Near-Hexaploid and Near-Tetraploid Aneuploid Progenies Derived from Backcrossing Tetraploid Parents Hibiscus syriacus × (H. syriacus × H. paramutabilis)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Pollen Staining Test and Size Measurement
2.3. Developing and Confirming Hybrids
2.4. Flower Size
2.5. Flow Cytometry
2.6. Chromosome Preparations
2.7. FISH Analysis
3. Results
3.1. Pollen Size Distribution
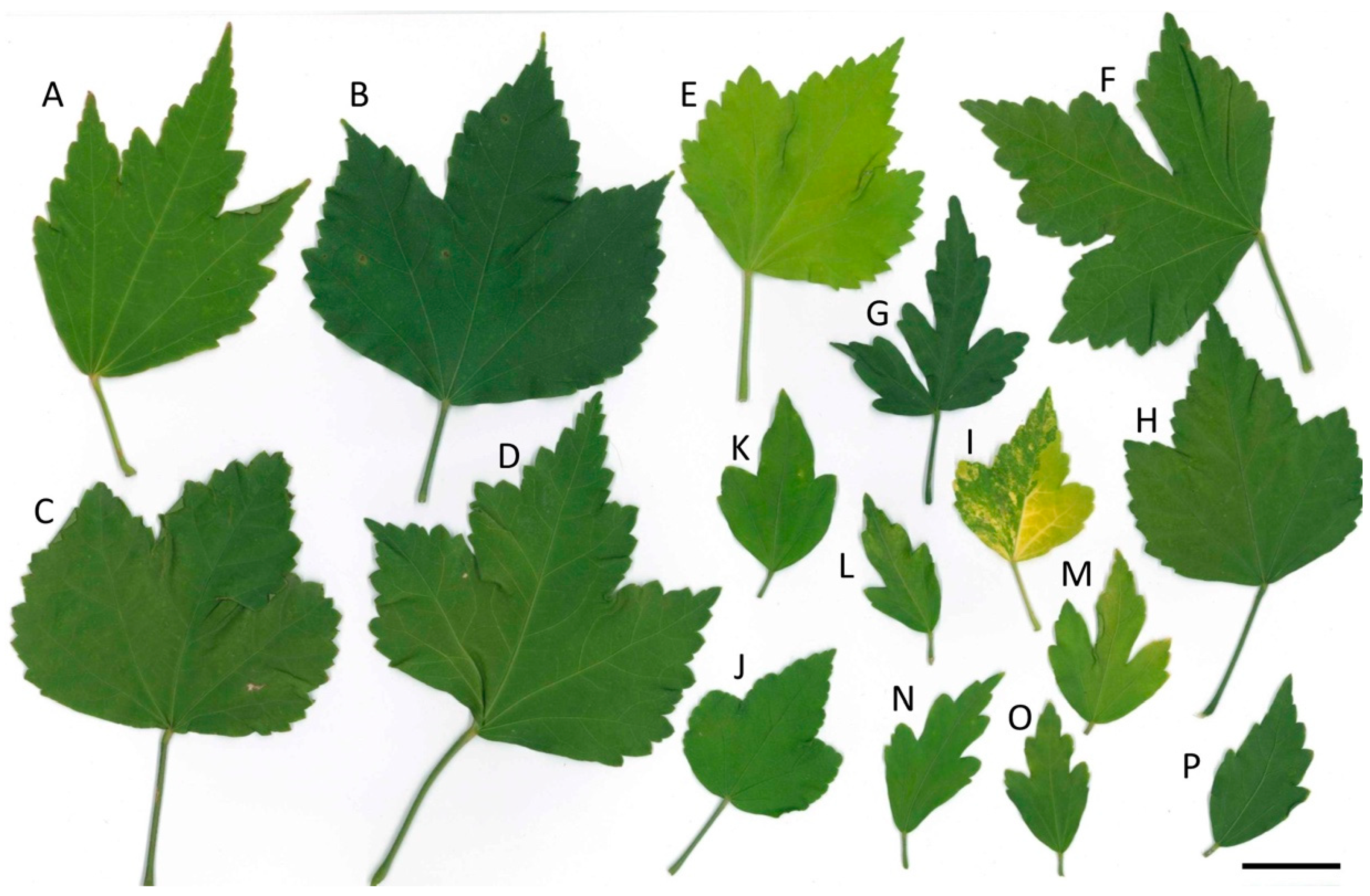
3.2. Hybrid Confirmed by Leaf Morphology and ISSR
3.3. Flow Cytometry Analysis for Ploidy
3.4. Flower Size Distribution and Other Phenotypes
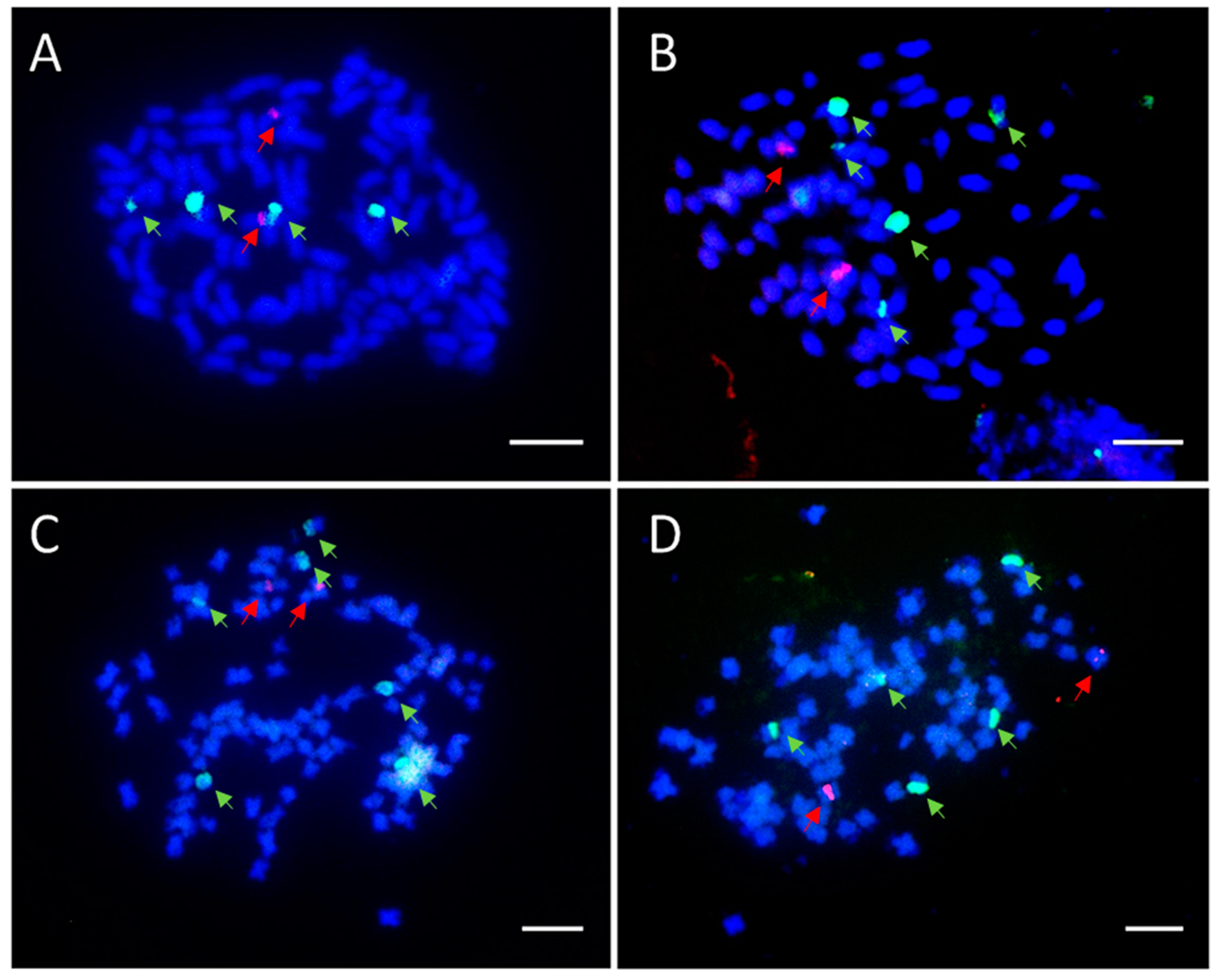
3.5. Chromosome Squash and FISH Analysis
4. Discussion
4.1. Pollen Size Distribution and Source of Unreduced Gametes
4.2. Breeding Potential of Hybrid Cultivars to Increase Flower Size
4.3. Diversity of rDNA Signals among BC1F1 Seedlings
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kuligowska, K.; Lütken, H.; Christensen, B.; Müller, R. Interspecific hybridization among cultivars of hardy Hibiscus species section Muenchhusia. Breed. Sci. 2016, 66, 300–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawton, B.P. Hibiscus: Hardy and Tropical Plants for the Garden; Timber Press: Portland, OR, USA, 2004. [Google Scholar]
- Dirr, M.A. Manual of Woody Landscape Plants: Their Identification, Ornamental Characteristics, Culture, Propagation and Uses, 6th ed.; Stipes Publishing Company: Champaign, IL, USA, 1998. [Google Scholar]
- Chen, H.; Xue, L.; Li, T.; Contreras, R.N. Quantile regression facilitates simultaneous selection of negatively correlated floral traits among BC1F1 progeny of male-fertile hybrid Hibiscus cultivars Lohengrin and Resi (H. syriacus × H. paramutabilis). J. Am. Soc. Hortic. Sci. 2019, 144, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Lattier, J.D.; Contreras, R.N. Segregation of flower color and eyespot in althea. J. Am. Soc. Hortic. Sci. 2020, 145, 215–227. [Google Scholar] [CrossRef] [Green Version]
- Van Laere, K.; Van Huylenbroeck, J.M.; Van Bockstaele, E. Interspecific hybridisation between Hibiscus syriacus, Hibiscus sinosyriacus and Hibiscus paramutabilis. Euphytica 2007, 155, 271–283. [Google Scholar] [CrossRef]
- Van Huylenbroeck, J.; De Riek, J.; De Loose, M. Genetic relationships among Hibiscus syriacus, Hibiscus sinosyriacus and Hibiscus paramutabilis revealed by AFLP, morphology and ploidy analysis. Genet. Resour. Crop Evol. 2000, 47, 335–343. [Google Scholar] [CrossRef]
- Pounders, C.T.; Sakhanokho, H.F. ‘Hapa White’,‘Hapa Pink’, and ‘Hapa Red’ interspecific hybrid Hibiscus cultivars. HortScience 2016, 51, 1616–1617. [Google Scholar] [CrossRef] [Green Version]
- Klips, R.A. Pollen competition as a reproductive isolating mechanism between two sympatric Hibiscus species (Malvaceae). Am. J. Bot. 1999, 86, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.M.; Shim, K.K.; Kang, H.C.; Lim, K.B. A new cultivar ‘Tohagol Red’ with unique flower shape and color through interspecific hybridization of Hibiscus species. Flower Res. J. 2014, 22, 278–282. [Google Scholar] [CrossRef]
- Ha, Y.M.; Lim, K.B.; Shim, K.K. Development of a new Hibiscus cultivar ‘Daewangchun’ with vigorous growth and unique red eye through interspecific hybridization. Hortic. Sci. Technol. 2015, 33, 453–458. [Google Scholar] [CrossRef] [Green Version]
- Van Laere, K.; Dewitte, A.; Van Huylenbroeck, J.; Van Bockstaele, E. Evidence for the occurrence of unreduced gametes in interspecific hybrids of Hibiscus. J. Hortic. Sci. Biotechnol. 2009, 84, 240–247. [Google Scholar] [CrossRef]
- Lattier, J.D.; Chen, H.; Contreras, R.N. Variation in genome size, ploidy, stomata, and rDNA signals in Althea. J. Am. Soc. Hortic. Sci. 2019, 144, 130–140. [Google Scholar] [CrossRef] [Green Version]
- Contreras, R.N.; Ruter, J.M.; Hanna, W.W. An oryzalin-induced autoallooctoploid of Hibiscus acetosella ‘Panama Red’. J. Am. Soc. Hortic. Sci. 2009, 134, 553–559. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Ruter, J.M. Development and evaluation of diploid and polyploid Hibiscus moscheutos. HortScience 2017, 52, 676–681. [Google Scholar] [CrossRef]
- Singh, F.; Khoshoo, T. Chromosomal polymorphism within the Hibiscus rosa-sinensis complex. Caryologia 1970, 23, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Lattier, J.; Lattier, J.D. Breeding and Genetics of Lilacs and Hardy Hibiscus. Ph.D. Thesis, Oregon State University, Corvallis, OR, USA, 2017. [Google Scholar]
- Lattier, J.D.; Contreras, R.N. Flower forms and ploidy levels impact fertility in althea. HortScience 2022, 57, 558–570. [Google Scholar] [CrossRef]
- Deepo, D.M.; Mazharul, I.M.; Hwang, Y.J.; Kim, H.Y.; Kim, C.K.; Lim, K.B. Chromosome and ploidy analysis of winter hardy Hibiscus Species by FISH and flow cytometry. Euphytica 2022, 218, 81. [Google Scholar] [CrossRef]
- Gan, Y.M.; Liu, F.; Chen, D.; Wu, Q.; Qin, Q.; Wang, C.Y.; Li, S.H.; Zhang, X.D.; Wang, Y.H.; Wang, K.B. Chromosomal locations of 5S and 45S rDNA in Gossypium genus and Its phylogenetic implications revealed by FISH. PLoS ONE 2013, 8, e68207. [Google Scholar] [CrossRef]
- Chung, M.Y.; Chung, J.D.; Ramanna, M.; van Tuyl, J.M.; Lim, K.B. Production of polyploids and unreduced gametes in Lilium auratum × L. henryi hybrid. Theor. Appl. Genet. 2013, 9, 693–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.T.; Yang, S.Q.; Li, Z.A.; Zhang, Y.X.; Wang, Y.Z.; Cheng, C.Y.; Li, J.; Chen, J.F.; Lou, Q.F. Comparative chromosomal localization of 45S and 5S rDNAs and implications for genome evolution in Cucumis. Genome 2016, 59, 449–457. [Google Scholar] [CrossRef] [Green Version]
- Van Laere, K.; Khrustaleva, L.; Van Huylenbroeck, J.; Van Bockstaele, E. Application of GISH to characterize woody ornamental hybrids with small genomes and chromosomes. Plant Breed. 2010, 129, 442–447. [Google Scholar] [CrossRef]
- Khafaga, A. Molecular genetic identification of some Egyptian Hibiscus samples. J. Am. Sci. 2013, 9, 28–35. [Google Scholar]
- Satya, P.; Karan, M.; Kar, C.; Mahapatra, A.; Mahapatra, B. Assessment of molecular diversity and evolutionary relationship of kenaf (Hibiscus cannabinus L.), roselle (H. sabdariffa L.) and their wild relatives. Plant Syst. Evol. 2013, 299, 619–629. [Google Scholar] [CrossRef]
- Yu, C.; Yin, Y.; Creech, D.L.; Lu, Z.; Xu, J. Morphological characters and SRAP analysis of two hybrids between Hibiscus dasycalyx and Hibiscus ‘Moy Grande’. Sci. Hortic. 2016, 198, 118–124. [Google Scholar] [CrossRef]
- Doležel, J.; Sgorbati, S.; Lucretti, S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 1992, 85, 625–631. [Google Scholar] [CrossRef]
- Doležel, J.; Dolezelova, M.; Novak, F.J. Flow cytometric estimation of nuclear-DNA amount in diploid bananas (Musa acuminata and M. balbisiana). Biol. Plant. 1994, 36, 351–357. [Google Scholar] [CrossRef]
- Lattier, J.D.; Chen, H.; Contreras, R.N. Improved method of enzyme digestion for root tip cytology. HortScience 2017, 52, 1029–1032. [Google Scholar] [CrossRef]
- Chang, Y.C.; Shii, C.T.; Chung, M.C. Variations in ribosomal RNA gene loci in spider lily (Lycoris spp.). J. Am. Soc. Hortic. Sci. 2009, 134, 567–573. [Google Scholar] [CrossRef] [Green Version]
- Gerlach, W.L.; Dyer, T.A. Sequence Organization of the repeating units in the nucleus of wheat which contain 5s ribosomal-RNA genes. Nucleic Acids Res. 1980, 8, 4851–4865. [Google Scholar] [CrossRef] [Green Version]
- Gerlach, W.; Bedbrook, J. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979, 7, 1869–1885. [Google Scholar] [CrossRef]
- Chen, H.; Chung, M.C.; Tsai, Y.C.; Wei, F.J.; Hsieh, J.S.; Hsing, Y.C. Distribution of new satellites and simple sequence repeats in annual and perennial Glycine species. Bot. Stud. 2015, 56, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, H.G.; Chen, X.; Chen, Z.; Ye, J.Q.; Li, K.M.; Liu, J.P. Induction of female 2n gametes and creation of tetraploids through sexual hybridization in cassava (Manihot esculenta). Euphytica 2015, 201, 265–273. [Google Scholar] [CrossRef]
- Fukuda, K.; Sakamoto, S. Cytological studies on unreduced male gamete formation in hybrids between tetraploid emmer wheats and Aegilops squarrosa L. Jpn. J. Breed. 1992, 42, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Nelson, M.N.; Mason, A.S.; Castello, M.C.; Thomson, L.; Yan, G.; Cowling, W.A. Microspore culture preferentially selects unreduced (2n) gametes from an interspecific hybrid of Brassica napus L.× Brassica carinata Braun. Theor. Appl. Genet. 2009, 119, 497–505. [Google Scholar] [CrossRef]
- Sora, D.; Kron, P.; Husband, B. Genetic and environmental determinants of unreduced gamete production in Brassica napus, Sinapis arvensis and their hybrids. Heredity 2016, 117, 440–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Tuyl, J.; De Jeu, M. Methods for overcoming interspecific crossing barriers. In Pollen Biotechnology for Crop Production and Improvement; Cambridge University Press: Cambridge, UK, 1997; pp. 273–292. [Google Scholar]
- Karlov, G.; Khrustaleva, L.; Lim, K.; Van Tuyl, J. Homoeologous recombination in 2 n-gametes producing interspecific hybrids of Lilium (Liliaceae) studied by genomic in situ hybridization (GISH). Genome 1999, 42, 681–686. [Google Scholar] [CrossRef]
- Williams, B.R.; Prabhu, V.R.; Hunter, K.E.; Glazier, C.M.; Whittaker, C.A.; Housman, D.E.; Amon, A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 2008, 322, 703–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, M.M. Chromosome numbers of Rubus cultivars at the national clonal germplasm repository. HortScience 1995, 30, 1453–1456. [Google Scholar] [CrossRef] [Green Version]
- Grivet, L.; DHont, A.; Roques, D.; Feldmann, P.; Lanaud, C.; Glaszmann, J.C. RFLP mapping in cultivated sugarcane (Saccharum spp.): Genome organization in a highly polyploid and aneuploid interspecific hybrid. Genetics 1996, 142, 987–1000. [Google Scholar] [CrossRef]
- Zhou, S.; Zhou, G.; Li, K. Euploid endosperm of triploid× diploid/tetraploid crosses results in aneuploid embryo survival in Lilium. HortScience 2011, 46, 558–562. [Google Scholar] [CrossRef]
- Crawford, S.; Rojas, B.M.; Crawford, E.; Otten, M.; Schoenenberger, T.A.; Garfinkel, A.R.; Chen, H. Characteristics of the diploid, triploid, and tetraploid versions of a cannabigerol-dominant F1 hybrid industrial hemp cultivar, Cannabis sativa ‘Stem Cell CBG’. Genes 2021, 12, 923. [Google Scholar] [CrossRef]




| Cultivar or Accession | Female Parent | Male Parent | Predicted Ploidy Level | Relative 2C Genome Size, Mean ± SE (pg) | Petal Area 2 (cm2) |
|---|---|---|---|---|---|
| H. syriacus ‘Fiji’ | 4x | 4.63 ± 0.05 | - | ||
| ‘Resi’ | 4x | 4.63 ± 0.08 | 35.5 | ||
| ‘Lohengrin’ | 4x | 4.58 ± 0.03 | 28.0 | ||
| H2015-019-08 | H. syriacus ‘Blue Chiffon’ | ‘Resi’ | 6x | 7.05 ± 0.01 | 53.0 * |
| H2015-019-09 | H. syriacus ‘Blue Chiffon’ | ‘Resi’ | 6x | 6.96 ± 0.05 | 54.9 * |
| H2015-024-01 | H. syriacus ‘Blue Chiffon’ | ‘Resi’ | 6x | 6.98 ± 0.02 | 40.0 * |
| H2015-024-05 | H. syriacus ‘Blue Chiffon’ | ‘Resi’ | 6x | 7.02 ± 0.05 | 34.1 |
| H2015-024-07 | H. syriacus ‘Blue Chiffon’ | ‘Resi’ | 6x | 6.97 ± 0.03 | 45.7 * |
| H2015-024-08 | H. syriacus ‘Blue Chiffon’ | ‘Resi’ | 6x | 6.99 ± 0.03 | 16.2 |
| H2015-016-01 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 7.05 ± 0.03 | 51.5 * |
| H2015-016-11 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 7.21 ± 0.09 | 42.6 * |
| H2015-016-02 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 7.07 ± 0.03 | 50.2 * |
| H2015-016-03 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 7.16 ± 0.08 | 35.2 |
| H2015-016-05 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 7.04 ± 0.08 | 46.5 * |
| H2015-016-08 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 7.03 ± 0.00 | 37.3 * |
| H2015-017-05 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 6.94 ± 0.07 | 42.6 * |
| H2015-109-01 | H. syriacus ‘Lavender Chiffon’ | ‘Resi’ | 6x | 7.09 ± 0.08 | 52.6 * |
| H2015-108-02 | H. syriacus ‘Lavender Chiffon’ | ‘Resi’ | 4x | 4.76 ± 0.07 | 36.6 * |
| H2015-122-01 | H. syriacus ‘Lavender Chiffon’ | ‘Resi’ | 4x | 4.82 ± 0.03 | 45.3 * |
| H2015-122-02 | H. syriacus ‘Lavender Chiffon’ | ‘Resi’ | 4x | 4.83 ± 0.07 | 32.3 |
| H2015-122-03 | H. syriacus ‘Lavender Chiffon’ | ‘Resi’ | 4x | 4.87 ± 0.05 | 26.4 |
| H2015-029-02 | H. syriacus ‘Strawberry Smoothie’ | ‘Lohengrin’ | 4x | 4.72 ± 0.03 | 25.9 |
| H2015-031-04 | H. syriacus ‘Blushing Bride’ | ‘Lohengrin’ | 4x | 4.72 ± 0.00 | 15.3 |
| H2015-043-06 | H. syriacus ‘Blushing Bride’ | ‘Lohengrin’ | 4x | 4.75 ± 0.02 | 19.7 |
| H2015-043-07 | H. syriacus ‘Blushing Bride’ | ‘Lohengrin’ | 4x | 4.83 ± 0.08 | 22.9 |
| H2015-044-17 | H. syriacus ‘Blushing Bride’ | ‘Lohengrin’ | 4x | 4.73 ± 0.07 | 18.7 |
| H2015-046-09 | H. syriacus ‘Strawberry Smoothie’ | ‘Lohengrin’ | 4x | 4.76 ± 0.05 | 17.2 |
| H2015-047-05 | H. syriacus ‘Strawberry Smoothie’ | ‘Lohengrin’ | 4x | 4.95 ± 0.13 | 25.4 |
| H2015-052-02 | H. syriacus ‘Blushing Bride’ | ‘Lohengrin’ | 4x | 4.83 ± 0.05 | 13.7 |
| H2015-052-05 | H. syriacus ‘Blushing Bride’ | ‘Lohengrin’ | 4x | 4.72 ± 0.02 | 4.8 |
| H2015-052-12 | H. syriacus ‘Blushing Bride’ | ‘Lohengrin’ | 4x | 4.81 ± 0.06 | 29.4 |
| H2015-052-X2 | H. syriacus ‘Blushing Bride’ | ‘Lohengrin’ | 4x | 4.79 ± 0.03 | 11.8 |
| H2015-060-23 | H. syriacus ‘White Chiffon’ | ‘Lohengrin’ | 4x | 4.78 ± 0.03 | 40.1 |
| H2015-061-03 | H. syriacus ‘Lavender Chiffon’ | ‘Lohengrin’ | 4x | 4.75 ± 0.01 | 31.9 |
| H2015-062-08 | H. syriacus ‘Lavender Chiffon’ | ‘Lohengrin’ | 4x | 4.83 ± 0.04 | 27.8 |
| H2015-064-10 | H. syriacus ‘Strawberry Smoothie’ | ‘Lohengrin’ | 4x | 4.76 ± 0.03 | 20.5 |
| H2015-068-01 | H. syriacus ‘White Chiffon’ | ‘Lohengrin’ | 4x | 4.63 ± 0.09 | 24.6 |
| H2015-072-10 | H. syriacus ‘Lavender Chiffon’ | ‘Lohengrin’ | 4x | 4.75 ± 0.04 | 35.6 |
| H2015-076-08 | H. syriacus ‘Blushing Bride’ | ‘Lohengrin’ | 4x | 4.75 ± 0.04 | 23.7 |
| H2015-081-01 | H. syriacus ‘Blushing Bride’ | ‘Lohengrin’ | 4x | 4.83 ± 0.02 | 27.6 |
| Cultivar Name or Accession | Female Parent | Male Parent | Predicted Ploidy Level | 45S rDNA | 5S rDNA |
|---|---|---|---|---|---|
| H. syriacus ‘Bali’ [13] | - | - | 4x | 4 | 2 |
| ‘Lohengrin’ | - | - | 4x | 5 | 2 |
| ‘Resi’ | - | - | 4x | 5 | 2 |
| H2015-029-02 | H. syriacus ‘Strawberry Smoothie’ | ‘Lohengrin’ | 4x | 4 | 2 |
| H2015-064-10 | H. syriacus ‘Strawberry Smoothie’ | ‘Lohengrin’ | 4x | 5 | 2 |
| H2015-061-03 | H. syriacus ‘Lavender Chiffon’ | ‘Lohengrin’ | 4x | 5 | 2 |
| H2015-052-X2 | H. syriacus ‘Blushing Bride’ | ‘Lohengrin’ | 4x | 6 | 2 |
| H2015-016-08 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 5 | 3 |
| H2015-016-03 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 6 | 3 |
| H2015-017-02 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 6 | 3 |
| H2015-017-05 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 6 | 3 |
| H2015-017-08 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 6 | 3 |
| H2015-019-09 | H. syriacus ‘Blue Chiffon’ | ‘Resi’ | 6x | 6 | 3 |
| H2015-024-07 | H. syriacus ‘Blue Chiffon’ | ‘Resi’ | 6x | 6 | 3 |
| H2015-109-01 | H. syriacus ‘Lavender Chiffon’ | ‘Resi’ | 6x | 6 | 3 |
| H2015-016-01 | H. syriacus ‘Blushing Bride’ | ‘Resi’ | 6x | 7 | 3 |
| H2015-024-01 | H. syriacus ‘Blue Chiffon’ | ‘Resi’ | 6x | 7 | 3 |
| H2015-104-05 | H. syriacus ‘Lavender Chiffon’ | ‘Resi’ | 6x | 8 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Contreras, R.N. Near-Hexaploid and Near-Tetraploid Aneuploid Progenies Derived from Backcrossing Tetraploid Parents Hibiscus syriacus × (H. syriacus × H. paramutabilis). Genes 2022, 13, 1022. https://doi.org/10.3390/genes13061022
Chen H, Contreras RN. Near-Hexaploid and Near-Tetraploid Aneuploid Progenies Derived from Backcrossing Tetraploid Parents Hibiscus syriacus × (H. syriacus × H. paramutabilis). Genes. 2022; 13(6):1022. https://doi.org/10.3390/genes13061022
Chicago/Turabian StyleChen, Hsuan, and Ryan N. Contreras. 2022. "Near-Hexaploid and Near-Tetraploid Aneuploid Progenies Derived from Backcrossing Tetraploid Parents Hibiscus syriacus × (H. syriacus × H. paramutabilis)" Genes 13, no. 6: 1022. https://doi.org/10.3390/genes13061022
APA StyleChen, H., & Contreras, R. N. (2022). Near-Hexaploid and Near-Tetraploid Aneuploid Progenies Derived from Backcrossing Tetraploid Parents Hibiscus syriacus × (H. syriacus × H. paramutabilis). Genes, 13(6), 1022. https://doi.org/10.3390/genes13061022






