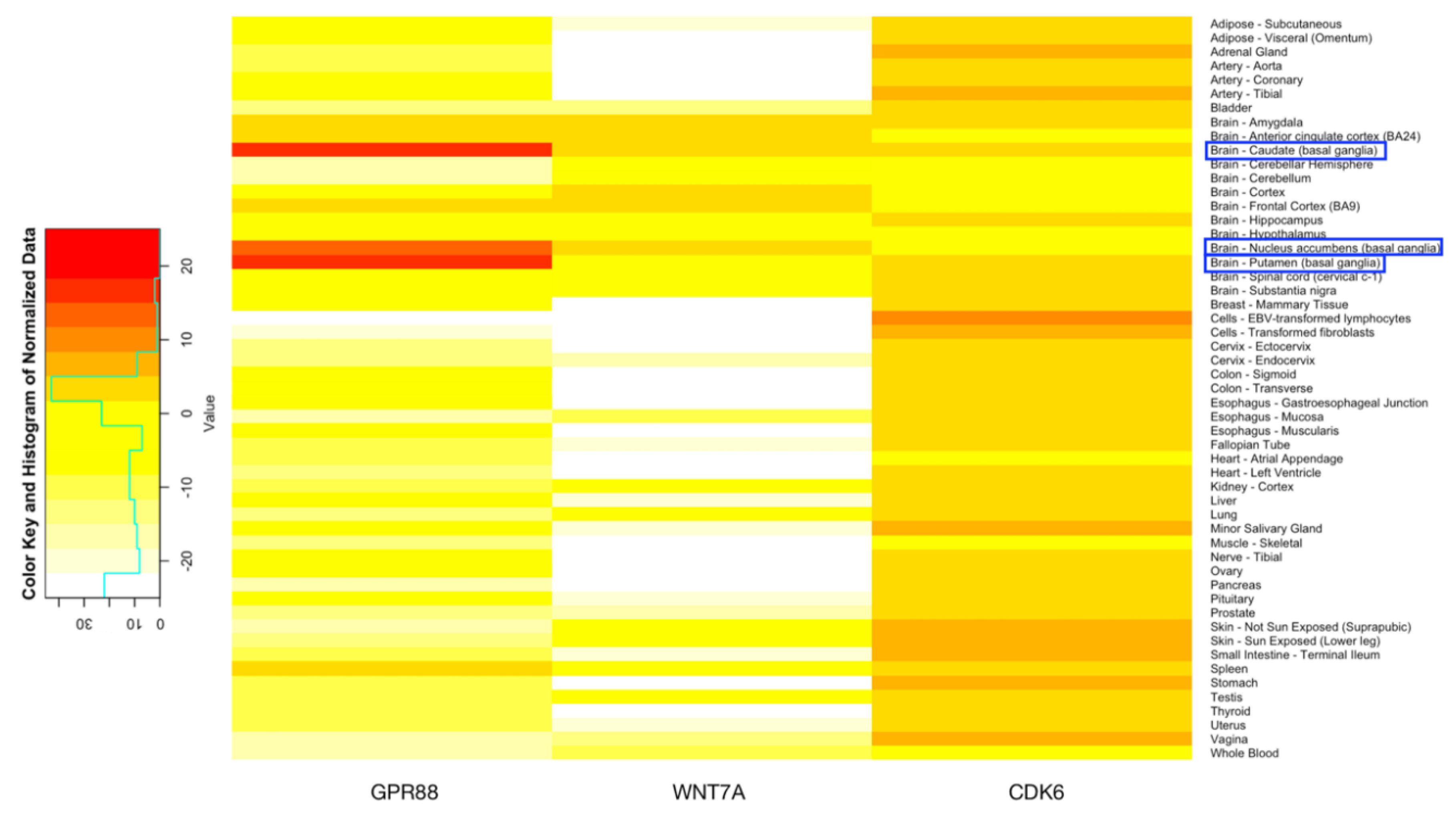
3.5. Expression Data of Genes Exclusively Expressed in 3′UTR Region
3.6. Pathway and Brain Diseases Association with Genes Exclusively Expressed in 3′UTR Region
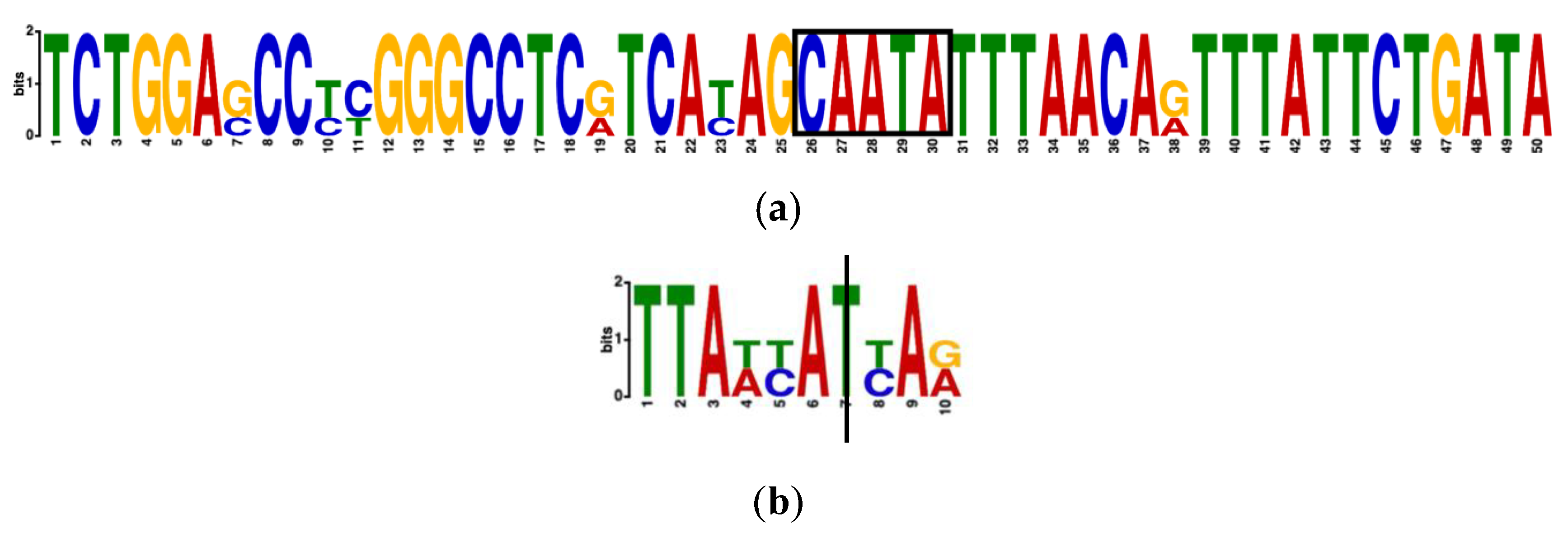
3.7. Conservation across Other Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ID | Intellectual disability |
| miR | MicroRNA |
| UTR | Untranslated region |
| SNV | Single variant nucleotide |
| GPCR | G protein-coupled receptors |
| PPI | Protein-protein interactions |
References
- Salvador-Carulla, L.; Reed, G.M.; Vaez-Azizi, L.M.; Cooper, S.A.; Martinez-Leal, R.; Bertelli, M.; Adnams, C.; Cooray, S.; Deb, S.; Akoury-Dirani, L.; et al. Intellectual developmental disorders: Towards a new name, definition and framework for "mental retardation/intellectual disability" in ICD-11. World Psychiatry 2011, 10, 175–180. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar]
- Khan, M.Z.; He, L. Neuro-psychopharmacological perspective of Orphan receptors of Rhodopsin (class A) family of G protein-coupled receptors. Psychopharmacology 2017, 234, 1181–1207. [Google Scholar] [CrossRef]
- De Giorgio, A. The roles of motor activity and environmental enrichment in intellectual disability. Somatosens. Mot. Res. 2017, 34, 34–43. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.B. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 2008, 31, 20–26. [Google Scholar] [CrossRef] [Green Version]
- Mendell, J.T. MicroRNAs: Critical regulators of development, cellular physiology and malignancy. Cell Cycle 2005, 4, 1179–1184. [Google Scholar] [CrossRef] [Green Version]
- Rylaarsdam, L.; Guemez-Gamboa, A. Genetic Causes and Modifiers of Autism Spectrum Disorder. Front. Cell Neurosci. 2019, 13, 385. [Google Scholar] [CrossRef]
- Gilissen, C.; Hehir-Kwa, J.Y.; Thung, D.T.; van de Vorst, M.; van Bon, B.W.; Willemsen, M.H.; Kwint, M.; Janssen, I.M.; Hoischen, A.; Schenck, A.; et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014, 511, 344–347. [Google Scholar] [CrossRef]
- Lelieveld, S.H.; Veltman, J.A.; Gilissen, C. Novel bioinformatic developments for exome sequencing. Hum. Genet. 2016, 135, 603–614. [Google Scholar] [CrossRef] [Green Version]
- Carvill, G.L.; Mefford, H.C. Next-Generation Sequencing in Intellectual Disability. J. Pediatr. Genet. 2015, 4, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vissers, L.E.; Gilissen, C.; Veltman, J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016, 17, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, N.; Zhang, Y.; Du, Y.; Zhang, T.; Li, Z.; Wu, J.; Wang, X. Prioritized High-Confidence Risk Genes for Intellectual Disability Reveal Molecular Convergence During Brain Development. Front. Genet. 2018, 9, 349. [Google Scholar] [CrossRef] [Green Version]
- de Ligt, J.; Willemsen, M.H.; van Bon, B.W.; Kleefstra, T.; Yntema, H.G.; Kroes, T.; Vulto-van Silfhout, A.T.; Koolen, D.A.; de Vries, P.; Gilissen, C.; et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012, 367, 1921–1929. [Google Scholar] [CrossRef] [Green Version]
- de Vries, B.B.; Pfundt, R.; Leisink, M.; Koolen, D.A.; Vissers, L.E.; Janssen, I.M.; Reijmersdal, S.; Nillesen, W.M.; Huys, E.H.; Leeuw, N.; et al. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 2005, 77, 606–616. [Google Scholar] [CrossRef] [Green Version]
- Rauch, A.; Wieczorek, D.; Graf, E.; Wieland, T.; Endele, S.; Schwarzmayr, T.; Albrecht, B.; Bartholdi, D.; Beygo, J.; Di Donato, N.; et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: An exome sequencing study. Lancet 2012, 380, 1674–1682. [Google Scholar] [CrossRef]
- Alkufri, F.; Shaag, A.; Abu-Libdeh, B.; Elpeleg, O. Deleterious mutation in GPR88 is associated with chorea, speech delay, and learning disabilities. Neurol. Genet. 2016, 2, e64. [Google Scholar] [CrossRef] [Green Version]
- Wanke, K.A.; Devanna, P.; Vernes, S.C. Understanding Neurodevelopmental Disorders: The Promise of Regulatory Variation in the 3′UTRome. Biol. Psychiatry 2018, 83, 548–557. [Google Scholar] [CrossRef] [Green Version]
- Brewster, B.L.; Rossiello, F.; French, J.D.; Edwards, S.L.; Wong, M.; Wronski, A.; Whiley, P.; Waddell, N.; Chen, X.; Bove, B.; et al. Identification of fifteen novel germline variants in the BRCA1 3′UTR reveals a variant in a breast cancer case that introduces a functional miR-103 target site. Hum. Mutat. 2012, 33, 1665–1675. [Google Scholar] [CrossRef]
- Juran, B.D.; Atkinson, E.J.; Schlicht, E.M.; Fridley, B.L.; Lazaridis, K.N. Primary biliary cirrhosis is associated with a genetic variant in the 3′ flanking region of the CTLA4 gene. Gastroenterology 2008, 135, 1200–1206. [Google Scholar] [CrossRef] [Green Version]
- Devanna, P.; van de Vorst, M.; Pfundt, R.; Gilissen, C.; Vernes, S.C. Genome-wide investigation of an ID cohort reveals de novo 3′UTR variants affecting gene expression. Hum. Genet. 2018, 137, 717–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Zhang, F.; Li, T.; Lu, M.; Wang, L.; Yue, W.; Zhang, D. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genom. 2012, 13, 661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmoudi, E.; Cairns, M.J. MiR-137: An important player in neural development and neoplastic transformation. Mol. Psychiatry 2017, 22, 44–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollins, S.; Tuffrey-Wijne, I. Improving hospital care for patients with intellectual disabilities. Br. J. Hosp. Med. (Lond.) 2014, 75, 304–305. [Google Scholar] [CrossRef]
- Kozaki, K.; Inazawa, J. Tumor-suppressive microRNA silenced by tumor-specific DNA hypermethylation in cancer cells. Cancer Sci. 2012, 103, 837–845. [Google Scholar] [CrossRef]
- Logue, S.F.; Grauer, S.M.; Paulsen, J.; Graf, R.; Taylor, N.; Sung, M.A.; Zhang, L.; Hughes, Z.; Pulito, V.L.; Liu, F.; et al. The orphan GPCR, GPR88, modulates function of the striatal dopamine system: A possible therapeutic target for psychiatric disorders? Mol. Cell Neurosci. 2009, 42, 438–447. [Google Scholar] [CrossRef]
- Quintana, A.; Sanz, E.; Wang, W.; Storey, G.P.; Guler, A.D.; Wanat, M.J.; Roller, B.A.; La Torre, A.; Amieux, P.S.; McKnight, G.S.; et al. Lack of GPR88 enhances medium spiny neuron activity and alters motor- and cue-dependent behaviors. Nat. Neurosci. 2012, 15, 1547–1555. [Google Scholar] [CrossRef]
- Ciani, L.; Boyle, K.A.; Dickins, E.; Sahores, M.; Anane, D.; Lopes, D.M.; Gibb, A.J.; Salinas, P.C. Wnt7a signaling promotes dendritic spine growth and synaptic strength through Ca(2)(+)/Calmodulin-dependent protein kinase II. Proc. Natl. Acad. Sci. USA 2011, 108, 10732–10737. [Google Scholar] [CrossRef] [Green Version]
- Gogolla, N.; Galimberti, I.; Deguchi, Y.; Caroni, P. Wnt signaling mediates experience-related regulation of synapse numbers and mossy fiber connectivities in the adult hippocampus. Neuron 2009, 62, 510–525. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Swartz, M.; Yu, B.; Bai, Y.; Liu, X. dbMTS: A comprehensive database of putative human microRNA target site SNVs and their functional predictions. bioRxiv 2019. [Google Scholar] [CrossRef]
- Asim, A.; Kumar, A.; Muthuswamy, S.; Jain, S.; Agarwal, S. Down syndrome: An insight of the disease. J. Biomed. Sci. 2015, 22, 41. [Google Scholar] [CrossRef] [Green Version]
- Chai, M.; Su, L.; Hao, X.; Zhang, M.; Zheng, L.; Bi, J.; Han, X.; Gao, C. Identification of a thymus microRNAmRNA regulatory network in Down syndrome. Mol. Med. Rep. 2019, 20, 2063–2072. [Google Scholar] [CrossRef] [Green Version]
- Franzoni, E.; Booker, S.A.; Parthasarathy, S.; Rehfeld, F.; Grosser, S.; Srivatsa, S.; Fuchs, H.R.; Tarabykin, V.; Vida, I.; Wulczyn, F.G. miR-128 regulates neuronal migration, outgrowth and intrinsic excitability via the intellectual disability gene Phf6. Elife 2015, 4. [Google Scholar] [CrossRef]
- Lim, J.H.; Han, Y.J.; Kim, H.J.; Kim, M.Y.; Park, S.Y.; Cho, Y.H.; Ryu, H.M. Integrative analyses of genes and microRNA expressions in human trisomy 21 placentas. BMC Med. Genom. 2018, 11, 46. [Google Scholar] [CrossRef] [Green Version]
- Siew, W.H.; Tan, K.L.; Babaei, M.A.; Cheah, P.S.; Ling, K.H. MicroRNAs and intellectual disability (ID) in Down syndrome, X-linked ID, and Fragile X syndrome. Front. Cell Neurosci. 2013, 7, 41. [Google Scholar] [CrossRef] [Green Version]
- Zablotskaya, A.; Van Esch, H.; Verstrepen, K.J.; Froyen, G.; Vermeesch, J.R. Mapping the landscape of tandem repeat variability by targeted long read single molecule sequencing in familial X-linked intellectual disability. BMC Med. Genom. 2018, 11, 123. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Ciufo, S.; Starchenko, E.; Darji, D.; Chlumsky, L.; Karsch-Mizrachi, I.; Schoch, C.L. The NCBI BioCollections Database. Database (Oxford) 2018, 2018. [Google Scholar] [CrossRef] [Green Version]
- Smedley, D.; Haider, S.; Durinck, S.; Pandini, L.; Provero, P.; Allen, J.; Arnaiz, O.; Awedh, M.H.; Baldock, R.; Barbiera, G.; et al. The BioMart community portal: An innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015, 43, W589–W598. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2018, 47, D155–D162. [Google Scholar] [CrossRef]
- Chen, C.; Chen, D.; Xue, H.; Liu, X.; Zhang, T.; Tang, S.; Li, W.; Xu, X. IDGenetics: A comprehensive database for genes and mutations of intellectual disability related disorders. Neurosci. Lett. 2018, 685, 96–101. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consortium, T.U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carithers, L.J.; Moore, H.M. The Genotype-Tissue Expression (GTEx) Project. Biopreserv. Biobank 2015, 13, 307–308. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Morgat, A.; Lombardot, T.; Coudert, E.; Axelsen, K.; Neto, T.B.; Gehant, S.; Bansal, P.; Bolleman, J.; Gasteiger, E.; de Castro, E.; et al. Enzyme annotation in UniProtKB using Rhea. Bioinformatics 2019. [Google Scholar] [CrossRef]
- Illumina BodyMap 2.0 Project. Available online: http://www.ensembl.info/2011/05/24/human-bodymap-2-0-data-from-illumina/ (accessed on 15 June 2020).
- Duff, M.O.; Olson, S.; Wei, X.; Garrett, S.C.; Osman, A.; Bolisetty, M.; Plocik, A.; Celniker, S.E.; Graveley, B.R. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature 2015, 521, 376–379. [Google Scholar] [CrossRef] [Green Version]
- Szabo, L.; Morey, R.; Palpant, N.J.; Wang, P.L.; Afari, N.; Jiang, C.; Parast, M.M.; Murry, C.E.; Laurent, L.C.; Salzman, J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015, 16, 126. [Google Scholar] [CrossRef] [Green Version]
- Alavi, M.S.; Shamsizadeh, A.; Azhdari-Zarmehri, H.; Roohbakhsh, A. Orphan G protein-coupled receptors: The role in CNS disorders. Biomed. Pharmacother. 2018, 98, 222–232. [Google Scholar] [CrossRef]
- Meirsman, A.C.; Le Merrer, J.; Pellissier, L.P.; Diaz, J.; Clesse, D.; Kieffer, B.L.; Becker, J.A. Mice Lacking GPR88 Show Motor Deficit, Improved Spatial Learning, and Low Anxiety Reversed by Delta Opioid Antagonist. Biol. Psychiatry 2016, 79, 917–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Zompo, M.; Deleuze, J.F.; Chillotti, C.; Cousin, E.; Niehaus, D.; Ebstein, R.P.; Ardau, R.; Mace, S.; Warnich, L.; Mujahed, M.; et al. Association study in three different populations between the GPR88 gene and major psychoses. Mol. Genet. Genom. Med. 2014, 2, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Wang, W.; Huang, Y.; Bu, X.; Zhao, C. Roles of Wnt7a in embryo development, tissue homeostasis, and human diseases. J. Cell Biochem. 2019, 120, 18588–18598. [Google Scholar] [CrossRef] [PubMed]
- Wallace, J.; Lutgen, V.; Avasarala, S.; St Croix, B.; Winn, R.A.; Al-Harthi, L. Wnt7a induces a unique phenotype of monocyte-derived macrophages with lower phagocytic capacity and differential expression of pro- and anti-inflammatory cytokines. Immunology 2018, 153, 203–213. [Google Scholar] [CrossRef] [Green Version]
- Curie, A.; Friocourt, G.; des Portes, V.; Roy, A.; Nazir, T.; Brun, A.; Cheylus, A.; Marcorelles, P.; Retzepi, K.; Maleki, N.; et al. Basal ganglia involvement in ARX patients: The reason for ARX patients very specific grasping? Neuroimage Clin. 2018, 19, 454–465. [Google Scholar] [CrossRef]
- Bronner, S.M.; Merrick, K.A.; Murray, J.; Salphati, L.; Moffat, J.G.; Pang, J.; Sneeringer, C.J.; Dompe, N.; Cyr, P.; Purkey, H.; et al. Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma. Bioorg. Med. Chem. Lett. 2019, 29, 2294–2301. [Google Scholar] [CrossRef]
- Silber, J.; Lim, D.A.; Petritsch, C.; Persson, A.I.; Maunakea, A.K.; Yu, M.; Vandenberg, S.R.; Ginzinger, D.G.; James, C.D.; Costello, J.F.; et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008, 6, 14. [Google Scholar] [CrossRef]
| Chr | Pos | Ref | Alt | VEP_Ensembl_Gene_Name |
|---|---|---|---|---|
| 1 | 100541492 | T | G | GPR88 |
| 3 | 13818428 | G | T | WNT7A |
| 3 | 13818646 | T | A | WNT7A |
| 3 | 13818646 | T | C | WNT7A |
| 3 | 13818646 | T | G | WNT7A |
| 3 | 13818647 | A | C | WNT7A |
| 3 | 13818647 | A | G | WNT7A |
| 3 | 13818647 | A | T | WNT7A |
| 3 | 13818648 | T | A | WNT7A |
| 3 | 13818648 | T | C | WNT7A |
| 3 | 13818648 | T | G | WNT7A |
| 3 | 13818649 | T | A | WNT7A |
| 3 | 13818649 | T | C | WNT7A |
| 3 | 13818649 | T | G | WNT7A |
| 3 | 13818650 | G | A | WNT7A |
| 3 | 13818650 | G | C | WNT7A |
| 3 | 13818650 | G | T | WNT7A |
| 7 | 92605125 | T | A | CDK6 |
| 7 | 92607925 | A | G | CDK6 |
| 7 | 92607993 | A | C | CDK6 |
| 7 | 92607993 | A | G | CDK6 |
| 7 | 92607993 | A | T | CDK6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Liu, A.; He, I.; Bai, Y. Bioinformatics Analysis Revealed Novel 3?UTR Variants Associated with Intellectual Disability. Genes 2020, 11, 998. https://doi.org/10.3390/genes11090998
Yang J, Liu A, He I, Bai Y. Bioinformatics Analysis Revealed Novel 3?UTR Variants Associated with Intellectual Disability. Genes. 2020; 11(9):998. https://doi.org/10.3390/genes11090998
Chicago/Turabian StyleYang, Junmeng, Anna Liu, Isabella He, and Yongsheng Bai. 2020. "Bioinformatics Analysis Revealed Novel 3?UTR Variants Associated with Intellectual Disability" Genes 11, no. 9: 998. https://doi.org/10.3390/genes11090998
APA StyleYang, J., Liu, A., He, I., & Bai, Y. (2020). Bioinformatics Analysis Revealed Novel 3?UTR Variants Associated with Intellectual Disability. Genes, 11(9), 998. https://doi.org/10.3390/genes11090998





