A Large-Scale Genome-Wide Association Study of Epistasis Effects of Production Traits and Daughter Pregnancy Rate in U.S. Holstein Cattle
Abstract
:1. Introduction
2. Materials and Methods
2.1. Holstein Populations and Genotyping Data
2.2. GWAS Analysis
3. Results
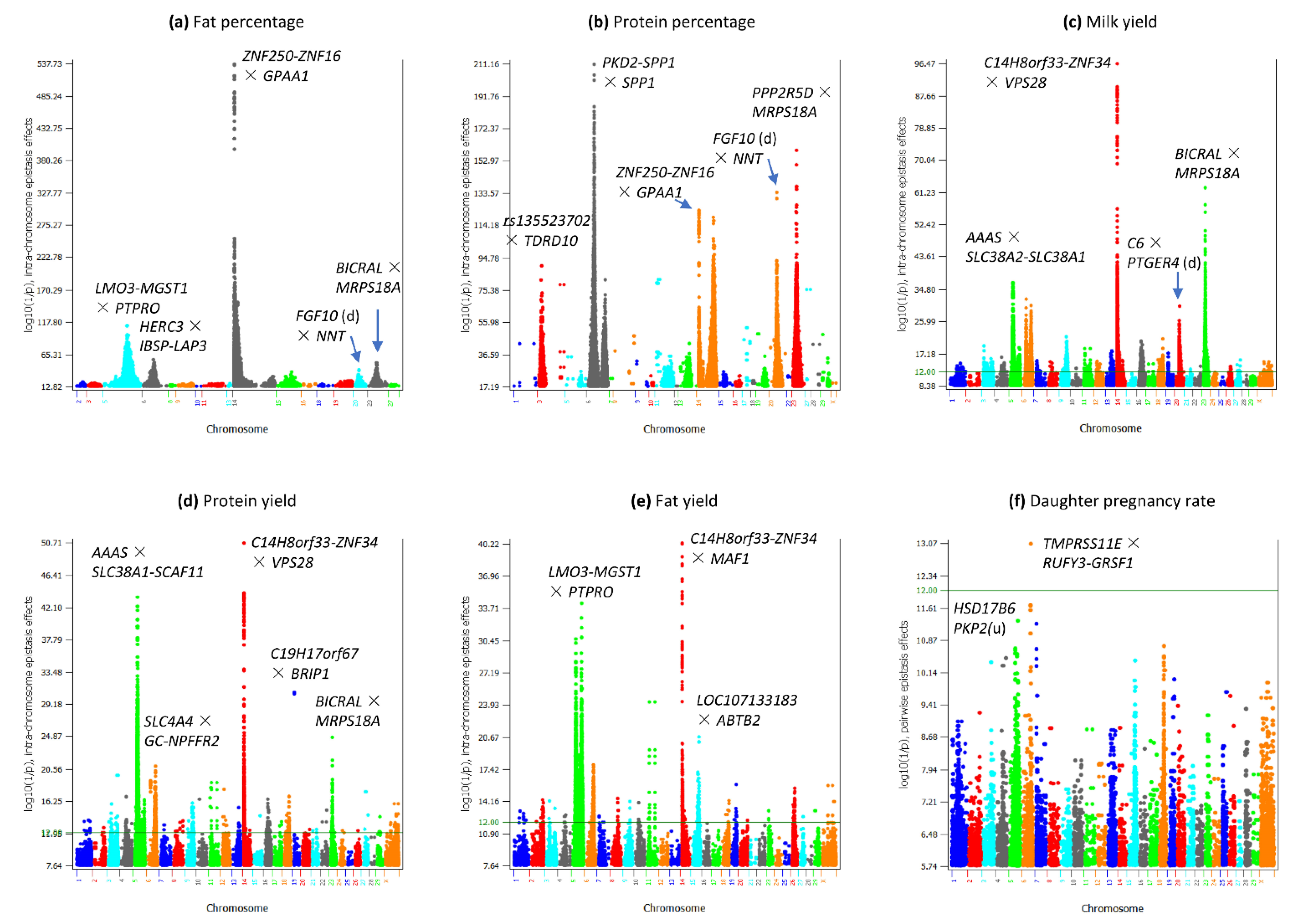
3.1. Overview of Epistasis Effects
3.2. Intra-Chromosome Epistasis Effects of Fat Percentage
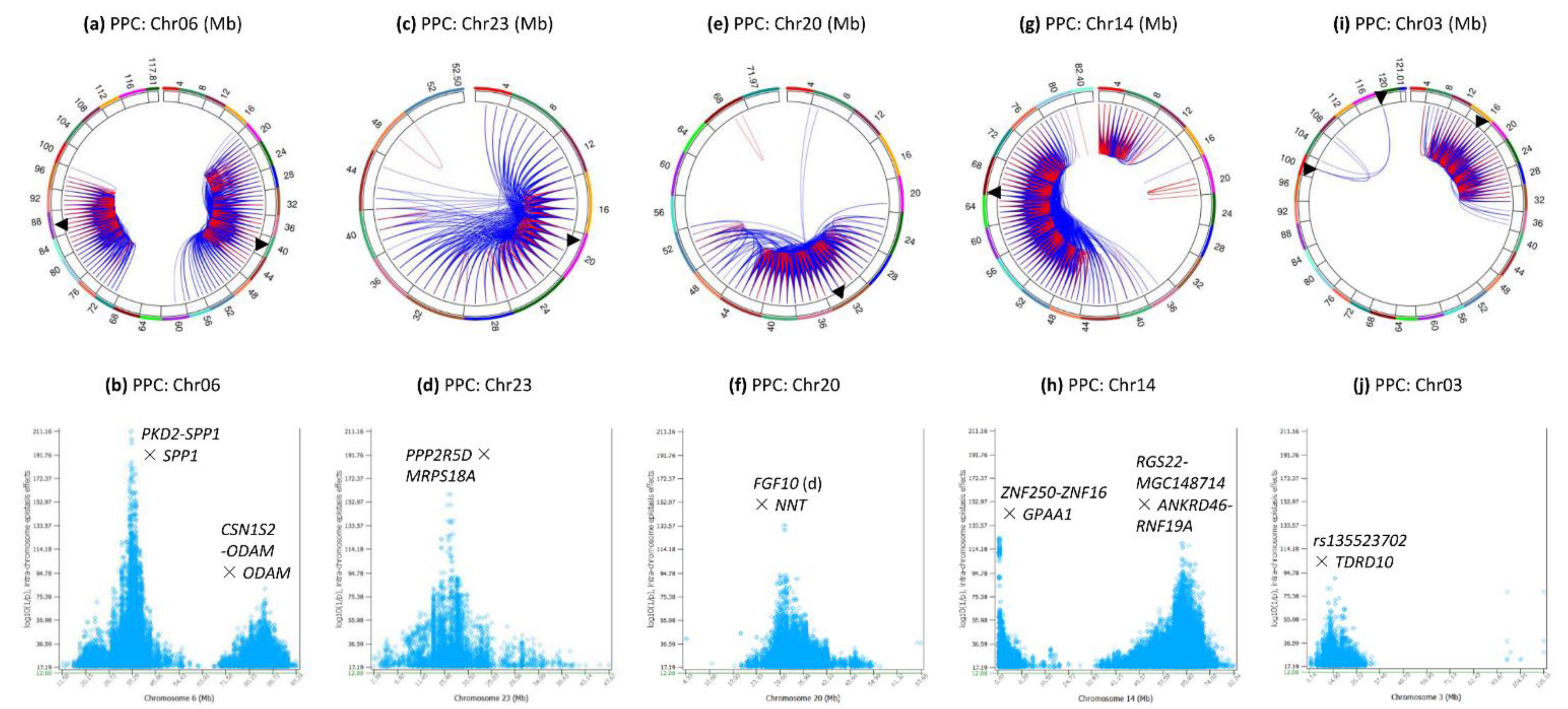
3.3. Intra-Chromosome Epistasis Effects of Protein Percentage
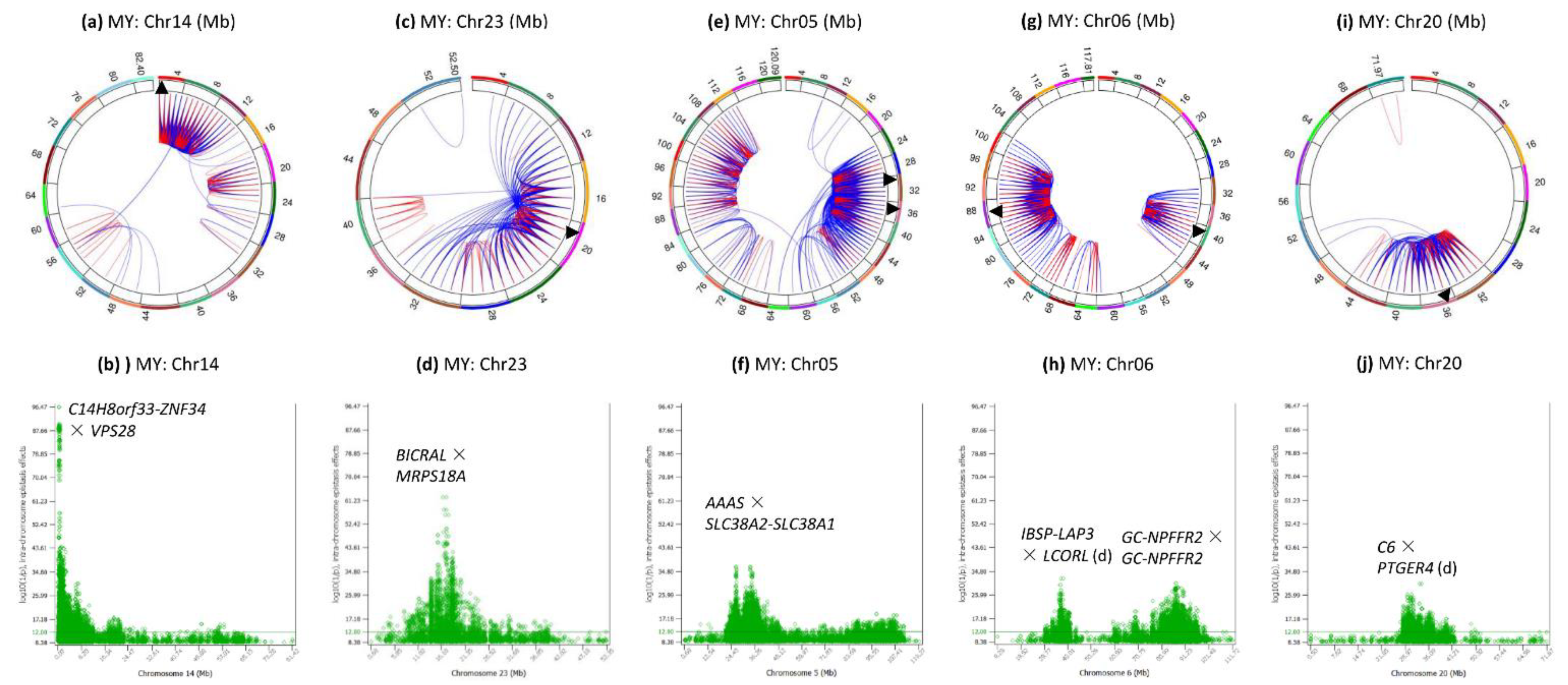
3.4. Intra-Chromosome Epistasis Effects of Milk Yield
3.5. Intra-Chromosome Epistasis Effects of Protein Yield
3.6. Intra-Chromosome Epistasis Effects of Fat Yield
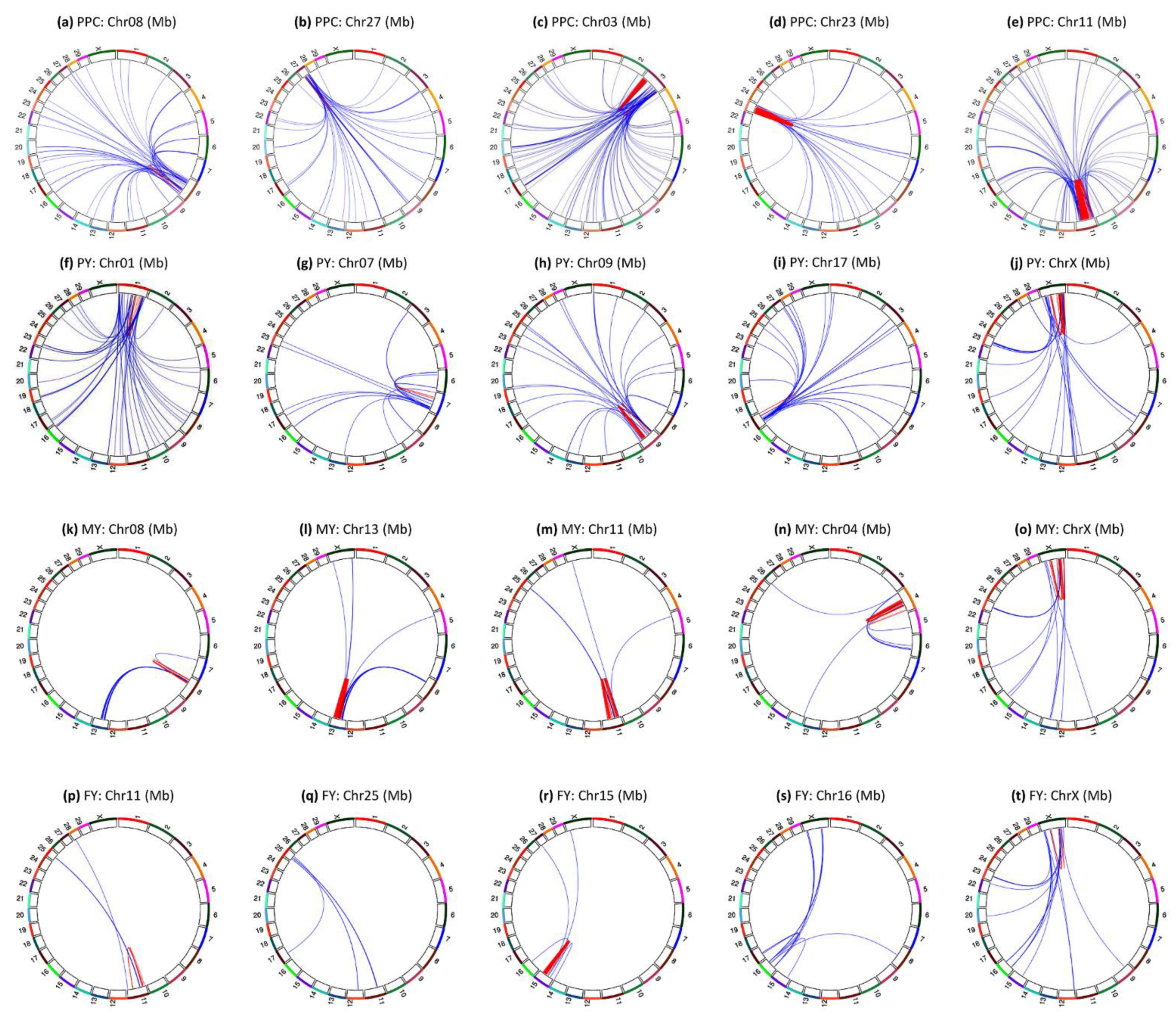
3.7. Inter-Chromosome Epistasis Effects of Fat Percentages
3.8. Inter-Chromosome Epistasis Effects of Protein Percentage and Yield Traits
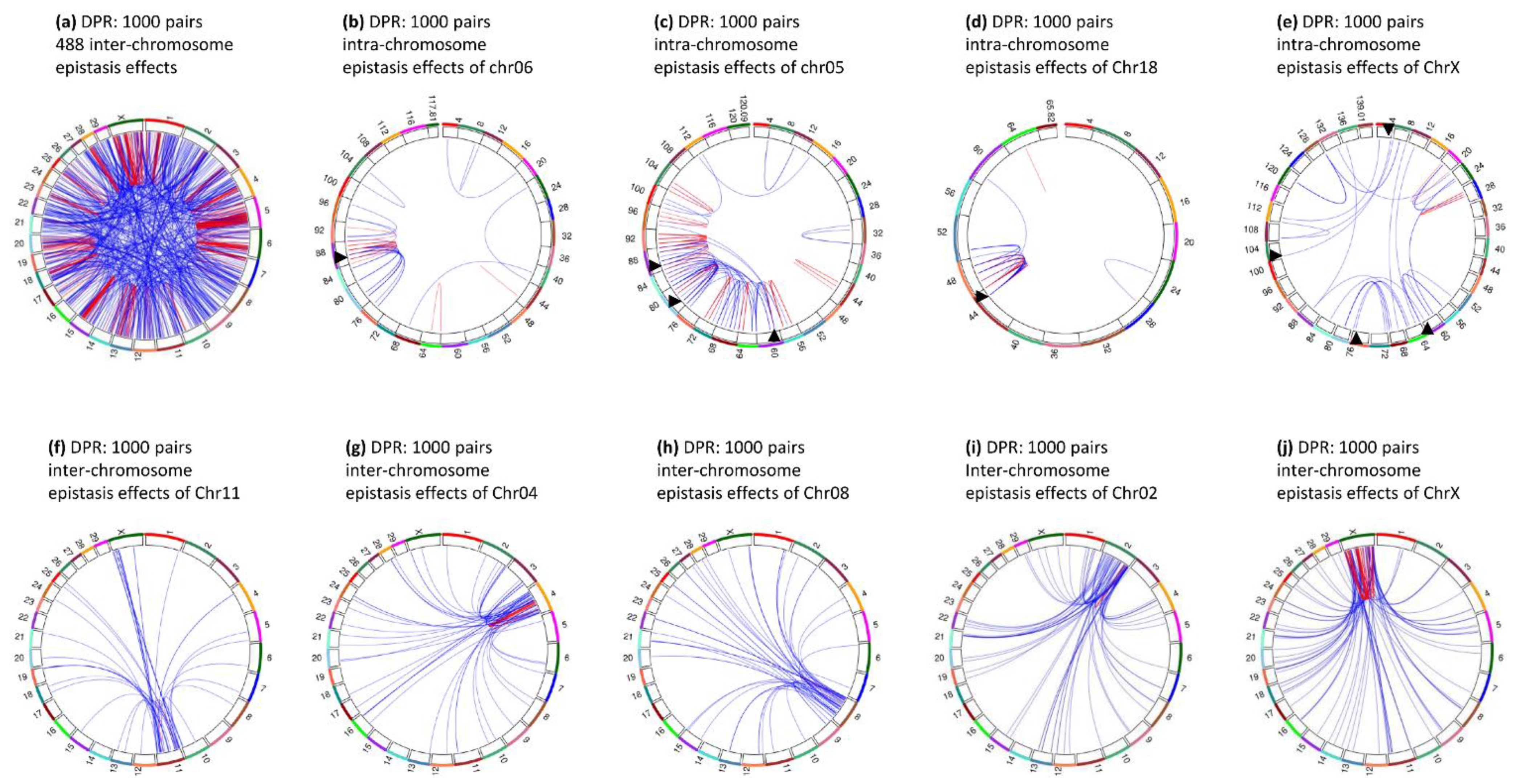
3.9. Epistasis Effects of Daughter Pregnancy Rate
4. Discussion
4.1. Complex Epistasis Effects Existed in U.S. Holstein Cattle
4.2. Genetic Selection Based on Genome-Wide SNP Additive Effects Likely Accounted for Most Intra-Chromosome A × A Effects
4.3. Inter-Chromosome Epistasis Effects Could Be a Genetic Mechanism for Lack of Selection Response and Low Heritability
4.4. Chr14-Specific Inter-Chromosome A × A Epistasis Effects Increase the Statistical Confidence of the Epistasis Results
4.5. An Intergenic Variant May Have an Important Role for Inter-Chromosome Epistasis Effects of Fat Percentage
4.6. Causal or Linked Epistasis Effects
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mackay, T.F. Epistasis and quantitative traits: Using model organisms to study gene–gene interactions. Nat. Rev. Genet. 2014, 15, 22–33. [Google Scholar] [CrossRef] [Green Version]
- Carlborg, Ö.; Haley, C.S. Epistasis: Too often neglected in complex trait studies? Nat. Rev. Genet. 2004, 5, 618–625. [Google Scholar] [CrossRef]
- Cordell, H.J. Epistasis: What it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 2002, 11, 2463–2468. [Google Scholar] [CrossRef] [Green Version]
- Phillips, P.C. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008, 9, 855–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.D.; Van Steen, K. The search for gene-gene interactions in genome-wide association studies: Challenges in abundance of methods, practical considerations, and biological interpretation. Ann. Transl. Med. 2018, 6, 157. [Google Scholar] [CrossRef] [PubMed]
- Pryce, J.; Bolormaa, S.; Chamberlain, A.; Bowman, P.; Savin, K.; Goddard, M.; Hayes, B. A validated genome-wide association study in 2 dairy cattle breeds for milk production and fertility traits using variable length haplotypes. J. Dairy Sci. 2010, 93, 3331–3345. [Google Scholar] [CrossRef] [Green Version]
- Cole, J.B.; Wiggans, G.R.; Ma, L.; Sonstegard, T.S.; Lawlor, T.J.; Crooker, B.A.; Van Tassell, C.P.; Yang, J.; Wang, S.; Matukumalli, L.K.; et al. Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary US Holstein cows. BMC Genom. 2011, 12, 408. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Shen, B.; O’Connell, J.R.; VanRaden, P.M.; Cole, J.B.; Ma, L. Dissection of additive, dominance, and imprinting effects for production and reproduction traits in Holstein cattle. BMC Genom. 2017, 18, 425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raven, L.-A.; Cocks, B.G.; Hayes, B.J. Multibreed genome wide association can improve precision of mapping causative variants underlying milk production in dairy cattle. BMC Genom. 2014, 15, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Wiggans, G.; Wang, S.; Sonstegard, T.; Yang, J.; Crooker, B.; Cole, J.; Van Tassell, C.; Lawlor, T.; Da, Y. Effect of sample stratification on dairy GWAS results. BMC Genom. 2012, 13, 536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouwman, A.C.; Daetwyler, H.D.; Chamberlain, A.J.; Ponce, C.H.; Sargolzaei, M.; Schenkel, F.S.; Sahana, G.; Govignon-Gion, A.; Boitard, S.; Dolezal, M. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 2018, 50, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Rothammer, S.; Seichter, D.; Förster, M.; Medugorac, I. A genome-wide scan for signatures of differential artificial selection in ten cattle breeds. BMC Genom. 2013, 14, 908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Jorjani, H.; Carlborg, Ö. A genome-wide association study using international breeding-evaluation data identifies major loci affecting production traits and stature in the Brown Swiss cattle breed. BMC Genom. 2012, 13, 82. [Google Scholar] [CrossRef] [Green Version]
- Bolormaa, S.; Pryce, J.; Hayes, B.; Goddard, M. Multivariate analysis of a genome-wide association study in dairy cattle. J. Dairy Sci. 2010, 93, 3818–3833. [Google Scholar] [CrossRef] [Green Version]
- Pryce, J.E.; Haile-Mariam, M.; Goddard, M.E.; Hayes, B.J. Identification of genomic regions associated with inbreeding depression in Holstein and Jersey dairy cattle. Genet. Sel. Evol. 2014, 46, 71. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, M.-P.; Govignon-Gion, A.; Croiseau, P.; Fritz, S.; Hozé, C.; Miranda, G.; Martin, P.; Barbat-Leterrier, A.; Letaïef, R.; Rocha, D. Within-breed and multi-breed GWAS on imputed whole-genome sequence variants reveal candidate mutations affecting milk protein composition in dairy cattle. Genet. Sel. Evol. 2017, 49, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, J.; Bickhart, D.; Wiggans, G.; Tooker, M.; O’Connell, J.; Jiang, J.; Ron, M.; VanRaden, P. Determination of quantitative trait nucleotides by concordance analysis between quantitative trait loci and marker genotypes of US Holsteins. J. Dairy Sci. 2018, 101, 9089–9107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Littlejohn, M.D.; Tiplady, K.; Fink, T.A.; Lehnert, K.; Lopdell, T.; Johnson, T.; Couldrey, C.; Keehan, M.; Sherlock, R.G.; Harland, C. Sequence-based association analysis reveals an MGST1 eQTL with pleiotropic effects on bovine milk composition. Sci. Rep. 2016, 6, 25376. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, L.; Prakapenka, D.; VanRaden, P.M.; Cole, J.B.; Da, Y. A large-scale genome-wide association study in US Holstein cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef]
- Jiang, J.; Cole, J.B.; Freebern, E.; Da, Y.; VanRaden, P.M.; Ma, L. Functional annotation and Bayesian fine-mapping reveals candidate genes for important agronomic traits in Holstein bulls. Commun. Biol. 2019, 2, 212. [Google Scholar] [CrossRef]
- Barendse, W.; Harrison, B.E.; Hawken, R.J.; Ferguson, D.M.; Thompson, J.M.; Thomas, M.B.; Bunch, R.J. Epistasis between calpain 1 and its inhibitor calpastatin within breeds of cattle. Genetics 2007, 176, 2601–2610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knaust, J.; Hadlich, F.; Weikard, R.; Kuehn, C. Epistatic interactions between at least three loci determine the “rat-tail” phenotype in cattle. Genet. Sel. Evol. 2016, 48, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suchocki, T.; Komisarek, J.; Szyda, J. Testing candidate gene effects on milk production traits in dairy cattle under various parameterizations and modes of inheritance. J. Dairy Sci. 2010, 93, 2703–2717. [Google Scholar] [CrossRef] [Green Version]
- Yao, C.; Spurlock, D.; Armentano, L.; Page Jr, C.; VandeHaar, M.; Bickhart, D.; Weigel, K. Random Forests approach for identifying additive and epistatic single nucleotide polymorphisms associated with residual feed intake in dairy cattle. J. Dairy Sci. 2013, 96, 6716–6729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiggans, G.R.; Cole, J.B.; Hubbard, S.M.; Sonstegard, T.S. Genomic selection in dairy cattle: The USDA experience. Annu. Rev. Anim. Biosci. 2017, 5, 309–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanRaden, P. Practical implications for genetic modeling in the genomics era. J. Dairy Sci. 2016, 99, 2405–2412. [Google Scholar] [CrossRef]
- Ma, L.; Jiang, J.; Prakapenka, D.; Cole, J.; Da, Y. Approximate generalized least squares method for large-scale genome-wide association study. J. Dairy Sci. 2019, 102, 30. [Google Scholar]
- Ma, L.; Runesha, H.B.; Dvorkin, D.; Garbe, J.; Da, Y. Parallel and serial computing tools for testing single-locus and epistatic SNP effects of quantitative traits in genome-wide association studies. BMC Bioinform. 2008, 9, 315. [Google Scholar] [CrossRef] [Green Version]
- Weeks, N.T.; Luecke, G.R.; Groth, B.M.; Kraeva, M.; Ma, L.; Kramer, L.M.; Koltes, J.E.; Reecy, J.M. High-performance epistasis detection in quantitative trait GWAS. Int. J. High Perform. Comput. Appl. 2016, 32, 321–336. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.; London, N.R.; Ma, L.; Dvorkin, D.; Da, Y. Detection of SNP epistasis effects of quantitative traits using an extended Kempthorne model. Physiol. Genom. 2007, 28, 46–52. [Google Scholar] [CrossRef]
- Wang, S.; Dvorkin, D.; Da, Y. SNPEVG: A graphical tool for GWAS graphing with mouse clicks. BMC Bioinform. 2012, 13, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnstone, O.; Lasko, P. Interaction with eIF5B is essential for Vasa function during development. Development 2004, 131, 4167–4178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Holzman, C.; Heng, Y.J.; Kibschull, M.; Lye, S.J.; Vazquez, A. EBF1 gene mRNA levels in maternal blood and spontaneous preterm birth. Reprod. Sci. 2020, 27, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Sasaki, S.; Gotoh, Y.; Nakamura, Y.; Aoyagi, Y.; Kawahara, T.; Sugimoto, Y. Genetic variants related to gap junctions and hormone secretion influence conception rates in cows. Proc. Natl. Acad. Sci. USA 2013, 110, 19495–19500. [Google Scholar] [CrossRef] [Green Version]
- Cochran, S.D.; Cole, J.B.; Null, D.J.; Hansen, P.J. Discovery of single nucleotide polymorphisms in candidate genes associated with fertility and production traits in Holstein cattle. BMC Genom. 2013, 14, 49. [Google Scholar] [CrossRef] [Green Version]
- Gu, X.; Li, H.; Chen, X.; Zhang, X.; Mei, F.; Jia, M.; Xiong, C. PEX10, SIRPA-SIRPG, and SOX5 gene polymorphisms are strongly associated with nonobstructive azoospermia susceptibility. J. Assist. Reprod. Genet. 2019, 36, 759–768. [Google Scholar] [CrossRef]






| Intra-Chromosome Epistasis (% of Intra-Chromosome SNP Pairs: 3.6) | Inter-Chromosome Epistasis (% of Inter-Chromosome SNP Pairs: 96.4) | |||||
|---|---|---|---|---|---|---|
| Freq (%) | Log10(1/p) | Pairs with log10(1/p) ≥ 12 | Freq (%) | Log10(1/p) | Pairs with log10(1/p) ≥ 12 | |
| FPC | 98.1 | 13–537 | 49,046 (98.1%) | 1.9 | 13–23 | 954 (1.9%) |
| PPC | 98.4 | 17–211 | 49,206 (98.4%) | 1.6 | 17–82 | 794 (1.6%) |
| MY | 89.4 | 9–97 | 10,440 (20.9%) | 10.6 | 13–19 | 68 (0.6%) |
| FY | 70.1 | 7–40 | 4715 (9.4%) | 29.9 | 7–14 | 48 (1.0%) |
| PY | 61.7 | 7–51 | 4325 (8.7%) | 39.3 | 7–21 | 312 (6.7%) |
| DPR | 15.8 | 6–13 | 1 (0.002%) | 84.2 | 6–11 | 0 |
| Intra-Chromosome Epistasis | Inter-Chromosome Epistasis | ||||||
|---|---|---|---|---|---|---|---|
| A × A | A × D and D × A | D × D | A × A | A × D and D × A | D × D | ||
| FPC | Count | 47,752 | 1023 | 271 | 952 | 0 | 1 |
| % | 95.5 | 2.1 | 0.5 | 1.9 | 0.0 | 0.0 | |
| PPC | Count | 45,379 | 2936 | 1116 | 10 | 3 | 780 |
| % | 90.8 | 5.9 | 2.2 | 0.0 | 0.0 | 1.6 | |
| MY | Count | 36,387 | 5978 | 2355 | 5197 | 35 | 48 |
| % | 72.8 | 12.0 | 4.7 | 10.4 | 0.1 | 0.1 | |
| FY | Count | 30,225 | 3767 | 1401 | 14,317 | 188 | 92 |
| % | 60.5 | 7.5 | 2.8 | 28.6 | 0.4 | 0.2 | |
| PY | Count | 20,582 | 7461 | 2840 | 17,468 | 200 | 1448 |
| % | 41.2 | 14.9 | 5.7 | 34.9 | 0.4 | 2.9 | |
| DPR | Count | 7122 | 602 | 196 | 26,430 | 10,531 | 5118 |
| % | 14.2 | 1.2 | 0.4 | 52.9 | 21.1 | 10.2 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prakapenka, D.; Liang, Z.; Jiang, J.; Ma, L.; Da, Y. A Large-Scale Genome-Wide Association Study of Epistasis Effects of Production Traits and Daughter Pregnancy Rate in U.S. Holstein Cattle. Genes 2021, 12, 1089. https://doi.org/10.3390/genes12071089
Prakapenka D, Liang Z, Jiang J, Ma L, Da Y. A Large-Scale Genome-Wide Association Study of Epistasis Effects of Production Traits and Daughter Pregnancy Rate in U.S. Holstein Cattle. Genes. 2021; 12(7):1089. https://doi.org/10.3390/genes12071089
Chicago/Turabian StylePrakapenka, Dzianis, Zuoxiang Liang, Jicai Jiang, Li Ma, and Yang Da. 2021. "A Large-Scale Genome-Wide Association Study of Epistasis Effects of Production Traits and Daughter Pregnancy Rate in U.S. Holstein Cattle" Genes 12, no. 7: 1089. https://doi.org/10.3390/genes12071089
APA StylePrakapenka, D., Liang, Z., Jiang, J., Ma, L., & Da, Y. (2021). A Large-Scale Genome-Wide Association Study of Epistasis Effects of Production Traits and Daughter Pregnancy Rate in U.S. Holstein Cattle. Genes, 12(7), 1089. https://doi.org/10.3390/genes12071089






