Abstract
Pericentromeric heterochromatin is mostly composed of repetitive DNA sequences prone to aberrant recombination. Cells have developed highly specialized mechanisms to enable ‘safe’ homologous recombination (HR) repair while preventing aberrant recombination in this domain. Understanding heterochromatin repair responses is essential to understanding the critical mechanisms responsible for genome integrity and tumor suppression. Here, we review the tools, approaches, and methods currently available to investigate double-strand break (DSB) repair in pericentromeric regions, and also suggest how technologies recently developed for euchromatin repair studies can be adapted to characterize responses in heterochromatin. With this ever-growing toolkit, we are witnessing exciting progress in our understanding of how the ‘dark matter’ of the genome is repaired, greatly improving our understanding of genome stability mechanisms.
1. Introduction
Pericentromeric heterochromatin (hereafter ‘heterochromatin’) occupies about 30% of fly and human genomes [1,2,3,4] (Figure 1), is enriched for silencing epigenetic marks (i.e., H3K9me2/3, and Drosophila HP1a [5] or mammalian HP1α/β, and is mostly composed of repeated DNA sequences. For example, in Drosophila, about half of these sequences are ‘satellite’ repeats (mostly 5-base-pair sequences repeated for hundreds of kilobases to megabases) and the rest are transposable elements, scrambled repeats, and only about 250 isolated genes [2,3,4] (reviewed in [6,7]).
Pericentromeric repeats are functionally and structurally distinct from lamina-associated domains (LADs) [8], which are distributed along the chromosome arms (Figure 1). In contrast to LADs, pericentromeric sequences are not usually associated with the nuclear periphery (see, for example, refs. [7,9,10,11,12,13,14,15]). Pericentromeric repeats are also distinct from Polycomb-mediated silenced regions, which are typically enriched for H3K27me3 and encompass LADs and other silenced domains [16] (reviewed in [17,18]). Pericentromeric heterochromatin is absent in budding yeast; thus, repair in these sequences has mostly been studied in Drosophila, mouse, and human cells.
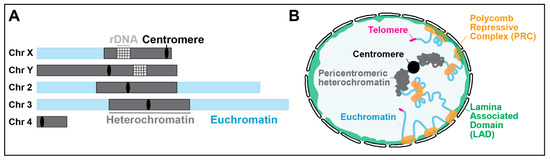
Figure 1.
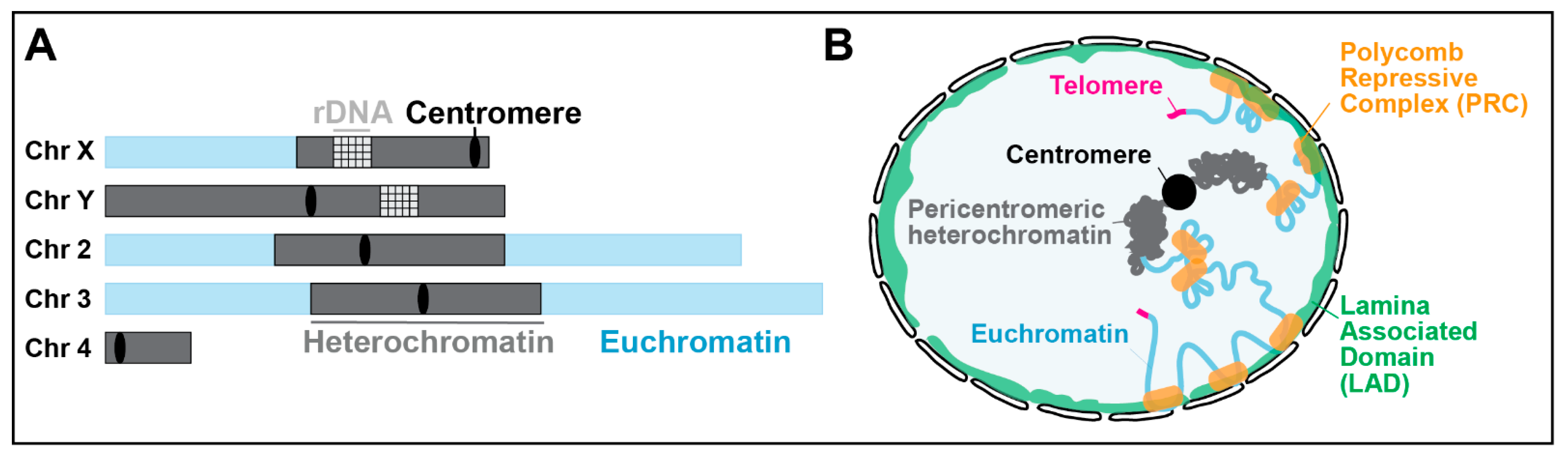
Chromosomal and nuclear position of pericentromeric heterochromatin. (A) Schematic view of Drosophila chromosomes showing the position and extent of pericentromeric heterochromatin (adapted from [5]). The Y chromosome is mostly heterochromatin. (B) Position of pericentromeric heterochromatin relative to the nuclear periphery in a Drosophila nucleus, which is distinct from LADs, telomeric repeats, and Polycomb-repressed domains (adapted from [7,18]).
Figure 1.
Chromosomal and nuclear position of pericentromeric heterochromatin. (A) Schematic view of Drosophila chromosomes showing the position and extent of pericentromeric heterochromatin (adapted from [5]). The Y chromosome is mostly heterochromatin. (B) Position of pericentromeric heterochromatin relative to the nuclear periphery in a Drosophila nucleus, which is distinct from LADs, telomeric repeats, and Polycomb-repressed domains (adapted from [7,18]).
Given the abundance of repeated sequences, heterochromatin poses unique challenges to homologous recombination (HR) repair of DNA double-strand breaks (DSBs) (reviewed in [6,7,19]). DSBs are mostly repaired by HR or non-homologous end joining (NHEJ) (Figure 2). NHEJ is characterized by the direct re-joining of the two ends, which frequently generates small mutations at the repair site [20]. Conversely, HR initiates with a resection to form 3′ single-stranded DNA (ssDNA) that invades ‘donor’ homologous sequences used as templates for DNA synthesis and repair [21]. In single-copy sequences, a unique donor is present on the sister chromatid or the homologous chromosome, and HR is largely ‘error free’. In heterochromatin, however, the availability of up to millions of potential donor sequences associated with different chromosomes can initiate unequal sister chromatid exchange, or intra/inter-chromosomal recombination, leading to deletions, duplications, translocations, release of extrachromosomal DNA circles, or the formation of dicentric or acentric chromosomes [10,15,22,23,24,25,26] (reviewed in [6,7,19,27]).
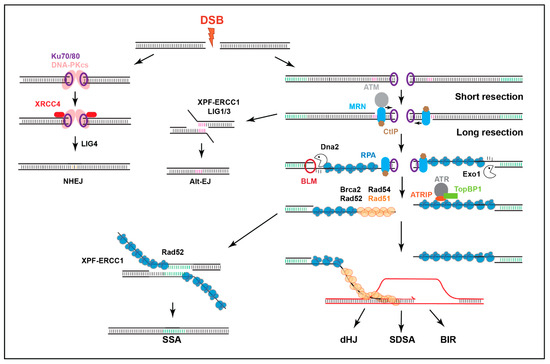
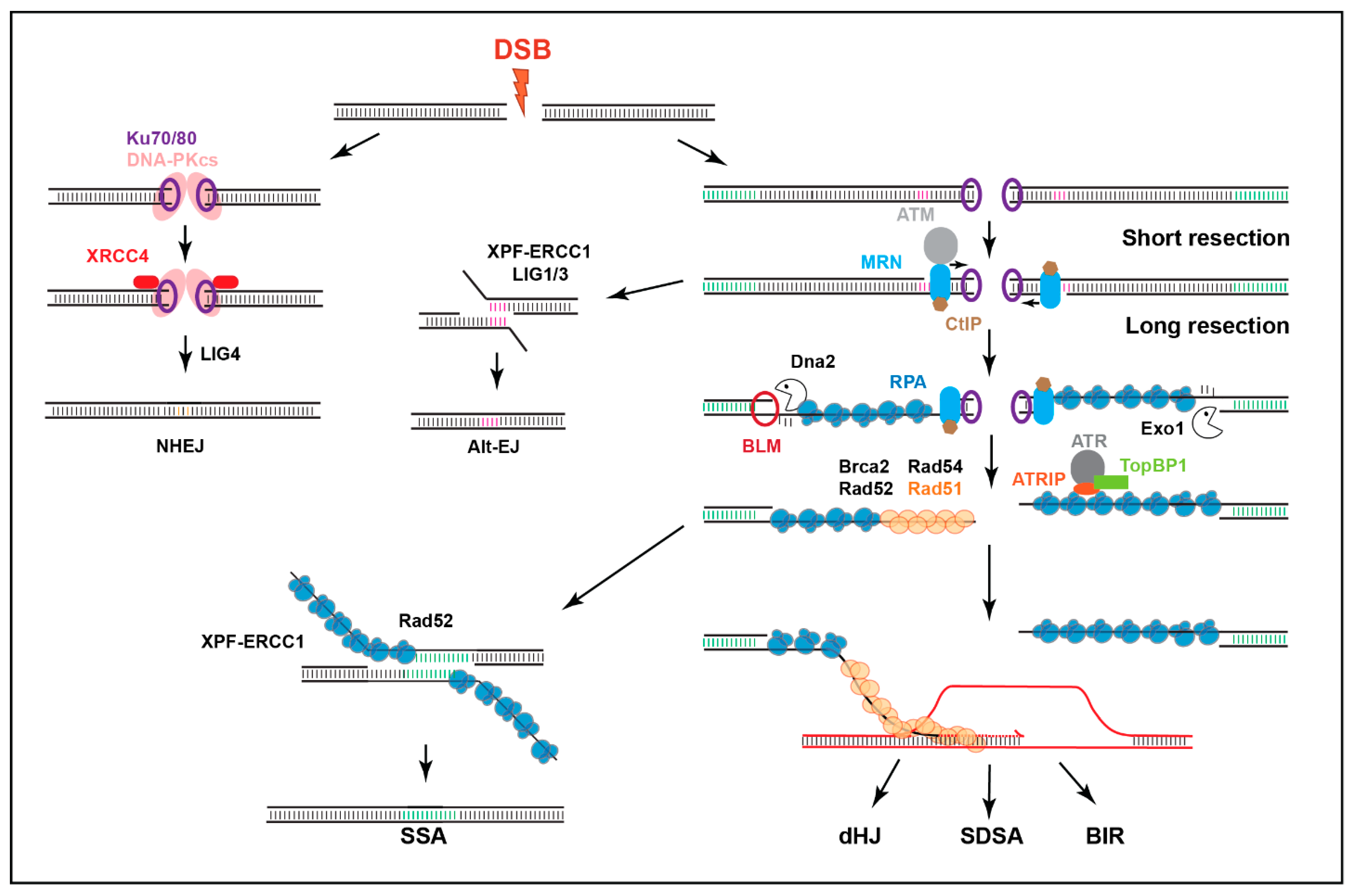
Figure 2.
Main DSB repair mechanisms. DSBs are mainly repaired by HR (mostly in S/G2) or NHEJ. NHEJ involves no or minimal processing of the DSB. HR relies on DSB resection to form ssDNA that ‘invades’ homologous sequences on the sister chromatid or homologous chromosomes to enable DNA synthesis and repair. In mitotic cells, synthesis-dependent strand annealing (SDSA) is the most common HR pathway, although break-induced replication (BIR) also frequently occurs near telomeres. Double-Holliday junction (dHJ) repair is better characterized in meiosis. Extensively resected DSBs can also utilize the Rad51-independent pathway of single strand annealing (SSA) when repeated sequences are available, while alternative-end joining pathways (Alt-EJ), such as MMEJ, involve limited resection to expose micro-homologies.
Despite the risk of aberrant recombination, HR appears to be a preferred pathway for heterochromatin repair in S/G2, i.e., when both HR and NHEJ are available [10,12,28]. Studies across different model systems have identified specialized mechanisms enabling ‘safe’ HR repair in heterochromatin, while preventing aberrant recombination. In Drosophila cells, where heterochromatin forms a distinct nuclear domain [10,14], HR repair of heterochromatin requires the relocalization of repair sites to the nuclear periphery before recruitment of the strand invasion component Rad51 [10,15,24,25,26] (reviewed in [6,29,30,31]). Similarly, in mouse cells, DSBs relocalize to the periphery of heterochromatin domains (called ‘chromocenters’), before Rad51 recruitment [11,12,25,32] (reviewed in [6,7]). In both contexts, relocalization likely promotes ‘safe’ HR repair while preventing aberrant recombination by moving the broken site and its repair templates (on the homologous chromosome or the sister chromatid [25,33]) away from the bulk of other repeated sequences before strand invasion (reviewed in [6]).
Various techniques have been developed to study DSB repair, as recently reviewed [34]. Specific approaches have also been applied to the study of DSB repair in other ‘silent’ genomic regions, such as telomeres [35], LADs [36], Polycomb-rich regions [37,38], other silenced repeats [39], and untranscribed loci (inactive genes or intergenic regions) [40] (reviewed in [18]). Additional studies have characterized the responses in pericentromeric regions to damage other than DSBs, such as from UV exposure [41], and heterochromatin replication or over-replication [42,43], and will not be covered by this review. Here, we focus on the main approaches that have been applied to the characterization of DSB repair in pericentromeric regions (Figure 3), including the main conclusions derived from different studies, and the strengths (+) or limitations (−) of different techniques. We also propose how other methods developed to study repair in euchromatin can be adapted to study heterochromatin repair.
2. Approaches to Study Heterochromatic DSB Responses
2.1. Imaging of Ionizing Radiation (IR)-Induced Repair Foci
HR repair can be followed cytologically because the recruitment of many HR repair components to DSBs gives rise to visible foci [10,44,45]. The formation and disappearance of different repair foci reflects HR progression, including: DSB detection and signaling (e.g., Mdc1 or γH2AX foci in mammalian cells, and Mu2 or γH2Av foci in Drosophila cells [10,15,25]); resection (e.g., RPA, ATRIP, TopBP1 [10]); Rad51 loading; and strand invasion (e.g., Brca2, Rad51, or Rad54 foci [10]) (see Glossary and Figure 2). Heterochromatin also forms distinct domains in Drosophila and mouse cells, which can be visualized by tagging or staining for heterochromatin-specific proteins or chromatin marks (e.g., HP1 proteins or H3K9me2/3 [9,10,15,25,26,46], Figure 3), enabling characterization of repair kinetics and dynamics relative to heterochromatin domains of these model systems.
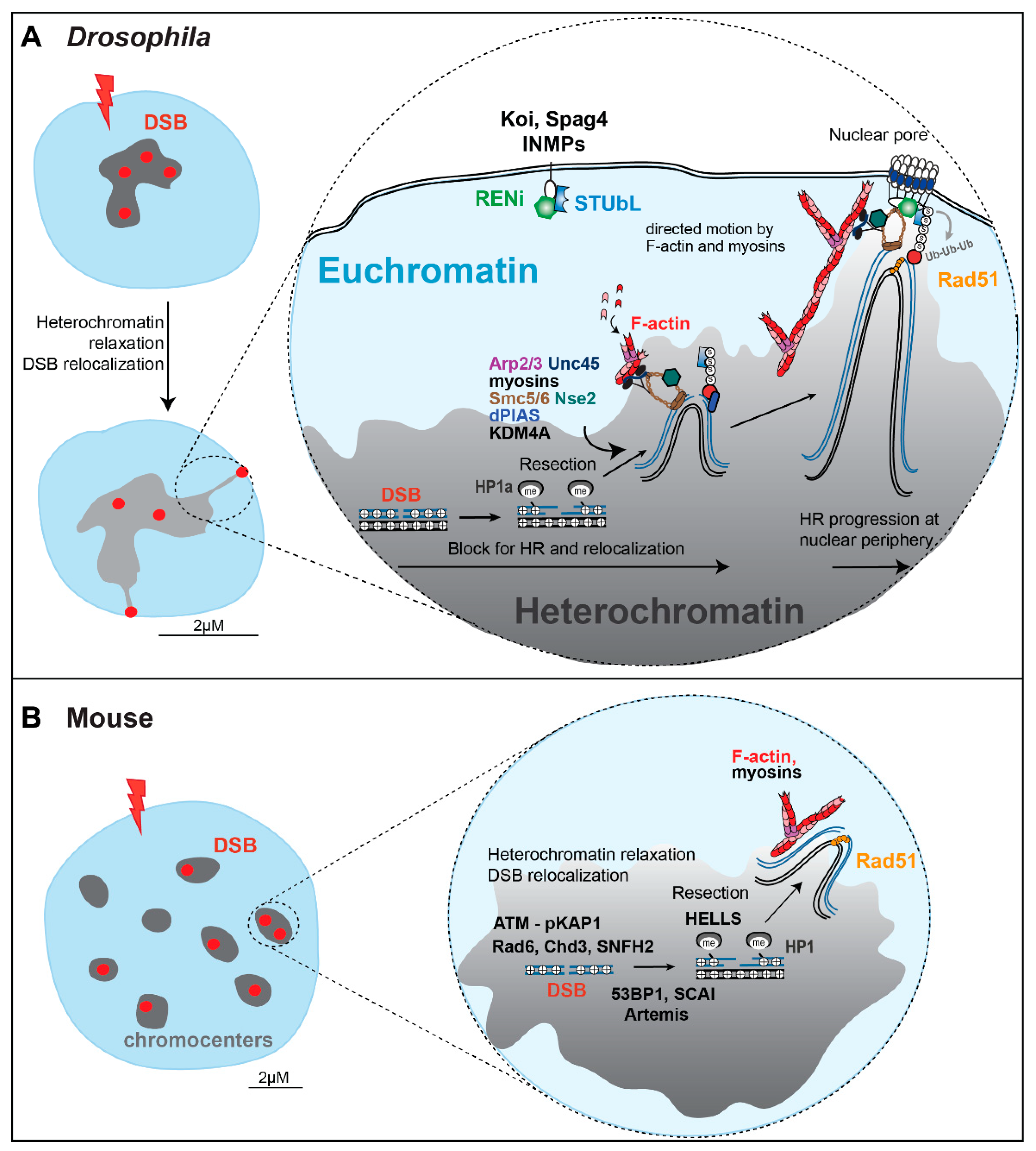
Live and fixed cell imaging of IR-induced repair foci has enabled major advances in our understanding of the molecular mechanisms involved in heterochromatin repair [9,10,11,15,24,25,26]. For example, live imaging of Drosophila cells revealed that Mdc1/Mu2 and ATRIP or TopBP1 foci form with high efficiency inside the heterochromatin domain [10,15,25], reversing the initial assumption that DNA damage detection, signaling, and resection are delayed by compaction and silencing inside the domain [47,48] (Figure 4). ATRIP and TopBP1 foci appear sooner and are brighter in heterochromatin relative to euchromatin, suggesting that the HR steps leading to resection occur even faster in heterochromatin [10]. These studies also revealed a striking relocalization pathway, with repair foci leaving the heterochromatin domain and reaching the nuclear periphery 1 h after IR, where Rad51 is recruited [10,15,24,25] (Figure 4).
A combination of live imaging and RNAi depletion studies in Drosophila cells established that SUMOylation by the heterochromatin-enriched complex Smc5/6, its SUMO-E3 ligases (Nse2/Cerv, and Nse2/Qjt), and the E3 SUMO ligase dPIAS, is required to halt HR progression inside the heterochromatin domain [10,15,24]. At the nuclear periphery, repair sites associate with nuclear pores and inner nuclear membrane proteins (e.g., the LINC complex subunits Koi and Spag4), where a SUMO-targeted ubiquitin ligase (STUbL)/RENi complex likely mediates the repair restart through the ubiquitination of SUMOylated proteins (whose identity is still unknown) (Figure 4), followed by their degradation or reactivation [15].
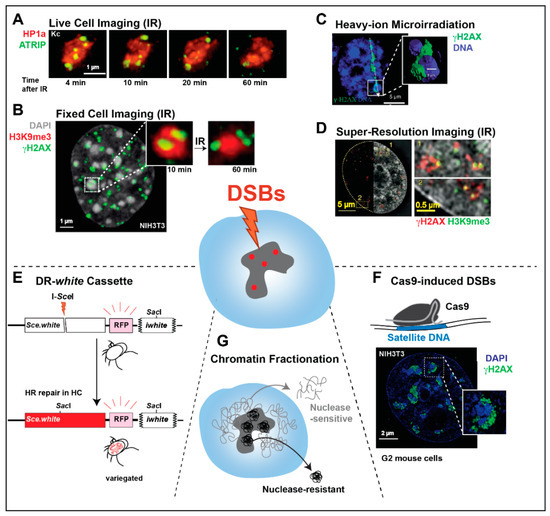
Figure 3.
Summary of the main approaches used to study heterochromatic DSB repair. (A) Live imaging of Drosophila Kc cells treated with a dose of 5 Gy X-rays shows the relocalization of ATRIP foci to outside the heterochromatic domain (marked by HP1a), at the indicated time points after ionizing radiation (IR) (image from [10]). (B) IF analysis of mouse NIH3T3 cells fixed 10 min after exposure to a dose of 5 Gy X-rays shows γH2AX foci inside DAPI-bright chromocenters. The magnified detail also shows foci predominantly outside the domain 60 min after IR. (C) Image and 3D reconstruction of a mouse embryonic fibroblast (MEF) cell 60 min after exposure to heavy-ion irradiation shows a linear damage streak (γH2AX) bent around a DAPI-bright chromocenter (image adapted from [32]). (D) IF analysis by 3D-SIM of γH2AX and H3K9me3 signals 24 h after exposure to a dose of 10 Gy X-rays shows damage foci associated with silenced sequences in HeLa cells (image from [49]). (E) HR repair of a DR-white cassette inserted in pericentromeric sequences of Drosophila results in variegated-red eyes in the progeny. (F) IF analysis of Cas9-induced DSBs in the major satellite DNA of mouse NIH3T3 cells shows γH2AX signals at the periphery of DAPI-bright chromocenters during HR repair (image from [12]). (G) Chromatin fractionation techniques can enrich nuclear extracts for MNase-resistant heterochromatin, enabling the characterization of heterochromatin-associated proteins during repair.
Figure 3.
Summary of the main approaches used to study heterochromatic DSB repair. (A) Live imaging of Drosophila Kc cells treated with a dose of 5 Gy X-rays shows the relocalization of ATRIP foci to outside the heterochromatic domain (marked by HP1a), at the indicated time points after ionizing radiation (IR) (image from [10]). (B) IF analysis of mouse NIH3T3 cells fixed 10 min after exposure to a dose of 5 Gy X-rays shows γH2AX foci inside DAPI-bright chromocenters. The magnified detail also shows foci predominantly outside the domain 60 min after IR. (C) Image and 3D reconstruction of a mouse embryonic fibroblast (MEF) cell 60 min after exposure to heavy-ion irradiation shows a linear damage streak (γH2AX) bent around a DAPI-bright chromocenter (image adapted from [32]). (D) IF analysis by 3D-SIM of γH2AX and H3K9me3 signals 24 h after exposure to a dose of 10 Gy X-rays shows damage foci associated with silenced sequences in HeLa cells (image from [49]). (E) HR repair of a DR-white cassette inserted in pericentromeric sequences of Drosophila results in variegated-red eyes in the progeny. (F) IF analysis of Cas9-induced DSBs in the major satellite DNA of mouse NIH3T3 cells shows γH2AX signals at the periphery of DAPI-bright chromocenters during HR repair (image from [12]). (G) Chromatin fractionation techniques can enrich nuclear extracts for MNase-resistant heterochromatin, enabling the characterization of heterochromatin-associated proteins during repair.
In addition, the histone demethylase Kdm4A is required to lower H3K56me3 levels in response to IR in Drosophila cells, and for the relocalization of heterochromatic repair foci, suggesting a role for Kdm4-mediated H3K56me3 reduction in these dynamics [50].
Live and fixed cell imaging studies also identified a striking network of transient nuclear actin filaments (F-actin) responsible for the directed motion of repair sites to the nuclear periphery [6,25,29,51,52] (Figure 4). Filaments are assembled at heterochromatic DSBs by the actin nucleator Arp2/3 and extend toward the nuclear periphery [25]. Relocalization is also mediated by nuclear myosins (i.e., Myo1A, Myo1B, and MyoV) that interact with Smc5/6, and are activated by Unc45 downstream from Smc5/6, to promote the directed motion of repair sites along the filaments [25]. Critically, extensive relocalization and directed motions are not typically detected at euchromatic DSBs [25]. Myosins are also not involved in the motion of euchromatic repair foci, revealing that long-range motions are specifically important for the repair of heterochromatin, and not of euchromatin [25].
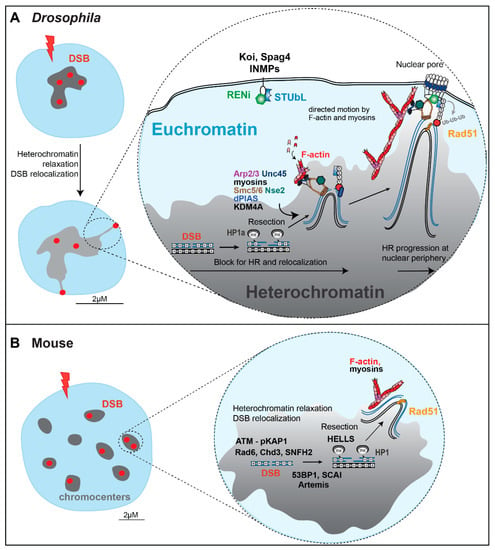
Figure 4.
Heterochromatin repair mechanisms in Drosophila and mouse cells. (A,B) Molecular mechanisms mediating HR repair in Drosophila and mouse cells, with some of the main factors regulating DSB resection, chromatin remodeling, heterochromatin relaxation, relocalization of repair sites, and HR progression (also reviewed in [6]). S: SUMOylation. Ub: Ubiquitinaltion. INMP: Inner nuclear membrane proteins. See details in the text.
Figure 4.
Heterochromatin repair mechanisms in Drosophila and mouse cells. (A,B) Molecular mechanisms mediating HR repair in Drosophila and mouse cells, with some of the main factors regulating DSB resection, chromatin remodeling, heterochromatin relaxation, relocalization of repair sites, and HR progression (also reviewed in [6]). S: SUMOylation. Ub: Ubiquitinaltion. INMP: Inner nuclear membrane proteins. See details in the text.
Importantly, for these studies live imaging needs to be tightly calibrated to enable long kinetics with sufficient time points for mean-square displacement (MSD) analyses. Successful execution of these experiments relies on careful selection of fluorescent tags and microscopy techniques that are particularly gentle on the samples, such as working with a bright-field fluorescent microscope and applying deconvolution processing to the images [25,51,52].
These studies and computer simulations also established that traditional MSD analyses of repair focus tracks are unable to detect directed motions in the context of mixed trajectories, in which directed motions start asynchronously for a population of foci, and in which directed motions are preceded and followed by sub-diffusive motion [6,25,30,52]. Instead, dedicated methods that identify the time intervals of directed motions within longer trajectories need to be applied to these contexts [25,30,52].
Fixed cell imaging studies in mouse NIH3T3 cells show that relocalization of IR-induced heterochromatic repair foci relies on actin polymerization and myosins also in this context [25], revealing conserved pathways between Drosophila and mammalian cells [25].
In addition, live imaging of HP1 proteins in response to IR identified a significant expansion of heterochromatin domains in Drosophila and mouse cells, corresponding to a volume increase of up to 50% in domain size [10,26,49]. This is dependent on checkpoint (particularly ATR) and resection in Drosophila cells [10]. While the significance of this expansion remains unclear, it might reflect a relaxation of the domain to promote damage signaling [46], repair protein accessibility, or increased dynamics [6,10].
Finally, fixed cell imaging analysis of γH2AX foci in mouse cells shows that heterochromatic DSBs are repaired with slower kinetics than euchromatic DSBs; thus, foci remaining in the nuclei 8 h after IR are mostly associated with chromocenters [53,54]. Repair kinetics are further delayed by ATM loss specifically in heterochromatin, revealing the importance of ATM in heterochromatin repair [9]. Cell imaging and genetic approaches have identified Kap1 as a critical phosphorylation target in this pathway, leading to Kap1 release from the chromatin, and chromatin relaxation [9]. Similar approaches have also identified the importance of SCAI in ATM signaling for heterochromatin repair [55].
Building on the discovery that persistent DSBs in response to IR are mostly heterochromatic, and that their resolution is facilitated in the absence of Kap1, several labs have identified additional heterochromatin repair components in human cells, using genetic approaches. These studies support a role for Cdh3 dispersal and Acf1-Snf2h activation in heterochromatin relaxation during repair [53,54], the importance of Artemis and 53BP1 in promoting HR in heterochromatin in G2 [28,56], and a role for HELLS in DSB resection in heterochromatin [57].
(+) A major advantage of using IR is that this treatment induces damage in a nearly synchronous manner, facilitating the detection of relocalization events that occur in relatively short time intervals (e.g., within 1 h from damage induction). IR also induces DSBs with high efficiency in both euchromatin and heterochromatin, and dose titration enables DSB induction at physiological levels. Using Drosophila or mouse cells provides unique advantages for imaging approaches, given the organization of heterochromatin in distinct nuclear domains that are easily detectable cytologically. Live cell imaging in response to IR enables the study of 3D dynamics, identifying relocalization pathways and directed motions. Responses to breaks in euchromatin and heterochromatin can be studied within the same cells, enabling direct comparisons and the detection of heterochromatin-specific responses. Another advantage is that the experiments can be performed from very few up to large numbers of cells, facilitating both genetic and biochemical approaches. For fixed and live cell imaging studies, immunofluorescence (IF) or time-lapse approaches are typically completed within a day, providing a quick readout. IR has also been coupled with fluorescent in-situ hybridization (FISH) to follow individual satellites [10]. In Drosophila, highly efficient and cost-effective RNAi techniques further facilitate a systematic exploration of the pathways involved in the relocalization of heterochromatic DSBs. Drosophila Kc cells are also mostly in S/G2, facilitating the study of HR repair. Large cell size facilitates studies in mouse cells, although individual chromocenters are smaller than the Drosophila heterochromatin domain.
(−) IR exposure induces a number of other damages in addition to DSBs [58]. Some repair proteins do not form foci, or form foci that are not visible in live imaging (including most NHEJ components), thus they cannot be studied with these approaches. The techniques described here are mostly applicable to cells with clearly defined ‘domains’ (e.g., mouse and Drosophila cells) [52], limiting what can be studied in human cells. Mouse fibroblasts used in the studies mentioned above are also particularly flat, resulting in low resolution along the Z stacks and more limited information on focus dynamics than what can be defined in Drosophila cells. It is also critical to choose the right imaging conditions to maximize image collection for focus tracking while minimizing photodamage and photobleaching [51,52]. Live imaging is particularly difficult with multiple channels, resulting in the use of fluorescent tags that are typically less bright and less photostable than GFP (e.g., YFP or BFP variants). This limits the number of components that can be simultaneously imaged in each experiment, reducing the amount of information that can be extracted. Live imaging is also more difficult in tissues [26], where more photo-damaging imaging techniques need to be applied. Further, IR exposure requires specific equipment (i.e., X-ray machine or 137Cs-irradiator), which might not be available to many laboratories. Once data is collected, quantification using manual or even semi-automated methods is time consuming [51,52]. Finally, random DSB induction does not enable the characterization of chromatin and DNA damage responses with chromatin immunoprecipitation (ChIP)-type approaches, limiting the resolution of these studies.
2.2. Imaging of Spatially Defined DSBs Induced by Laser or Heavy-Ion Irradiation
Live and fixed cell imaging have also been applied to DSBs induced along linear paths by heavy-ion irradiation or laser beams crossing heterochromatin domains, where knowing the spatial positioning of the damage facilitates the characterization of kinetic and dynamics of the damage response. For example, when heavy ions traverse mouse chromocenters or human perinucleolar heterochromatin, linear γH2AX streaks bend around the heterochromatin domains, consistent with damage relocalization [32] (Figure 3). TUNEL, γH2AX, and RPA IF signals are visible inside the domain shortly after irradiation, indicating that early DSB formation, signaling, and resection, occur quickly inside the chromocenters [32], similar to what is observed in Drosophila cells after IR treatment. This study also confirmed the importance of ATM in resolving heterochromatic DSBs in mouse cells, as ATM−/− MEFs display persistent DSBs associated with the chromocenters. However, similar to what is observed in Drosophila cells, ATM is not required for relocalization [32]. Additionally, the progressive reduction in DNA signals at the sites hit by ion radiation suggests local de-condensation of the chromatin at repair sites [32].
Laser beams present an alternative method to induce spatially-defined DSBs in heterochromatin. DSBs are typically induced using a UV laser mounted on the microscope, after cell sensitization with a DNA intercalator (e.g., Hoechst) or a nucleotide analog (e.g., BrdU) [59]. Alternatively, damage can be directly induced without cell sensitization using a near-infrared (NIR) pulsed laser [60]. Live imaging of mouse cells, in response to UV laser-targeting of the chromocenters (marked by HP1β-GFP), revealed heterochromatin expansion and the importance of HP1β phosphorylation by casein kinase 2 (CK2) in this response [46]. It also established the function of HP1β phosphorylation in γH2AX induction, consistent with a role for heterochromatin relaxation in DSB signaling [46]. Chromocenters have also been imaged with energy-filtering transmission electron microscopy after laser-induced damage in mouse cells, which detected heterochromatin relaxation as a 30–40% reduction in the density of chromatin fibers proximal to DSBs [61].
In Drosophila cells, DSBs induced inside heterochromatin domains by NIR laser treatment result in γH2Av foci inside the domain shortly after damage [10]. At later time points, sites enriched for Rad51 are mutually exclusive with HP1a signals, suggesting local HP1a release or heterochromatin de-condensation during HR progression [10]. However, surprisingly, laser-induced DSBs do not relocalize to outside heterochromatin domains in this context, suggesting that either breaks induced by laser radiation do not relocalize efficiently, or the high energy associated with laser radiation disrupts some aspect of the relocalization pathway [10].
(+) A clear advantage of these approaches is that the damage is directed across the domain or outside the domain in a precise manner, facilitating the identification of heterochromatin-specific responses. Comparisons between responses in euchromatin and heterochromatin can be conducted within the same cells, reducing confounding effects from cell-to-cell variability, and highlighting heterochromatin specific responses. Relocalization of DNA breaks in response to heavy-ion irradiation is easy to quantify as the beam ‘bends’ around the chromocenter. Generating a high number of DSBs within a short distance also amplifies damage signals, thus facilitating detection of low-abundance proteins by IF or live imaging [59]. Microscope-mounted laser radiation enables fast post-damage imaging and detection of very early repair steps or transient protein recruitment. Finally, as mentioned in the previous section, detection techniques with live or fixed cell imaging are fast, providing a quick readout of experiments.
(−) The high energy associated with heavy ions or lasers can affect other components, including proteins responsible for relocalization [10,62]. Similar to IR, heavy ions and lasers induce multiple types of DNA lesions in addition to DSBs, complicating the analysis of DSB-specific responses [59,63]. Additionally, both lasers and ion irradiation induce multiple DSBs in close proximity [59,63,64,65] (clustered DSBs), which behave differently from sparsely-induced DSBs, potentially inducing non-physiological responses [59,64,66]. For example, the complex damage and clustering of multiple breaks induced by high-LET results in lower usage of NHEJ and higher reliance on HR (reviewed in [66]). Further, the sensitization of cells with a thymidine analog or a DNA intercalator for UV laser treatment [59,64] can potentially affect the DNA and chromatin structure, altering DNA damage responses [67,68]. In these techniques, damage is induced without sequence specificity and at low resolution, preventing the application of ChIP-like approaches to the characterization of damage-induced changes in chromatin responses and protein recruitment. Laser or heavy-ion irradiation are also difficult to apply to small nuclei (i.e., Drosophila), given the relatively large size of the beam. Finally, specialized equipment is required for heavy-ion radiation (synchrotron), or for applying a laser mounted on a microscope, which might not be readily available in many laboratories.
2.3. Super-Resolution Imaging of Repair Responses in Human Heterochromatin
Heterochromatin repair is particularly difficult to study with cytological approaches in human cells, where pericentromeric regions are not clustered into cytologically distinct domains (e.g., [49]). In addition, transposons and other repeated sequences are abundantly present between genes and in intronic sequences in human cells, generating a large amount of constitutively silent chromatin along the chromosome arms, including within genes [1,2,3,4]. To probe repair in heterochromatin, super resolution techniques have been applied to detect DSBs associated with H3K9me3-rich regions in fixed cells. For example, the Cardoso lab used super-resolution imaging techniques (3D-SIM and STED microscopy) to visualize repair in H3K9me3-rich regions in HeLa cells [49] (Figure 3). Consistent with studies in mouse cells, they detected γH2AX associated with ‘silent’ histone marks at later repair steps following IR, suggesting the delayed completion of repair in heterochromatin [49]. This was confirmed by ChIP-seq analysis of γH2AX in response to IR, which showed a broad enrichment of γH2AX peaks in pericentromeric and other H3K9me3-enriched regions. They also estimated a reduction of DAPI intensity at later repair steps in these regions, consistent with heterochromatin de-condensation, while most silent marks were retained [49]. Similarly, single-molecule localization microscopy (SMLM) of heterochromatin and repair foci identified γH2AX signals proximal to heterochromatin marks, consistent with a local relocalization of repair sites to outside of the local heterochromatic regions [69].
(+) Super resolution analyses allow the probing of heterochromatin repair responses in human cells, where heterochromatin does not form distinct domains, identifying conserved relocalization pathways. A combination of IF and FISH experiments can potentially be used to detect the behavior of specific satellites.
(−) The major caveat of the approach described here is that using H3K9me2/3 as a marker for heterochromatin does not allow discernment of different silent regions. Thus, LADs, telomeres, ribosomal sequences, silenced genes, or intragenic regions of active genes are not distinguishable from pericentromeric heterochromatin in these studies. Further, equipment and expertise required for super resolution imaging are not available in all laboratories. Finally, live imaging of multiple markers is more difficult with super-resolution techniques, limiting the dynamic information that can be extracted.
2.4. Chemically Induced DSBs
Not many studies have applied chemical treatments to investigate DSB repair in heterochromatin. This is likely because of the disadvantage of asynchronous damage induction and the lack of specificity coming from these approaches. A notable exception is a study in Arabidopsis which showed that the heterochromatin-enriched histone variant H2A.W.7 is phosphorylated by ATM to mediate the DNA damage response in heterochromatin, in response to the radiomimetic treatments Bleomycin or Zeocin [70]. Conversely, H2AX phosphorylation contributed to damage signaling mostly in euchromatin [70]). This is a unique example of differentiated damage signaling for euchromatin and heterochromatin, although H2A.W.7 and its phosphorylation are not limited to pericentromeric regions but encompass most silenced chromatin.
(+) Chemical treatments are easy to deliver as they do not require any specialized equipment.
(−) Chemical treatments induce different types of breaks [71], and they damage both euchromatin and heterochromatin. Additionally, damage is created asynchronously, complicating the study of repair dynamics.
2.5. DR-White Repair Cassette Inserted in Heterochromatin
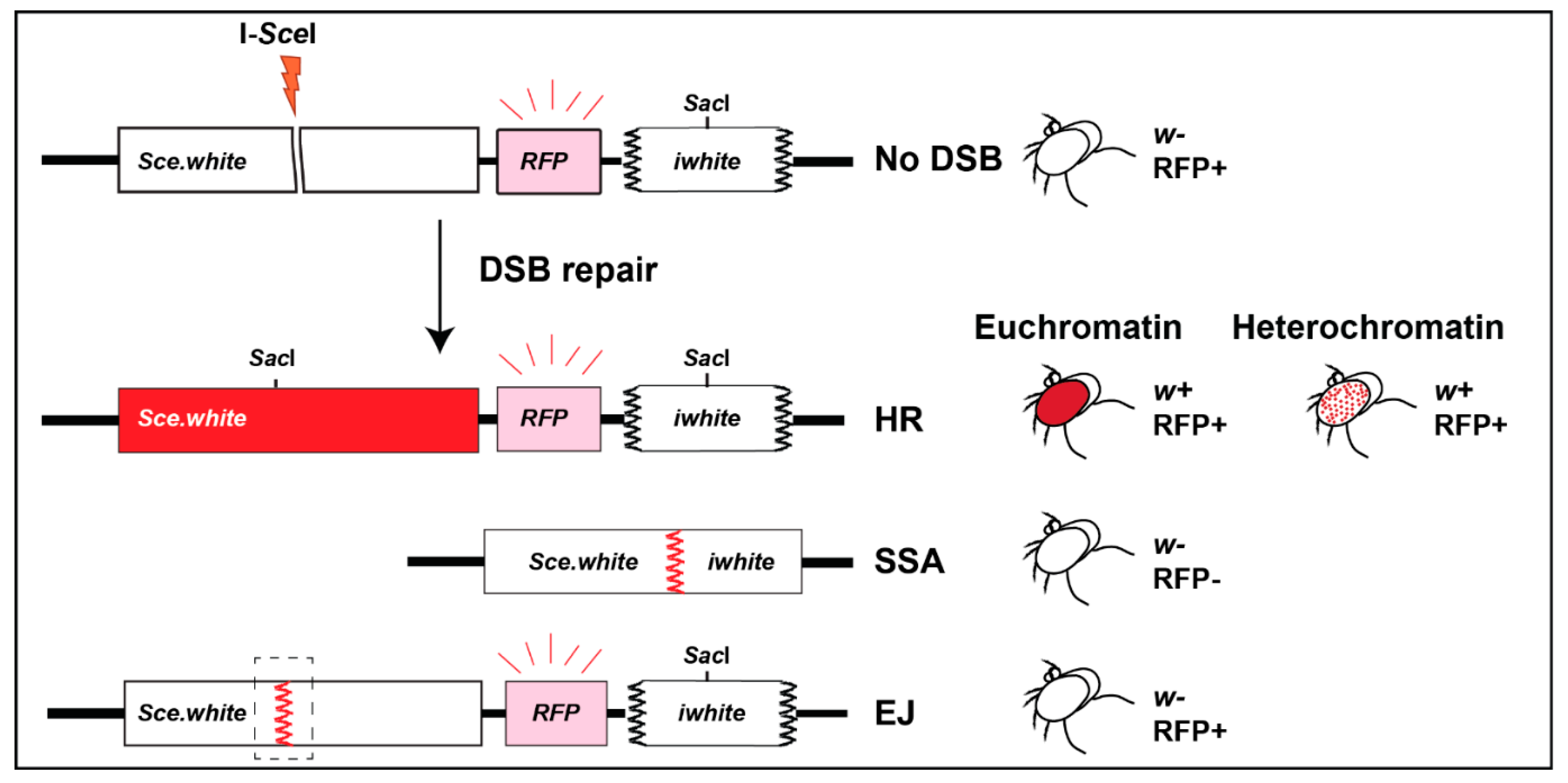
Repair cassettes have been widely used to characterize repair outcomes in response to site-specific DSBs from yeast to human cells. In flies, the LaRocque lab established the direct repeat (DR)-white repair cassette, where HR repair of a mutated white gene containing an I-SceI cut site (Sce.white), using donor sequences from a downstream truncated white gene (iwhite), results in a wild-type white sequence [72] (Figure 5). When I-SceI is expressed in the germ line, repair products can be uniquely detected in the progeny, and HR repair leads to red-eyed flies (Figure 5). Alternatively, single-strand annealing (SSA) repair (Figure 2) results in the deletion of a gene placed between iwhite and Sce.white (e.g., a red fluorescence marker, RFP), and the loss of fluorescence in white-eyed progeny [72]. When placed in heterochromatin, the cassette is partially silenced, resulting in a mosaic phenotype [33] (variegated-red eye, Figure 3 and Figure 5). End-joining (EJ) repair outcomes (reviewed in [73,74], Figure 2) can also be characterized by sequencing across the cut site of white-eyed flies [33] (Figure 5). Finally, repair outcomes can be studied in somatic cells by PCR and sequence analyses [33].
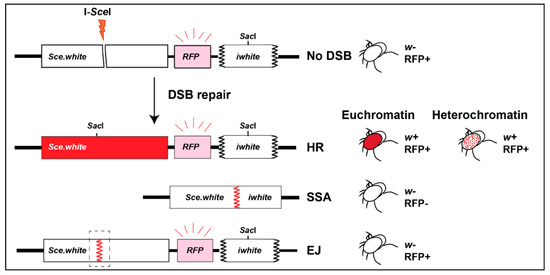
Figure 5.
Schematic representation of DR-white cassette and repair outcomes. The DR-white reporter is composed of an upstream nonfunctional white gene (Sce.white) containing an I-SceI target sequence, a red fluorescent marker (RFP), and a downstream truncated nonfunctional white gene (iwhite) (Adapted from [75]). Sce.white contains a premature stop codon as part of the I-SceI recognition sequence. Repair of DSBs induced by I-SceI in the germline of male flies can be detected in the progeny. HR repair of euchromatic insertions results in red eyes, and HR of heterochromatic insertions results in variegated-red eyes [33]. SSA results in white-, RFP- flies. Uncut, NHEJ, or Alt-EJ result in white-, RFP+ progeny, and different repair products can be distinguished by sequencing across the cut site.
The Karpen and LaRocque labs combined DR-white insertions in heterochromatin or euchromatin with genetic and live imaging experiments to establish heterochromatin repair pathways at the organismal level, including repair pathway choice in euchromatin vs. heterochromatin [33]. Using this system, they showed that NHEJ occurs with high frequency in the heterochromatin of fly tissues (which are enriched for G1 cells). Additionally, ChIP analyses of histone modifications at the repair sites and genetic approaches support the conclusion that Kdm4A demethylates H3K9me2/3 and H3K56me3 specifically at heterochromatic DSBs to promote NHEJ repair, while silencing promotes HR [76]. Importantly, Kdm4A-induced histone demethylation specifically affects heterochromatic DSBs and not euchromatic sites [76]. Finally, using a modified version of this cassette with a donor on the homologous chromosome [33,77], these studies showed that both homologous chromosomes and sister chromatids are used for HR repair in heterochromatin, albeit with a preference for the sister chromatid [33]. Notably, in fly cells, homologous chromosomes are paired in somatic cells [78], providing a readily available template for repair in addition to the sister chromatid.
(+) This system is very versatile and is applied to a genetically tractable whole organism. I-SceI cleavage creates a clean and readily reparable DSB, similar to endogenous DNA breaks that arise during replication (e.g., different from complex breaks generated by exposure to radiation). Responses to breaks in euchromatin and heterochromatin can be directly compared across different strains, enabling the detection of heterochromatin-specific responses. The DR-white cassette can be inserted in different epigenetic contexts to study the relationship between pre-existing chromatin state and repair outcomes. Further, the use of tissue-specific promoters for I-SceI or flies of different ages enables the analysis of repair outcomes across different tissues or during aging [79]. The DR-white system enables simultaneous study of the balance between SSA, HR, NHEJ, and alternative-EJ (Alt-EJ) in both somatic and germline cells. Additionally, the insertion of polymorphisms in the donor sequence enables the measurement of DSB repair outcomes between divergent sequences or to assess the extent of gene conversion tracts [72,80,81]. Finally, similar repair cassettes can be designed to provide a red-eyed phenotype associated with different repair pathways, as previously done for GFP in human cells [82,83,84], providing a quick readout of the pathway of interest.
(−) A potential limitation of this approach is that inserting an exogenous cassette into a genomic location might alter the behavior of that locus. For example, DR-white needs to be expressed in order to provide a ‘red eye’ or ‘red fluorescence’ readout, indicating that even insertions in highly silenced heterochromatin are at least partially expressed in order for the assay to work [33]. Assessing the extent to which cassette insertion affects the pre-existing chromatin state is an important step to validate the use of these constructs and provide a proper interpretation of the results. Further, DR-white insertions have been applied to a few genomic locations, limiting the statistical power of this approach. Satellite sequences have also not been interrogated with this system. Additionally, the impact of the DNA sequence environment on repair outcomes cannot be detected as each I-SceI site is surrounded by the same sequence carried by the cassette.
Further, DR-white is designed to work in the whole organism, limiting the number of mutations that can be tested to investigate genetic pathways due to the relatively long time required to perform the necessary crosses. This is particularly true for live imaging experiments, where flies need to express fluorescent markers for damage (e.g., GFP-Mdc1/Mu2) and heterochromatin (e.g., mCh-HP1a), in addition to the repair cassette, the I-SceI endonuclease, and the mutation of interest.
Additional challenges for the live imaging of the I-SceI-induced response to DSBs inside the DR-white cassette relate to the higher difficulty of imaging tissues vs. cultured cells, with tissues typically requiring more cell-damaging imaging techniques and more Z-stacks, increasing the risk of photobleaching and phototoxicity [26]. For these reasons, most studies have been done in thin and transparent tissues, such as imaginal discs [33].
The asynchronous nature of I-SceI-induced damage results in long imaging experiments needed for capturing relocalization events, increasing the possibility of photodamage. Further, all heterochromatic DR-white insertions generated so far are already positioned at the heterochromatin domain periphery at the beginning of the live imaging experiment [33]. This could be because these sites were more available for insertion of the MiMIC system used to create the DR-white insertions in the first place [85], or because of the difficulty of capturing early time points after I-SceI induced breaks in asynchronous damage inductions. Regardless of the reason, the peripheral position of damage foci relative to the heterochromatin domain precludes the analysis of early repair dynamics.
Finally, tissues are typically characterized by significant variability in cell cycle phases, cell differentiation, and epigenetic states, all of which influence repair outcomes, and these differences cannot be easily captured at the population level. Most somatic cells are also in G0/G1, complicating the study of HR repair pathways. Indeed, cell cycle differences in whole larvae versus premeiotic germline cells might account for the different frequencies of repair outcomes observed across different studies [33].
2.6. Cas9-Induced DSBs in Heterochromatic Satellites
The discovery of the CRISPR/Cas9 system has sparked great progress in the field of DNA damage and repair. Primarily used for genome editing, Cas9 has also emerged as a tool of great utility to induce site-specific DSBs in euchromatin and heterochromatin, which are targeted using a single-guide RNA (sgRNA) complementary to the region of interest [86,87].
The Soutoglou lab has successfully applied Cas9 to induce DSBs in mouse chromocenters, by targeting it to the major satellite [12,88] (Figure 3). Using this system, and a combination of genetic and fixed cell imaging studies, they showed that NHEJ prevails in heterochromatin when damage is induced in G1, while HR prevails when damage is induced in S/G2 [12]. Similar to IR-induced damage in Drosophila cells, repair starts efficiently inside the heterochromatin domain, and this response in mouse cells is ATM-dependent [12] (Figure 4). Further, DSBs repaired by HR relocalize to the periphery of the chromocenters, forming distinct γH2AX signals surrounding the DAPI-bright regions, and relocalization requires resection [12]. While resection occurs inside the domain (marked by RPA foci), Rad51 is recruited only after DSB relocalization to the periphery of the chromocenters (Figure 4). The heterochromatin domain also expands in response to Cas9-induced DSBs [12]. Finally, repair by EJ or SSA (marked by Rad52 foci), does not require relocalization [12].
Similarly, the Karpen lab used Cas9 to induce DSBs in the dodeca satellite of Drosophila cells [76]. They showed that Kdm4A mutation results in a higher number of foci of resection proteins (CtIP and Tosca/Exo1) inside the damaged satellite, supporting a role for Kdm4A in promoting NHEJ while inhibiting HR in these sequences [76].
(+) This is the only approach currently available to directly investigate heterochromatin repair in large satellite sequences and has the additional advantage of being available in cultured cells, facilitating live imaging, genetic approaches, and cell cycle studies. A major advantage of this system is also that a large number of breaks are induced only inside the DAPI-bright regions, so the relocalization of damage signals to outside these regions is easy to detect and quantify. Further, NHEJ proteins that do not form foci in response to IR are detectable in response to Cas9-induced breaks, given that the large amount of damage in the satellite amplifies the signal. Other repeated sequences can be targeted with similar approaches, including centromeres [12,89] or rDNA [90,91,92], enabling the exploration of several domains characterized by repeated DNAs.
(−) Similar to other endonuclease-based systems, Cas9-based systems induce damage asynchronously. Thus, they are less suitable for focus kinetic and dynamic analyses relative to IR-induced breaks. Breaks are only induced in satellite DNAs, so direct comparisons with responses in euchromatin have not been done with this system. Error-free repair results in re-cleavage of the damage by Cas9, potentially affecting the frequency at which each repair pathway is utilized. Using Cas9 to induce damage might also affect repair kinetics and outcomes. For example, microhomology-mediated end joining (MMEJ) pathways seem to require unusually long kinetics when damage is induced with Cas9 [93]. Similarly, the Fanconi Anemia pathway is involved in repairing Cas9-induced damage, while this pathway is typically required only in the presence of inter-strand crosslinks or stalled replication forks [94]. In yeast, repair of Cas9-induced breaks at the boundary between single-copy and CTG repeats results in more extensive resection and a higher frequency of SSA repair on the side containing repeated sequences, leading to unexpectedly large chromosomal deletions of the CTG repeat, and suggesting specific effects of Cas9 on repair in repeated sequences [95].
2.7. Biochemical Fractionation of Heterochromatin
Biochemical approaches have been broadly applied to the study of heterochromatin repair, for example for identifying components that co-immunoprecipitate with heterochromatin histone marks or associated proteins in response to DNA damage [10,96,97]. Some of the most appealing techniques use fractionation approaches to separate heterochromatin from euchromatin and isolate heterochromatin-specific repair components. For example, the Jeggo and Goodarzi labs took advantage of the resistance of heterochromatin to micrococcal nuclease (MNase) digestion to isolate chromatin fractions containing silent histone marks and associated proteins [9,53]. They also discovered that damaged heterochromatin is more susceptible to MNase digestion than undamaged heterochromatin, reflecting a relaxation of the domain [53]. Similar fractionation studies have identified a role for Rad6 in HP1α degradation to promote heterochromatin relaxation and HR repair [98]. Biochemical approaches have also established that phosphorylated HP1β is released from damaged chromatin, promoting the recruitment of the histone acetyltransferase Tip60 and ATM activation [97].
Of note, MNase-resistant fractions are not only enriched for pericentromeric heterochromatin, but also for other silenced regions of the genome (e.g., LADs and silenced genes). Thus, the use of chromatin fractionation with MNase digestion does not enable a direct characterization of repair in pericentromeric sequences. Nevertheless, this technique provided important complementary approaches to live and fixed cell imaging studies for the characterization of repair components acting in pericentromeric regions.
(+) Chromatin fractionation and other biochemical approaches have been successfully applied to identify components that associate with heterochromatin in response to damage, heterochromatin relaxation, and the components mediating this response.
(−) Nuclease-resistant fractions are largely free of euchromatic contamination in the absence of DNA damage, but fractions that are slightly more sensitive to MNase are typically enriched for both euchromatic and heterochromatic histone marks [53]. Additionally, even the most MNase-resistant fractions contain different silenced sequences, not just pericentromeric heterochromatin. Thus, MNase-dependent fractionation methods are not robust enough to clearly separate euchromatin and heterochromatin biochemically, and thus to distinguish between repair components uniquely associated with each of these domains.
3. Perspectives
The variety of techniques discussed above have driven the heterochromatin repair research in the past decade, leading to exciting new discoveries (Table 1). Live and fixed cell imaging after irradiation, the use of repair cassettes inserted into heterochromatin, and Cas9-induced DSBs, have enabled the identification of heterochromatin-specific responses. Biochemical approaches have provided important complementary tools for these discoveries. Here, we reflect on some of the limitations of these techniques and suggest possible areas of development to further deepen our understanding of this field in future studies.

Table 1.
Summary of techniques available to study heterochromatin repair, including main advantages and disadvantages.
3.1. Live and Fixed Cell Imaging
Cell imaging techniques are rapidly evolving, with advances in instrumentation and new tools for biomolecule visualization. For example, the application of extrinsically fluorescent protein tags (i.e., SNAP [99], Halo [100], and CLIP [101] (reviewed in [102]) provides signals that are typically brighter and more photostable than those detected with intrinsically fluorescent tags (e.g., GFP, mCherry). Applying extrinsically fluorescent tags to the study of heterochromatin and repair proteins would enable faster and longer kinetics, with a more extensive exploration of heterochromatin dynamics. Collecting more time points during focus relocalization would also enable the tracking of cells for multiple hours after IR, and, for example, establish what happens to the repair site position or heterochromatin decondensation once repair is complete. More frequent imaging would also facilitate the automated tracking of repair foci by reducing the frequency of ambiguous tracks [51,52], and enable the study of local chromatin movements [30]. Additionally, small X-ray sources can be directly mounted on a microscope to enable the detection of very early repair responses [103,104].
In fixed cell imaging, a proximity ligation assay (PLA) can be applied to detect low abundance proteins or specific post-translational modifications at heterochromatic repair sites [105].
Further, both live and fixed cell imaging studies would greatly benefit from the development of fully automated methods to detect repair focus positions relative to the heterochromatin domain, dramatically reducing the processing time for image/movie analyses.
In addition, several studies have identified phase separation properties associated with the heterochromatin domain and repair foci [106,107,108,109,110,111], but the impact of these properties on heterochromatin repair is unknown. Future studies can combine live imaging approaches with fluorescent recovery after photobleaching (FRAP) or optogenetic tools that induce phase separation (reviewed in [112,113]) to better characterize the phase separation properties of the heterochromatin domain and repair foci, and the role(s) of these properties in repair.
Finally, non-coding RNAs (ncRNAs) have been associated with both heterochromatin establishment [114] and DSB responses (reviewed in [115,116,117]), but their roles in the DSB repair of pericentromeric regions remains unexplored. The application of recently-developed tools to investigate the role of ncRNAs in repair, such as antisense oligonucleotides (ASOs) [118,119,120] coupled with site-specific DSB systems described in this review, provides new opportunities to explore these responses in heterochromatin.
3.2. Site-Specific DSB Induction Systems
The use of restriction enzymes that directly cut the genome in different positions revolutionized our understanding of repair responses with genome-wide approaches. For example, in the DIvA (DSB inducible via AsiSI) cell system developed by the Legube lab [121], the restriction endonuclease AsiSI cuts the genome at more than a hundred sites. Site-specific DSBs have been used for studying: DSB resection, using quantitative real time PCR or END-seq [122,123,124]; repair pathway choice, using ChIP-seq analysis of HR and NHEJ components [38,40,125]; and the spreading of γH2AX signaling by loop extrusion, using Hi-C approaches [126]. However, the DIvA system has not been applied to pericentromeric heterochromatin, because DSBs are not efficiently induced by AsiSI in these sequences due to DNA methylation and chromatin compaction [121,123]. An important goal for future studies is to extend site-specific DSB systems to pericentromeric sequences, such as by working in model systems that lack DNA or using approaches that overcome the barrier to digestion.
3.3. Repair Cassettes
In addition to generating new repair cassettes for Drosophila repair studies (such as to explore canonical end-joining and Alt-EJ pathways with ‘red-eyed’ readouts), it would be important to develop repair cassettes that do not rely on transcription for a readout.
Applying these tools to multiple genomic sites simultaneously would also greatly improve the statistical power of these approaches. For example, in a recent study, the van Steensel lab integrated a multiplexed reporter system in thousands of genomic loci with Cas9, and explored the relative contribution of three DSB repair pathways (NHEJ, MMEJ, or single strand template repair (SSTR)), in different chromatin contexts, including in silenced chromatin [37]. These studies identified a preferential usage of MMEJ in facultative heterochromatin (marked by H3K27me3), although pericentromeric regions were not directly investigated [37]. Extending similar tools to pericentromeric heterochromatin would provide new opportunities for discovery.
3.4. Exploring the Heterochromatin Damage Proteome with Biochemical Approaches
The isolation of damaged heterochromatin with biochemical approaches would enable the identification of new components required for heterochromatin repair. Chromatin fractionation techniques have greatly helped dissect heterochromatic specific regulators and study chromatin decompaction. However, the current methods do not achieve a ‘clean’ separation between euchromatin and heterochromatin. As an alternative approach, immunoprecipitation techniques targeting HP1 have been successfully applied to isolate heterochromatin proteins in different model systems [127,128,129,130,131,132]. Similarly, proximity-dependent biotin identification (BioID) techniques [133], have been applied to identify components associated with silenced chromatin domains, including LADs [133,134], telomeres [135,136], and H3K9me3-enriched-regions [137]. While these approaches did not (or not exclusively) address repair in pericentromeric heterochromatin, applying similar purifications in response to DNA damage can help reveal the heterochromatin-specific proteome for repair. An alternative chromatin fractionation method to MNase digestion took advantage of the sensitivity of different types of chromatins to sonication, and successfully separated H3K9me3 from H3K27me3-enriched fractions for proteomic and sequencing analysis [138].
Additionally, purification techniques can be theoretically applied to Cas9-induced damage specifically in euchromatin or heterochromatin satellites, isolating the components that uniquely contribute to repair of pericentromeric regions. Together, biochemical fractionation and immunoprecipitation approaches are attractive options to isolate novel components participating in heterochromatin repair and can be combined with complementary live and fixed cell imaging studies described in this review, and further dissect the contribution of different components to repair in pericentromeric regions.
4. Conclusions
The ability to induce DSBs in heterochromatin with different approaches, and to detect repair with a variety of techniques, has significantly enhanced our understanding of heterochromatin repair mechanisms in the past decade. These studies mostly relied on; (i) live and fixed imaging after irradiation in systems with distinct heterochromatin domains; (ii) the use of reporter cassettes inserted in heterochromatin; (iii) the induction of DSBs in satellite sequences with Cas9; and (iv) biochemical approaches to enrich for heterochromatin fractions (Figure 3 and Table 1). While these techniques enabled major advances in our understanding of heterochromatin repair, many open questions remain. Improving these approaches, along with adapting the tools recently developed for euchromatin repair studies to heterochromatin domains, will enable a deeper understanding of heterochromatin repair mechanisms. Urgent unanswered questions include understanding chromatin responses in heterochromatin by next-generation sequencing, establishing repair outcomes across multiple heterochromatic sites, identifying the role of phase separation and ncRNAs in heterochromatin repair, and advancing our understanding of the spatial dynamics of heterochromatin repair in human cells. With this ever-growing toolbox, we expect to see exciting progress in our understanding of heterochromatin repair mechanisms across different model systems, damage sources, epigenetic states, and sequence composition, in the coming years.
Author Contributions
Conceptualization, I.C. and C.C.R.; writing—original draft preparation, C.C.R., N.L.B., A.M. and I.C.; writing—review and editing, C.C.R., N.L.B., A.M. and I.C.; supervision, I.C.; funding acquisition, I.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by NIH R01GM117376 and NSF Career 1751197 to I.C.
Acknowledgments
We thank J. Miller, C. Merigliano and S. Keagy for useful comments on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Glossary
Abbreviations for proteins and methods not explained in the main text. Protein functions are briefly introduced in the context of DSB repair.
| Proteins | |
| 53BP1 (P53-binding protein 1) | Promotes DSB repair and checkpoint signaling. |
| Acf1-Snf2h (ATP-dependent chromatin assembly factor complex containing the SWI/SNF related 2h subunit) | Chromatin remodeler |
| Artemis | Structure-specific endonuclease that promotes DSB repair |
| ATM (Ataxia telangiectasia mutated) | DNA damage checkpoint kinase. |
| ATR (Ataxia telangiectasia and Rad3 related) | DNA damage checkpoint kinase. |
| ATRIP (ATR interacting protein) | Protein that mediates ATR recruitment in response to DSBs |
| Brca2 (Breast Cancer 2) | Protein that facilitates Rad51 recruitment onto resected DNA |
| Chd3 (Chromodomain Helicase DNA Binding Protein 3) | Chromatin remodeler. |
| CtIP (C-terminal binding protein (CtBP)-interacting protein | Protein that promotes resection. |
| HELLS (Helicase, Lymphoid Specific) | Chromatin remodeler. |
| HP1 (Heterochromatin protein 1) | Heterochromatin-specific protein associated with H3K9me2/3. |
| KAP1 (Kruppel-associated box (KRAB)-associated protein 1) | Protein that promotes silencing. |
| LINC (Linker of nucleoskeleton and cytoskeleton complex) | Complex spanning the nuclear envelope, composed of a SUN protein across the inner membrane and a KASH protein across the outer membrane. |
| Mdc1 (Mediator of DNA damage checkpoint 1) | Protein that interacts with γH2AX to mediate DSB repair and checkpoint activation. |
| Mu2 (Mutator 2) | Drosophila homologue of Mdc1. |
| Rad54 (Radiation-sensitive mutant protein 54) | Protein that promotes Rad51-mediated strand invasion. |
| Rad6 (Radiation-sensitive mutant protein 6) | E2 ubiquitin-conjugating enzyme that promotes DNA repair progression. |
| RENi (Rad60-Esc2-NIP45 protein family) | Protein family with SUMO-like domains that work in concert with STUbL for DSB repair. |
| RPA (Replication Protein A) | Trimeric complex that binds to ssDNA after resection. |
| SCAI (Suppressor of cancer cell invasion) | Protein that promotes DSB repair. |
| Smc5/6 (Structural maintenance of chromosomes complex 5/6) | Complex that promotes HR. |
| SUMO (Small Ubiquitin-like Modifier) | Peptide covalently attached to other proteins to modify their function. |
| TopBP1 (DNA Topoisomerase II Binding Protein 1) | Protein that promotes ATR activation. |
| Tosca | Drosophila homologue of the exonuclease Exo1 that contributes to resection. |
| Unc45 (Uncoordinated mutant protein 45) | Myosin chaperone and activator. |
| γH2AX/γH2Av | Phosphorylated form of the histone H2A variants H2AX or H2Av. Involved in DSB signaling in mammalian or fly cells, respectively. |
| Methods | |
| 3D-SIM microscopy (3D structured illumination microscopy) | Super resolution microscopy technique. |
| BrdU (5–bromo–2′–deoxyuridine) | Nucleotide analog. |
| CLIP tag | Extrinsically fluorescent protein tag that can be labeled with synthetic probes. |
| CRISPR/Cas9 system (Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 system) | Technology that can be used to induce site-specific DSBs by the Cas9 endonuclease directed to a sequence of interest. |
| Halo tag | Extrinsically fluorescent protein tag that can be labeled with synthetic probes. |
| Hi-C | Chromatin conformation capture technique used to identify the proximity of genomic regions. |
| I-SceI (Intron-encoded Saccaromyces cerevisiae endonuclease 1) | Endonuclease used to induce site-specific DSBs. |
| MiMIC (Minos-mediated integration cassette) | Collection of insertions of transposon for Drosophila genetic manipulation |
| SNAP tag | Extrinsically fluorescent protein tag that can be labeled with synthetic probes. |
| STED microscopy (Stimulated emission depletion microscopy) | Super resolution microscopy technique. |
| TUNEL (Terminal deoxynucleotidyl transferase (TdT) dUTP nick-end labeling) | Assay to detect DNA breaks. |
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [PubMed] [Green Version]
- Hoskins, R.A.; Carlson, J.W.; Kennedy, C.; Acevedo, D.; Evans-Holm, M.; Frise, E.; Wan, K.H.; Park, S.; Mendez-Lago, M.; Rossi, F.; et al. Sequence Finishing and Mapping of Drosophila Melanogaster Heterochromatin. Science 2007, 316, 1625–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, J.W.K.; Jung, Y.L.; Liu, T.; Alver, B.H.; Lee, S.; Ikegami, K.; Sohn, K.-A.; Minoda, A.; Tolstorukov, M.Y.; Appert, A.; et al. Comparative Analysis of Metazoan Chromatin Organization. Nature 2014, 512, 449–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoskins, R.A.; Carlson, J.W.; Wan, K.H.; Park, S.; Mendez, I.; Galle, S.E.; Booth, B.W.; Pfeiffer, B.D.; George, R.A.; Svirskas, R.; et al. The Release 6 Reference Sequence of the Drosophila Melanogaster Genome. Genome Res. 2015, 25, 445–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riddle, N.C.; Minoda, A.; Kharchenko, P.V.; Alekseyenko, A.A.; Schwartz, Y.B.; Tolstorukov, M.Y.; Gorchakov, A.A.; Jaffe, J.D.; Kennedy, C.; Linder-Basso, D.; et al. Plasticity in Patterns of Histone Modifications and Chromosomal Proteins in Drosophila Heterochromatin. Genome Res. 2011, 21, 147–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawal, C.C.; Caridi, C.P.; Chiolo, I. Actin’ between Phase Separated Domains for Heterochromatin Repair. DNA Repair 2019, 81, 102646. [Google Scholar] [CrossRef] [PubMed]
- Caridi, P.C.; Delabaere, L.; Zapotoczny, G.; Chiolo, I. And Yet, It Moves: Nuclear and Chromatin Dynamics of a Heterochromatic Double-Strand Break. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160291. [Google Scholar] [CrossRef] [Green Version]
- Pickersgill, H.; Kalverda, B.; de Wit, E.; Talhout, W.; Fornerod, M.; van Steensel, B. Characterization of the Drosophila Melanogaster Genome at the Nuclear Lamina. Nat. Genet. 2006, 38, 1005–1014. [Google Scholar] [CrossRef]
- Goodarzi, A.A.; Noon, A.T.; Deckbar, D.; Ziv, Y.; Shiloh, Y.; Löbrich, M.; Jeggo, P.A. ATM Signaling Facilitates Repair of DNA Double-Strand Breaks Associated with Heterochromatin. Mol. Cell 2008, 31, 167–177. [Google Scholar] [CrossRef]
- Chiolo, I.; Minoda, A.; Colmenares, S.U.; Polyzos, A.; Costes, S.V.; Karpen, G.H. Double-Strand Breaks in Heterochromatin Move outside of a Dynamic HP1a Domain to Complete Recombinational Repair. Cell 2011, 144, 732–744. [Google Scholar] [CrossRef] [Green Version]
- Chiolo, I.; Tang, J.; Georgescu, W.; Costes, S.V. Nuclear Dynamics of Radiation-Induced Foci in Euchromatin and Heterochromatin. Mutat. Res. 2013, 750, 56–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsouroula, K.; Furst, A.; Rogier, M.; Heyer, V.; Maglott-Roth, A.; Ferrand, A.; Reina-San-Martin, B.; Soutoglou, E. Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol. Cell 2016, 63, 293–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tjong, H.; Li, W.; Kalhor, R.; Dai, C.; Hao, S.; Gong, K.; Zhou, Y.; Li, H.; Zhou, X.J.; Le Gros, M.A.; et al. Population-Based 3D Genome Structure Analysis Reveals Driving Forces in Spatial Genome Organization. Proc. Natl. Acad. Sci. USA 2016, 113, E1663–E1672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Tjong, H.; Li, X.; Gong, K.; Zhou, X.J.; Chiolo, I.; Alber, F. The Three-Dimensional Genome Organization of Drosophila Melanogaster through Data Integration. Genome Biol. 2017, 18, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, T.; Spatola, B.; Delabaere, L.; Bowlin, K.; Hopp, H.; Kunitake, R.; Karpen, G.H.; Chiolo, I. Heterochromatic Breaks Move to the Nuclear Periphery to Continue Recombinational Repair. Nat. Cell Biol. 2015, 17, 1401–1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harr, J.C.; Luperchio, T.R.; Wong, X.; Cohen, E.; Wheelan, S.J.; Reddy, K.L. Directed Targeting of Chromatin to the Nuclear Lamina Is Mediated by Chromatin State and A-Type Lamins. J. Cell Biol. 2015, 208, 33–52. [Google Scholar] [CrossRef] [PubMed]
- Briand, N.; Collas, P. Lamina-Associated Domains: Peripheral Matters and Internal Affairs. Genome Biol. 2020, 21, 85. [Google Scholar] [CrossRef] [Green Version]
- Kendek, A.; Wensveen, M.R.; Janssen, A. The Sound of Silence: How Silenced Chromatin Orchestrates the Repair of Double-Strand Breaks. Genes 2021, 12, 1415. [Google Scholar] [CrossRef]
- Peng, J.C.; Karpen, G.H. Epigenetic Regulation of Heterochromatic DNA Stability. Curr. Opin. Genet. Dev. 2008, 18, 204–211. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-Homologous DNA End Joining and Alternative Pathways to Double-Strand Break Repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Cejka, P.; Symington, L.S. DNA End Resection: Mechanism and Control. Annu. Rev. Genet. 2021, 55, 285–307. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.C.; Karpen, G.H. H3K9 Methylation and RNA Interference Regulate Nucleolar Organization and Repeated DNA Stability. Nat. Cell Biol. 2007, 9, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.C.; Karpen, G.H. Heterochromatic Genome Stability Requires Regulators of Histone H3 K9 Methylation. PLoS Genet. 2009, 5, e1000435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, T.; Bonner, M.R.; Chiolo, I. Cervantes and Quijote Protect Heterochromatin from Aberrant Recombination and Lead the Way to the Nuclear Periphery. Nucleus 2016, 7, 485–497. [Google Scholar] [CrossRef] [Green Version]
- Caridi, C.P.; D’Agostino, C.; Ryu, T.; Zapotoczny, G.; Delabaere, L.; Li, X.; Khodaverdian, V.Y.; Amaral, N.; Lin, E.; Rau, A.R.; et al. Nuclear F-Actin and Myosins Drive Relocalization of Heterochromatic Breaks. Nature 2018, 559, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Dialynas, G.; Delabaere, L.; Chiolo, I. Arp2/3 and Unc45 Maintain Heterochromatin Stability in Polytene Chromosomes. Exp. Biol. Med. 2019, 244, 1362–1371. [Google Scholar] [CrossRef]
- Amaral, N.; Ryu, T.; Li, X.; Chiolo, I. Nuclear Dynamics of Heterochromatin Repair. Trends Genet. 2017, 33, 86–100. [Google Scholar] [CrossRef] [Green Version]
- Beucher, A.; Birraux, J.; Tchouandong, L.; Barton, O.; Shibata, A.; Conrad, S.; Goodarzi, A.A.; Krempler, A.; Jeggo, P.A.; Löbrich, M. ATM and Artemis Promote Homologous Recombination of Radiation-Induced DNA Double-Strand Breaks in G2. EMBO J. 2009, 28, 3413–3427. [Google Scholar] [CrossRef] [Green Version]
- Caridi, C.P.; Plessner, M.; Grosse, R.; Chiolo, I. Nuclear Actin Filaments in DNA Repair Dynamics. Nat. Cell Biol. 2019, 21, 1068–1077. [Google Scholar] [CrossRef]
- Miné-Hattab, J.; Chiolo, I. Complex Chromatin Motions for DNA Repair. Front. Genet. 2020, 11, 800. [Google Scholar] [CrossRef]
- Merigliano, C.; Chiolo, I. Multi-Scale Dynamics of Heterochromatin Repair. Curr. Opin. Genet. Dev. 2021, 71, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Jakob, B.; Splinter, J.; Conrad, S.; Voss, K.-O.; Zink, D.; Durante, M.; Löbrich, M.; Taucher-Scholz, G. DNA Double-Strand Breaks in Heterochromatin Elicit Fast Repair Protein Recruitment, Histone H2AX Phosphorylation and Relocation to Euchromatin. Nucleic Acids Res. 2011, 39, 6489–6499. [Google Scholar] [CrossRef]
- Janssen, A.; Breuer, G.A.; Brinkman, E.K.; van der Meulen, A.I.; Borden, S.V.; van Steensel, B.; Bindra, R.S.; LaRocque, J.R.; Karpen, G.H. A Single Double-Strand Break System Reveals Repair Dynamics and Mechanisms in Heterochromatin and Euchromatin. Genes Dev. 2016, 30, 1645–1657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vítor, A.C.; Huertas, P.; Legube, G.; de Almeida, S.F. Studying DNA Double-Strand Break Repair: An Ever-Growing Toolbox. Front. Mol. Biosci. 2020, 7, 24. [Google Scholar] [CrossRef] [Green Version]
- Mao, P.; Liu, J.; Zhang, Z.; Zhang, H.; Liu, H.; Gao, S.; Rong, Y.S.; Zhao, Y. Homologous Recombination-Dependent Repair of Telomeric DSBs in Proliferating Human Cells. Nat. Commun. 2016, 7, 12154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemaître, C.; Grabarz, A.; Tsouroula, K.; Andronov, L.; Furst, A.; Pankotai, T.; Heyer, V.; Rogier, M.; Attwood, K.M.; Kessler, P.; et al. Nuclear Position Dictates DNA Repair Pathway Choice. Genes Dev. 2014, 28, 2450–2463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schep, R.; Brinkman, E.K.; Leemans, C.; Vergara, X.; van der Weide, R.H.; Morris, B.; van Schaik, T.; Manzo, S.G.; Peric-Hupkes, D.; van den Berg, J.; et al. Impact of Chromatin Context on Cas9-Induced DNA Double-Strand Break Repair Pathway Balance. Mol. Cell 2021, 81, 2216–2230.e10. [Google Scholar] [CrossRef] [PubMed]
- Clouaire, T.; Rocher, V.; Lashgari, A.; Arnould, C.; Aguirrebengoa, M.; Biernacka, A.; Skrzypczak, M.; Aymard, F.; Fongang, B.; Dojer, N.; et al. Comprehensive Mapping of Histone Modifications at DNA Double-Strand Breaks Deciphers Repair Pathway Chromatin Signatures. Mol. Cell 2018, 72, 250–262.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgess, R.C.; Burman, B.; Kruhlak, M.J.; Misteli, T. Activation of DNA Damage Response Signaling by Condensed Chromatin. Cell Rep. 2014, 9, 1703–1717. [Google Scholar] [CrossRef] [Green Version]
- Aymard, F.; Bugler, B.; Schmidt, C.K.; Guillou, E.; Caron, P.; Briois, S.; Iacovoni, J.S.; Daburon, V.; Miller, K.M.; Jackson, S.P.; et al. Transcriptionally Active Chromatin Recruits Homologous Recombination at DNA Double-Strand Breaks. Nat. Struct. Mol. Biol. 2014, 21, 366–374. [Google Scholar] [CrossRef] [Green Version]
- Fortuny, A.; Chansard, A.; Caron, P.; Chevallier, O.; Leroy, O.; Renaud, O.; Polo, S.E. Imaging the Response to DNA Damage in Heterochromatin Domains Reveals Core Principles of Heterochromatin Maintenance. Nat. Commun. 2021, 12, 2428. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Hale, C.J.; Over, R.S.; Cokus, S.J.; Jacobsen, S.E.; Michaels, S.D. Large-Scale Heterochromatin Remodeling Linked to Overreplication-Associated DNA Damage. Proc. Natl. Acad. Sci. USA 2017, 114, 406–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Yamin, B.; Ahmed-Seghir, S.; Tomida, J.; Despras, E.; Pouvelle, C.; Yurchenko, A.; Goulas, J.; Corre, R.; Delacour, Q.; Droin, N.; et al. DNA Polymerase Zeta Contributes to Heterochromatin Replication to Prevent Genome Instability. EMBO J. 2021, 40, e104543. [Google Scholar] [CrossRef] [PubMed]
- Lisby, M.; Rothstein, R. Choreography of Recombination Proteins during the DNA Damage Response. DNA Repair 2009, 8, 1068–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costes, S.V.; Chiolo, I.; Pluth, J.M.; Barcellos-Hoff, M.H.; Jakob, B. Spatiotemporal Characterization of Ionizing Radiation Induced DNA Damage Foci and Their Relation to Chromatin Organization. Mutat. Res. 2010, 704, 78–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayoub, N.; Jeyasekharan, A.D.; Bernal, J.A.; Venkitaraman, A.R. HP1-β Mobilization Promotes Chromatin Changes That Initiate the DNA Damage Response. Nature 2008, 453, 682–686. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, A.A.; Jeggo, P.A. The Heterochromatic Barrier to DNA Double Strand Break Repair: How to Get the Entry Visa. Int. J. Mol. Sci. 2012, 13, 11844–11860. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-A.; Kruhlak, M.; Dotiwala, F.; Nussenzweig, A.; Haber, J.E. Heterochromatin Is Refractory to γ-H2AX Modification in Yeast and Mammals. J. Cell Biol. 2007, 178, 209–218. [Google Scholar] [CrossRef] [Green Version]
- Natale, F.; Rapp, A.; Yu, W.; Maiser, A.; Harz, H.; Scholl, A.; Grulich, S.; Anton, T.; Hörl, D.; Chen, W.; et al. Identification of the Elementary Structural Units of the DNA Damage Response. Nat. Commun. 2017, 8, 15760. [Google Scholar] [CrossRef]
- Colmenares, S.U.; Swenson, J.M.; Langley, S.A.; Kennedy, C.; Costes, S.V.; Karpen, G.H. Drosophila Histone Demethylase KDM4A Has Enzymatic and Non-Enzymatic Roles in Controlling Heterochromatin Integrity. Dev. Cell 2017, 42, 156–169.e5. [Google Scholar] [CrossRef] [Green Version]
- See, C.; Arya, D.; Lin, E.; Chiolo, I. Live Cell Imaging of Nuclear Actin Filaments and Heterochromatic Repair Foci in Drosophila and Mouse Cells. Methods Mol. Biol. 2021, 2153, 459–482. [Google Scholar] [PubMed]
- Caridi, C.P.; Delabaere, L.; Tjong, H.; Hopp, H.; Das, D.; Alber, F.; Chiolo, I. Quantitative Methods to Investigate the 4D Dynamics of Heterochromatic Repair Sites in Drosophila Cells. Methods Enzymol. 2018, 601, 359–389. [Google Scholar] [PubMed]
- Klement, K.; Luijsterburg, M.S.; Pinder, J.B.; Cena, C.S.; Del Nero, V.; Wintersinger, C.M.; Dellaire, G.; van Attikum, H.; Goodarzi, A.A. Opposing ISWI- and CHD-Class Chromatin Remodeling Activities Orchestrate Heterochromatic DNA Repair. J. Cell Biol. 2014, 207, 717–733. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, A.A.; Kurka, T.; Jeggo, P.A. KAP-1 Phosphorylation Regulates CHD3 Nucleosome Remodeling during the DNA Double-Strand Break Response. Nat. Struct. Mol. Biol. 2011, 18, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Hansen, R.K.; Mund, A.; Poulsen, S.L.; Sandoval, M.; Klement, K.; Tsouroula, K.; Tollenaere, M.A.X.; Räschle, M.; Soria, R.; Offermanns, S.; et al. SCAI Promotes DNA Double-Strand Break Repair in Distinct Chromosomal Contexts. Nat. Cell Biol. 2016, 18, 1357–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakarougkas, A.; Ismail, A.; Klement, K.; Goodarzi, A.A.; Conrad, S.; Freire, R.; Shibata, A.; Lobrich, M.; Jeggo, P.A. Opposing Roles for 53BP1 during Homologous Recombination. Nucleic Acids Res. 2013, 41, 9719–9731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kollárovič, G.; Topping, C.E.; Shaw, E.P.; Chambers, A.L. The Human HELLS Chromatin Remodelling Protein Promotes End Resection to Facilitate Homologous Recombination and Contributes to DSB Repair within Heterochromatin. Nucleic Acids Res. 2020, 48, 1872–1885. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M. Measurement of Radiation-Induced Damage to DNA at the Molecular Level. Int. J. Radiat. Biol. 1992, 61, 175–183. [Google Scholar] [CrossRef]
- Kong, X.; Mohanty, S.K.; Stephens, J.; Heale, J.T.; Gomez-Godinez, V.; Shi, L.Z.; Kim, J.-S.; Yokomori, K.; Berns, M.W. Comparative Analysis of Different Laser Systems to Study Cellular Responses to DNA Damage in Mammalian Cells. Nucleic Acids Res. 2009, 37, e68. [Google Scholar] [CrossRef]
- Mari, P.-O.; Florea, B.I.; Persengiev, S.P.; Verkaik, N.S.; Brüggenwirth, H.T.; Modesti, M.; Giglia-Mari, G.; Bezstarosti, K.; Demmers, J.A.A.; Luider, T.M.; et al. Dynamic Assembly of End-Joining Complexes Requires Interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. USA 2006, 103, 18597–18602. [Google Scholar] [CrossRef] [Green Version]
- Kruhlak, M.J.; Celeste, A.; Dellaire, G.; Fernandez-Capetillo, O.; Müller, W.G.; McNally, J.G.; Bazett-Jones, D.P.; Nussenzweig, A. Changes in Chromatin Structure and Mobility in Living Cells at Sites of DNA Double-Strand Breaks. J. Cell Biol. 2006, 172, 823–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Yu, Z.; Su, W. Ion irradiation induced direct damage to proteins and their components. arXiv 2008, arXiv:0807.0084v1. [Google Scholar]
- Hada, M.; Georgakilas, A.G. Formation of Clustered DNA Damage after High-LET Irradiation: A Review. J. Radiat. Res. 2008, 49, 203–210. [Google Scholar] [CrossRef]
- Muster, B.; Rapp, A.; Cristina Cardoso, M. Systematic Analysis of DNA Damage Induction and DNA Repair Pathway Activation by Continuous Wave Visible Light Laser Micro-Irradiation. AIMS Genet. 2017, 04, 047–068. [Google Scholar] [CrossRef]
- Timm, S.; Lorat, Y.; Jakob, B.; Taucher-Scholz, G.; Rübe, C.E. Clustered DNA Damage Concentrated in Particle Trajectories Causes Persistent Large-Scale Rearrangements in Chromatin Architecture. Radiother. Oncol. 2018, 129, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.; Stanley, F.K.T.; Goodarzi, A.A. The Repair of Environmentally Relevant DNA Double Strand Breaks Caused by High Linear Energy Transfer Irradiation--No Simple Task. DNA Repair 2014, 17, 64–73. [Google Scholar] [CrossRef]
- Solarczyk, K.J.; Zarębski, M.; Dobrucki, J.W. Inducing Local DNA Damage by Visible Light to Study Chromatin Repair. DNA Repair 2012, 11, 996–1002. [Google Scholar] [CrossRef]
- Dinant, C.; de Jager, M.; Essers, J.; van Cappellen, W.A.; Kanaar, R.; Houtsmuller, A.B.; Vermeulen, W. Activation of Multiple DNA Repair Pathways by Sub-Nuclear Damage Induction Methods. J. Cell Sci. 2007, 120, 2731–2740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hausmann, M.; Wagner, E.; Lee, J.-H.; Schrock, G.; Schaufler, W.; Krufczik, M.; Papenfuß, F.; Port, M.; Bestvater, F.; Scherthan, H. Super-Resolution Localization Microscopy of Radiation-Induced Histone H2AX-Phosphorylation in Relation to H3K9-Trimethylation in HeLa Cells. Nanoscale 2018, 10, 4320–4331. [Google Scholar] [CrossRef]
- Lorković, Z.J.; Park, C.; Goiser, M.; Jiang, D.; Kurzbauer, M.-T.; Schlögelhofer, P.; Berger, F. Compartmentalization of DNA Damage Response between Heterochromatin and Euchromatin Is Mediated by Distinct H2A Histone Variants. Curr. Biol. 2017, 27, 1192–1199. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Ghorai, M.K.; Kenney, G.; Stubbe, J. Mechanistic Studies on Bleomycin-Mediated DNA Damage: Multiple Binding Modes Can Result in Double-Stranded DNA Cleavage. Nucleic Acids Res. 2008, 36, 3781–3790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Do, A.T.; LaRocque, J.R. The Role of Drosophila Mismatch Repair in Suppressing Recombination between Diverged Sequences. Sci. Rep. 2015, 5, 17601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodgers, K.; McVey, M. Error-Prone Repair of DNA Double-Strand Breaks. J. Cell. Physiol. 2016, 231, 15–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanscom, T.; McVey, M. Regulation of Error-Prone DNA Double-Strand Break Repair and Its Impact on Genome Evolution. Cells 2020, 9, 1657. [Google Scholar] [CrossRef] [PubMed]
- Do, A.T.; Brooks, J.T.; Le Neveu, M.K.; LaRocque, J.R. Double-Strand Break Repair Assays Determine Pathway Choice and Structure of Gene Conversion Events in Drosophila Melanogaster. G3 Genes Genomes Genet. 2014, 4, 425–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, A.; Colmenares, S.U.; Lee, T.; Karpen, G.H. Timely Double-Strand Break Repair and Pathway Choice in Pericentromeric Heterochromatin Depend on the Histone Demethylase dKDM4A. Genes Dev. 2019, 33, 103–115. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, J.; Bloomer, H.; Kellam, N.; LaRocque, J.R. Chromosome Preference During Homologous Recombination Repair of DNA Double-Strand Breaks in. G3 Genes Genomes Genet. 2019, 9, 3773–3780. [Google Scholar] [CrossRef] [Green Version]
- McKee, B.D. Homologous Pairing and Chromosome Dynamics in Meiosis and Mitosis. Biochim. Biophys. Acta 2004, 1677, 165–180. [Google Scholar] [CrossRef]
- Delabaere, L.; Ertl, H.A.; Massey, D.J.; Hofley, C.M.; Sohail, F.; Bienenstock, E.J.; Sebastian, H.; Chiolo, I.; LaRocque, J.R. Aging Impairs Double-Strand Break Repair by Homologous Recombination in Drosophila Germ Cells. Aging Cell 2017, 16, 320–328. [Google Scholar] [CrossRef]
- Chen, C.-C.; Avdievich, E.; Zhang, Y.; Zhang, Y.; Wei, K.; Lee, K.; Edelmann, W.; Jasin, M.; LaRocque, J.R. EXO1 Suppresses Double-Strand Break Induced Homologous Recombination between Diverged Sequences in Mammalian Cells. DNA Repair 2017, 57, 98–106. [Google Scholar] [CrossRef]
- Ertl, H.A.; Russo, D.P.; Srivastava, N.; Brooks, J.T.; Dao, T.N.; LaRocque, J.R. The Role of Blm Helicase in Homologous Recombination, Gene Conversion Tract Length, and Recombination Between Diverged Sequences in. Genetics 2017, 207, 923–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinstock, D.M.; Nakanishi, K.; Helgadottir, H.R.; Jasin, M. Assaying Double-Strand Break Repair Pathway Choice in Mammalian Cells Using a Targeted Endonuclease or the RAG Recombinase. Methods Enzymol. 2006, 409, 524–540. [Google Scholar] [PubMed] [Green Version]
- Gunn, A.; Stark, J.M. I-SceI-Based Assays to Examine Distinct Repair Outcomes of Mammalian Chromosomal Double Strand Breaks. Methods Mol. Biol. 2012, 920, 379–391. [Google Scholar] [PubMed]
- van de Kooij, B.; van Attikum, H. Genomic Reporter Constructs to Monitor Pathway-Specific Repair of DNA Double-Strand Breaks. Front. Genet. 2022, 12, 809832. [Google Scholar] [CrossRef] [PubMed]
- Venken, K.J.T.; Schulze, K.L.; Haelterman, N.A.; Pan, H.; He, Y.; Evans-Holm, M.; Carlson, J.W.; Levis, R.W.; Spradling, A.C.; Hoskins, R.A.; et al. MiMIC: A Highly Versatile Transposon Insertion Resource for Engineering Drosophila Melanogaster Genes. Nat. Methods 2011, 8, 737–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-Guided Genetic Silencing Systems in Bacteria and Archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Mitrentsi, I.; Soutoglou, E. CRISPR/Cas9-Induced Breaks in Heterochromatin, Visualized by Immunofluorescence. Methods Mol. Biol. 2021, 2153, 439–445. [Google Scholar] [PubMed]
- Yilmaz, D.; Furst, A.; Meaburn, K.; Lezaja, A.; Wen, Y.; Altmeyer, M.; Reina-San-Martin, B.; Soutoglou, E. Activation of Homologous Recombination in G1 Preserves Centromeric Integrity. Nature 2021, 600, 748–753. [Google Scholar] [CrossRef]
- van Sluis, M.; McStay, B. A Localized Nucleolar DNA Damage Response Facilitates Recruitment of the Homology-Directed Repair Machinery Independent of Cell Cycle Stage. Genes Dev. 2015, 29, 1151–1163. [Google Scholar] [CrossRef] [Green Version]
- Korsholm, L.M.; Gál, Z.; Lin, L.; Quevedo, O.; Ahmad, D.A.; Dulina, E.; Luo, Y.; Bartek, J.; Larsen, D.H. Double-Strand Breaks in Ribosomal RNA Genes Activate a Distinct Signaling and Chromatin Response to Facilitate Nucleolar Restructuring and Repair. Nucleic Acids Res. 2019, 47, 8019–8035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marnef, A.; Finoux, A.-L.; Arnould, C.; Guillou, E.; Daburon, V.; Rocher, V.; Mangeat, T.; Mangeot, P.E.; Ricci, E.P.; Legube, G. A cohesin/HUSH- and LINC-Dependent Pathway Controls Ribosomal DNA Double-Strand Break Repair. Genes Dev. 2019, 33, 1175–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkman, E.K.; Chen, T.; de Haas, M.; Holland, H.A.; Akhtar, W.; van Steensel, B. Kinetics and Fidelity of the Repair of Cas9-Induced Double-Strand DNA Breaks. Mol. Cell 2018, 70, 801–813.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, C.D.; Kazane, K.R.; Feng, S.J.; Zelin, E.; Bray, N.L.; Schäfer, A.J.; Floor, S.N.; Corn, J.E. CRISPR-Cas9 Genome Editing in Human Cells Occurs via the Fanconi Anemia Pathway. Nat. Genet. 2018, 50, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Mosbach, V.; Viterbo, D.; Descorps-Declère, S.; Poggi, L.; Vaysse-Zinkhöfer, W.; Richard, G.-F. Resection and Repair of a Cas9 Double-Strand Break at CTG Trinucleotide Repeats Induces Local and Extensive Chromosomal Deletions. PLoS Genet. 2020, 16, e1008924. [Google Scholar] [CrossRef] [PubMed]
- Noon, A.T.; Shibata, A.; Rief, N.; Löbrich, M.; Stewart, G.S.; Jeggo, P.A.; Goodarzi, A.A. 53BP1-Dependent Robust Localized KAP-1 Phosphorylation Is Essential for Heterochromatic DNA Double-Strand Break Repair. Nat. Cell Biol. 2010, 12, 177–184. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, X.; Xu, Y.; Ayrapetov, M.K.; Moreau, L.A.; Whetstine, J.R.; Price, B.D. Histone H3 Methylation Links DNA Damage Detection to Activation of the Tumour Suppressor Tip60. Nat. Cell Biol. 2009, 11, 1376–1382. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Wang, C.; Sun, L.; Wang, D.-L.; Chen, L.; Huang, Z.; Yang, Q.; Gao, J.; Yang, X.-B.; Chang, J.-F.; et al. RAD6 Promotes Homologous Recombination Repair by Activating the Autophagy-Mediated Degradation of Heterochromatin Protein HP1. Mol. Cell. Biol. 2015, 35, 406–416. [Google Scholar] [CrossRef] [Green Version]
- Keppler, A.; Gendreizig, S.; Gronemeyer, T.; Pick, H.; Vogel, H.; Johnsson, K. A General Method for the Covalent Labeling of Fusion Proteins with Small Molecules in Vivo. Nat. Biotechnol. 2003, 21, 86–89. [Google Scholar] [CrossRef]
- Los, G.V.; Encell, L.P.; McDougall, M.G.; Hartzell, D.D.; Karassina, N.; Zimprich, C.; Wood, M.G.; Learish, R.; Ohana, R.F.; Urh, M.; et al. HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem. Biol. 2008, 3, 373–382. [Google Scholar] [CrossRef]
- Gautier, A.; Juillerat, A.; Heinis, C.; Corrêa, I.R., Jr.; Kindermann, M.; Beaufils, F.; Johnsson, K. An Engineered Protein Tag for Multiprotein Labeling in Living Cells. Chem. Biol. 2008, 15, 128–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoelzel, C.A.; Zhang, X. Visualizing and Manipulating Biological Processes by Using HaloTag and SNAP-Tag Technologies. Chembiochem 2020, 21, 1935–1946. [Google Scholar] [CrossRef] [PubMed]
- Jakob, B.; Dubiak-Szepietowska, M.; Janiel, E.; Schmidt, A.; Durante, M.; Taucher-Scholz, G. Differential Repair Protein Recruitment at Sites of Clustered and Isolated DNA Double-Strand Breaks Produced by High-Energy Heavy Ions. Sci. Rep. 2020, 10, 1443. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, W.; Osseiran, A.; Rojec, M.; Liu, Y.; Bombrun, M.; Tang, J.; Costes, S.V. Characterizing the DNA Damage Response by Cell Tracking Algorithms and Cell Features Classification Using High-Content Time-Lapse Analysis. PLoS ONE 2015, 10, e0129438. [Google Scholar]
- Gullberg, M.; Fredriksson, S.; Taussig, M.; Jarvius, J.; Gustafsdottir, S.; Landegren, U. A Sense of Closeness: Protein Detection by Proximity Ligation. Curr. Opin. Biotechnol. 2003, 14, 82–86. [Google Scholar] [CrossRef]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase Separation Drives Heterochromatin Domain Formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Pessina, F.; Giavazzi, F.; Yin, Y.; Gioia, U.; Vitelli, V.; Galbiati, A.; Barozzi, S.; Garre, M.; Oldani, A.; Flaus, A.; et al. Functional Transcription Promoters at DNA Double-Strand Breaks Mediate RNA-Driven Phase Separation of Damage-Response Factors. Nat. Cell Biol. 2019, 21, 1286–1299. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.A.; Doolittle, L.K.; Schneider, M.W.G.; Jensen, L.E.; Gamarra, N.; Henry, L.; Gerlich, D.W.; Redding, S.; Rosen, M.K. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179, 470–484.e21. [Google Scholar] [CrossRef] [PubMed]
- Kilic, S.; Lezaja, A.; Gatti, M.; Bianco, E.; Michelena, J.; Imhof, R.; Altmeyer, M. Phase Separation of 53BP1 Determines Liquid-like Behavior of DNA Repair Compartments. EMBO J. 2019, 38, e101379. [Google Scholar] [CrossRef] [PubMed]
- Oshidari, R.; Huang, R.; Medghalchi, M.; Tse, E.Y.W.; Ashgriz, N.; Lee, H.O.; Wyatt, H.; Mekhail, K. DNA Repair by Rad52 Liquid Droplets. Nat. Commun. 2020, 11, 695. [Google Scholar] [CrossRef] [Green Version]
- Frattini, C.; Promonet, A.; Alghoul, E.; Vidal-Eychenie, S.; Lamarque, M.; Blanchard, M.-P.; Urbach, S.; Basbous, J.; Constantinou, A. TopBP1 Assembles Nuclear Condensates to Switch on ATR Signaling. Mol. Cell 2021, 81, 1231–1245.e8. [Google Scholar] [CrossRef] [PubMed]
- Spegg, V.; Altmeyer, M. Biomolecular Condensates at Sites of DNA Damage: More than Just a Phase. DNA Repair 2021, 106, 103179. [Google Scholar] [CrossRef] [PubMed]
- Ganser, L.R.; Myong, S. Methods to Study Phase-Separated Condensates and the Underlying Molecular Interactions. Trends Biochem. Sci. 2020, 45, 1004–1005. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Eickbush, D.G.; Speece, I.; Larracuente, A.M. Heterochromatin-Dependent Transcription of Satellite DNAs in the Female Germline. Elife 2021, 10, e62375. [Google Scholar] [CrossRef] [PubMed]
- d’Adda di Fagagna, F. A Direct Role for Small Non-Coding RNAs in DNA Damage Response. Trends Cell Biol. 2014, 24, 171–178. [Google Scholar] [CrossRef]
- Michelini, F.; Jalihal, A.P.; Francia, S.; Meers, C.; Neeb, Z.T.; Rossiello, F.; Gioia, U.; Aguado, J.; Jones-Weinert, C.; Luke, B.; et al. From “Cellular” RNA to “Smart” RNA: Multiple Roles of RNA in Genome Stability and Beyond. Chem. Rev. 2018, 118, 4365–4403. [Google Scholar] [CrossRef]
- Pessina, F.; Gioia, U.; Brandi, O.; Farina, S.; Ceccon, M.; Francia, S.; d’Adda di Fagagna, F. DNA Damage Triggers a New Phase in Neurodegeneration. Trends Genet. 2021, 37, 337–354. [Google Scholar] [CrossRef] [PubMed]
- Rossiello, F.; Aguado, J.; Sepe, S.; Iannelli, F.; Nguyen, Q.; Pitchiaya, S.; Carninci, P.; d’Adda di Fagagna, F. DNA Damage Response Inhibition at Dysfunctional Telomeres by Modulation of Telomeric DNA Damage Response RNAs. Nat. Commun. 2017, 8, 13980. [Google Scholar] [CrossRef] [Green Version]
- Michelini, F.; Pitchiaya, S.; Vitelli, V.; Sharma, S.; Gioia, U.; Pessina, F.; Cabrini, M.; Wang, Y.; Capozzo, I.; Iannelli, F.; et al. Damage-Induced lncRNAs Control the DNA Damage Response through Interaction with DDRNAs at Individual Double-Strand Breaks. Nat. Cell Biol. 2017, 19, 1400–1411. [Google Scholar] [CrossRef] [Green Version]
- Aguado, J.; Sola-Carvajal, A.; Cancila, V.; Revêchon, G.; Ong, P.F.; Jones-Weinert, C.W.; Arzt, E.W.; Lattanzi, G.; Dreesen, O.; Tripodo, C.; et al. Inhibition of DNA Damage Response at Telomeres Improves the Detrimental Phenotypes of Hutchinson–Gilford Progeria Syndrome. Nat. Commun. 2019, 10, 4990. [Google Scholar] [CrossRef]
- Iacovoni, J.S.; Caron, P.; Lassadi, I.; Nicolas, E.; Massip, L.; Trouche, D.; Legube, G. High-Resolution Profiling of gammaH2AX around DNA Double Strand Breaks in the Mammalian Genome. EMBO J. 2010, 29, 1446–1457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Caron, P.; Legube, G.; Paull, T.T. Quantitation of DNA Double-Strand Break Resection Intermediates in Human Cells. Nucleic Acids Res. 2014, 42, e19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canela, A.; Sridharan, S.; Sciascia, N.; Tubbs, A.; Meltzer, P.; Sleckman, B.P.; Nussenzweig, A. DNA Breaks and End Resection Measured Genome-Wide by End Sequencing. Mol. Cell 2016, 63, 898–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, N.; John, S.; Nussenzweig, A.; Canela, A. END-Seq: An Unbiased, High-Resolution, and Genome-Wide Approach to Map DNA Double-Strand Breaks and Resection in Human Cells. Methods Mol. Biol. 2021, 2153, 9–31. [Google Scholar] [PubMed]
- Clouaire, T.; Legube, G. DNA Double Strand Break Repair Pathway Choice: A Chromatin Based Decision? Nucleus 2015, 6, 107–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnould, C.; Rocher, V.; Finoux, A.-L.; Clouaire, T.; Li, K.; Zhou, F.; Caron, P.; Mangeot, P.E.; Ricci, E.P.; Mourad, R.; et al. Loop Extrusion as a Mechanism for Formation of DNA Damage Repair Foci. Nature 2021, 590, 660–665. [Google Scholar] [CrossRef]
- Lechner, M.S.; Schultz, D.C.; Negorev, D.; Maul, G.G.; Rauscher, F.J., 3rd. The Mammalian Heterochromatin Protein 1 Binds Diverse Nuclear Proteins through a Common Motif That Targets the Chromoshadow Domain. Biochem. Biophys. Res. Commun. 2005, 331, 929–937. [Google Scholar] [CrossRef]
- Rosnoblet, C.; Vandamme, J.; Völkel, P.; Angrand, P.-O. Analysis of the Human HP1 Interactome Reveals Novel Binding Partners. Biochem. Biophys. Res. Commun. 2011, 413, 206–211. [Google Scholar] [CrossRef]
- Motamedi, M.R.; Hong, E.-J.E.; Li, X.; Gerber, S.; Denison, C.; Gygi, S.; Moazed, D. HP1 Proteins Form Distinct Complexes and Mediate Heterochromatic Gene Silencing by Nonoverlapping Mechanisms. Mol. Cell 2008, 32, 778–790. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, N.; Paulo, J.A.; Tatarakis, A.; Wang, X.; Edwards, A.L.; Bhanu, N.V.; Garcia, B.A.; Haas, W.; Gygi, S.P.; Moazed, D. Native Chromatin Proteomics Reveals a Role for Specific Nucleoporins in Heterochromatin Organization and Maintenance. Mol. Cell 2020, 77, 51–66.e8. [Google Scholar] [CrossRef]
- Alekseyenko, A.A.; Gorchakov, A.A.; Zee, B.M.; Fuchs, S.M.; Kharchenko, P.V.; Kuroda, M.I. Heterochromatin-Associated Interactions of Drosophila HP1a with dADD1, HIPP1, and Repetitive RNAs. Genes Dev. 2014, 28, 1445–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, H.-W.; Lee, D.H.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Kwon, S.H. Analysis of the Heterochromatin Protein 1 (HP1) Interactome in Drosophila. J. Proteom. 2014, 102, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A Promiscuous Biotin Ligase Fusion Protein Identifies Proximal and Interacting Proteins in Mammalian Cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef] [Green Version]
- Chojnowski, A.; Ong, P.F.; Wong, E.S.M.; Lim, J.S.Y.; Mutalif, R.A.; Navasankari, R.; Dutta, B.; Yang, H.; Liow, Y.Y.; Sze, S.K.; et al. Progerin Reduces LAP2α-Telomere Association in Hutchinson-Gilford Progeria. Elife 2015, 4, e07759. [Google Scholar] [CrossRef] [PubMed]
- Schmidtmann, E.; Anton, T.; Rombaut, P.; Herzog, F.; Leonhardt, H. Determination of Local Chromatin Composition by CasID. Nucleus 2016, 7, 476–484. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Meng, Y.; Wang, Y.; Li, J.; Lam, M.; Wang, L.; Di, L.-J. Nuclear Localization of Desmoplakin and Its Involvement in Telomere Maintenance. Int. J. Biol. Sci. 2019, 15, 2350–2362. [Google Scholar] [CrossRef] [PubMed]
- Santos-Barriopedro, I.; van Mierlo, G.; Vermeulen, M. Off-the-Shelf Proximity Biotinylation for Interaction Proteomics. Nat. Commun. 2021, 12, 5015. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.S.; McCarthy, R.L.; Sidoli, S.; Donahue, G.; Kaeding, K.E.; He, Z.; Lin, S.; Garcia, B.A.; Zaret, K.S. Genomic and Proteomic Resolution of Heterochromatin and Its Restriction of Alternate Fate Genes. Mol. Cell 2017, 68, 1023–1037.e15. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).