Comprehensive Analysis of the lncRNA–miRNA–mRNA Regulatory Network for Intramuscular Fat in Pigs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Sample Collection
2.2. RNA Extraction and Sequencing
2.3. Mapping and Assembly of Sequenced RNA Data
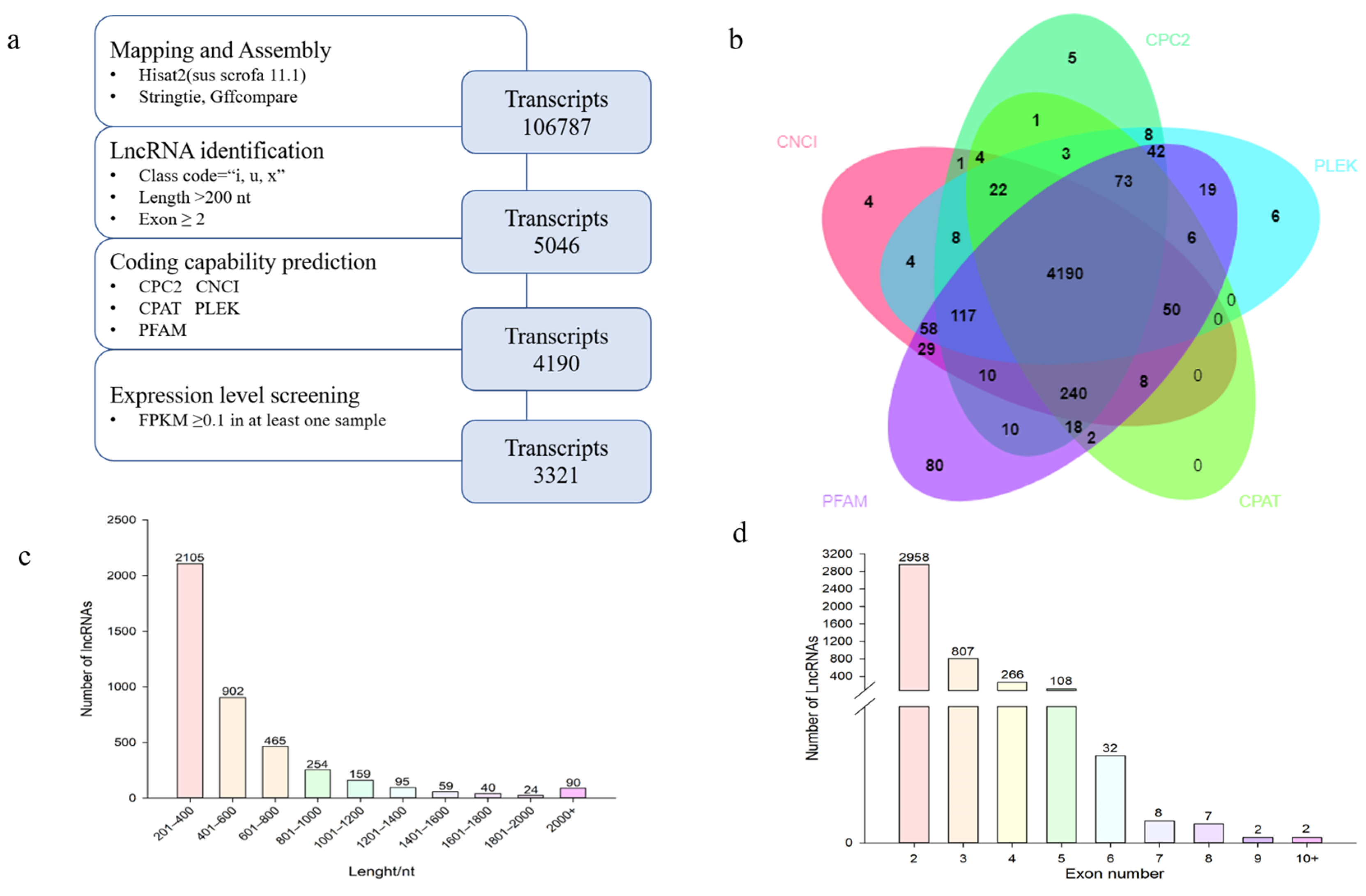
2.4. Pipeline for lncRNA Identification
2.5. Screening of Differentially Expressed RNA
2.6. Gene Ontology and Pathway Enrichment Analysis
2.7. Target Gene Prediction and Functional Analysis
3. Results
3.1. Overview of RNA Sequencing Data
3.2. Identification and Characterization of lncRNA
3.3. Differential Expression RNA Analysis
3.4. GO and KEGG Functional Enrichment Analysis of DEGs
3.5. Target Gene Prediction of Differentially Expressed miRNA and lncRNA
3.6. Analysis of PPI Network and lncRNA-miRNA-mRNA Regulatory Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poklukar, K.; Candek-Potokar, M.; Batorek Lukac, N.; Tomazin, U.; Skrlep, M. Lipid Deposition and Metabolism in Local and Modern Pig Breeds: A Review. Animals 2020, 10, 424. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, X.; Monin, G.; Talmant, A.; Mourot, J.; Lebret, B. Influence of intramuscular fat content on the quality of pig meat—2. Consumer acceptability of m. longissimus lumborum. Meat Sci. 1999, 53, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Renaudeau, D.; Mourot, J. A comparison of carcass and meat quality characteristics of Creole and Large White pigs slaughtered at 90 kg BW. Meat Sci. 2007, 76, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Fortin, A.; Robertson, W.M.; Tong, A.K. The eating quality of Canadian pork and its relationship with intramuscular fat. Meat Sci. 2005, 69, 297–305. [Google Scholar] [CrossRef]
- Hocquette, J.F.; Gondret, F.; Baeza, E.; Medale, F.; Jurie, C.; Pethick, D.W. Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Animal 2010, 4, 303–319. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Cai, R.; Wang, Y.; Zhao, R.; Qin, J.; Pang, W. A Newly Identified LncRNA LncIMF4 Controls Adipogenesis of Porcine Intramuscular Preadipocyte through Attenuating Autophagy to Inhibit Lipolysis. Animals 2020, 10, 926. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, X.; Qin, J.; Liu, S.; Zhao, R.; Yu, T.; Chu, G.; Yang, G.; Pang, W. Comparative Analysis of Long Noncoding RNAs Expressed during Intramuscular Adipocytes Adipogenesis in Fat-Type and Lean-Type Pigs. J. Agric. Food Chem. 2018, 66, 12122–12130. [Google Scholar] [CrossRef]
- An, X.; Ma, K.; Zhang, Z.; Zhao, T.; Zhang, X.; Tang, B.; Li, Z. miR-17, miR-21, and miR-143 Enhance Adipogenic Differentiation from Porcine Bone Marrow-Derived Mesenchymal Stem Cells. DNA Cell Biol. 2016, 35, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chen, Y.; Zhang, Y.; Zhang, Y.; Chen, L.; Mo, D. Up-regulated miR-145 expression inhibits porcine preadipocytes differentiation by targeting IRS1. Int. J. Biol. Sci. 2012, 8, 1408–1417. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, F.F.; Ge, J.; Zhu, J.Y.; Shi, X.E.; Li, X.; Yu, T.Y.; Chu, G.Y.; Yang, G.S. miR-429 Inhibits Differentiation and Promotes Proliferation in Porcine Preadipocytes. Int. J. Mol. Sci. 2016, 17, 2047. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, K.; Zhu, F.; Zhai, L.; Liu, H.; Wang, Z.; Hou, Z.; Wang, C. The liver transcriptome of two full-sibling Songliao black pigs with extreme differences in backfat thickness. J. Anim. Sci. Biotechnol. 2014, 5, 32. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Nolan, T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. 2004, 15, 155–166. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC A Quality Control Tool for High Throughput Sequence Data; 2010. Available online: https://github.com/s-andrews/FastQC (accessed on 4 January 2021).
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [Green Version]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef] [Green Version]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Li, J.H.; Shao, P.; Zhou, H.; Chen, Y.Q.; Qu, L.H. starBase: A database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011, 39, D202–D209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Yu, K.; Shu, G.; Yuan, F.; Zhu, X.; Gao, P.; Wang, S.; Wang, L.; Xi, Q.; Zhang, S.; Zhang, Y.; et al. Fatty acid and transcriptome profiling of longissimus dorsi muscles between pig breeds differing in meat quality. Int. J. Biol. Sci. 2013, 9, 108–118. [Google Scholar] [CrossRef]
- Xu, J.; Wang, C.; Jin, E.; Gu, Y.; Li, S.; Li, Q. Identification of differentially expressed genes in longissimus dorsi muscle between Wei and Yorkshire pigs using RNA sequencing. Genes Genom. 2018, 40, 413–421. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Q.; Chamba, Y.; Zhang, B.; Shang, P.; Zhang, H.; Wu, C. Identification of Genes Related to Growth and Lipid Deposition from Transcriptome Profiles of Pig Muscle Tissue. PLoS ONE 2015, 10, e0141138. [Google Scholar] [CrossRef] [Green Version]
- Miao, W.; Ma, Z.; Tang, Z.; Yu, L.; Liu, S.; Huang, T.; Wang, P.; Wu, T.; Song, Z.; Zhang, H.; et al. Integrative ATAC-seq and RNA-seq Analysis of the Longissimus Muscle of Luchuan and Duroc Pigs. Front. Nutr. 2021, 8, 742672. [Google Scholar] [CrossRef]
- Yamakawa, T.; Whitson, R.H.; Li, S.L.; Itakura, K. Modulator recognition factor-2 is required for adipogenesis in mouse embryo fibroblasts and 3T3-L1 cells. Mol. Endocrinol. 2008, 22, 441–453. [Google Scholar] [CrossRef] [Green Version]
- Whitson, R.H.; Tsark, W.; Huang, T.H.; Itakura, K. Neonatal mortality and leanness in mice lacking the ARID transcription factor Mrf-2. Biochem. Biophys. Res. Commun. 2003, 312, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Gao, M.; Yang, R.; Zhao, Z.; Mi, J.; Sun, H.; Xiao, H.; Fang, X. The effect of CPT1B gene on lipid metabolism and its polymorphism analysis in Chinese Simmental cattle. Anim. Biotechnol. 2021, 1–13. [Google Scholar] [CrossRef]
- Tang, Y.; Zhou, J.; Hooi, S.C.; Jiang, Y.M.; Lu, G.D. Fatty acid activation in carcinogenesis and cancer development: Essential roles of long-chain acyl-CoA synthetases. Oncol. Lett. 2018, 16, 1390–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkes, H.A.; Preston, E.; Wilks, D.; Ballesteros, M.; Carpenter, L.; Wood, L.; Kraegen, E.W.; Furler, S.M.; Cooney, G.J. Overexpression of acyl-CoA synthetase-1 increases lipid deposition in hepatic (HepG2) cells and rodent liver in vivo. Am. J. Physiol. Endocrinol. Metab. 2006, 291, E737–E744. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, K.; Shan, B.; Wei, S.; Li, D.; Han, H.; Wei, W.; Chen, J.; Liu, H.; Zhang, L. A genome-wide landscape of mRNAs, lncRNAs, and circRNAs during subcutaneous adipogenesis in pigs. J. Anim. Sci. Biotechnol. 2018, 9, 76. [Google Scholar] [CrossRef] [Green Version]
- Straub, B.K.; Gyoengyoesi, B.; Koenig, M.; Hashani, M.; Pawella, L.M.; Herpel, E.; Mueller, W.; Macher-Goeppinger, S.; Heid, H.; Schirmacher, P. Adipophilin/perilipin-2 as a lipid droplet-specific marker for metabolically active cells and diseases associated with metabolic dysregulation. Histopathology 2013, 62, 617–631. [Google Scholar] [CrossRef]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef] [Green Version]
- Tsai, T.H.; Chen, E.; Li, L.; Saha, P.; Lee, H.J.; Huang, L.S.; Shelness, G.S.; Chan, L.; Chang, B.H. The constitutive lipid droplet protein PLIN2 regulates autophagy in liver. Autophagy 2017, 13, 1130–1144. [Google Scholar] [CrossRef] [Green Version]
- Imamura, M.; Inoguchi, T.; Ikuyama, S.; Taniguchi, S.; Kobayashi, K.; Nakashima, N.; Nawata, H. ADRP stimulates lipid accumulation and lipid droplet formation in murine fibroblasts. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E775–E783. [Google Scholar] [CrossRef] [Green Version]
- Yu, P.; Wang, Y.; Yang, W.T.; Li, Z.; Zhang, X.J.; Zhou, L.; Gui, J.F. Upregulation of the PPAR signaling pathway and accumulation of lipids are related to the morphological and structural transformation of the dragon-eye goldfish eye. Sci. China Life Sci. 2021, 64, 1031–1049. [Google Scholar] [CrossRef]
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018, 34, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, H.; Yuan, S.; Chen, T.; Ji, P. Development of an immune-related lncRNA-miRNA-mRNA network based on competing endogenous RNA in periodontitis. J. Clin. Periodontol. 2021, 48, 1470–1479. [Google Scholar] [CrossRef] [PubMed]
- Braga, E.A.; Fridman, M.V.; Moscovtsev, A.A.; Filippova, E.A.; Dmitriev, A.A.; Kushlinskii, N.E. LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: ceRNA and Alternative Mechanisms. Int. J. Mol. Sci. 2020, 21, 8855. [Google Scholar] [CrossRef] [PubMed]
- Raziq, K.; Cai, M.; Dong, K.; Wang, P.; Afrifa, J.; Fu, S. Competitive endogenous network of lncRNA, miRNA, and mRNA in the chemoresistance of gastrointestinal tract adenocarcinomas. Biomed. Pharmacother. 2020, 130, 110570. [Google Scholar] [CrossRef]
- Phan, J.; Peterfy, M.; Reue, K. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J. Biol. Chem. 2004, 279, 29558–29564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterfy, M.; Phan, J.; Xu, P.; Reue, K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 2001, 27, 121–124. [Google Scholar] [CrossRef] [PubMed]
- He, X.P.; Xu, X.W.; Zhao, S.H.; Fan, B.; Yu, M.; Zhu, M.J.; Li, C.C.; Peng, Z.Z.; Liu, B. Investigation of Lpin1 as a candidate gene for fat deposition in pigs. Mol. Biol. Rep. 2009, 36, 1175–1180. [Google Scholar] [CrossRef]
- Wang, Q.; Ji, C.; Huang, J.; Yang, F.; Zhang, H.; Liu, L.; Yin, J. The mRNA of lipin1 and its isoforms are differently expressed in the longissimus dorsi muscle of obese and lean pigs. Mol. Biol. Rep. 2011, 38, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Chao, X.; Guo, L.; Wang, Q.; Huang, W.; Liu, M.; Luan, K.; Jiang, J.; Lin, S.; Nie, Q.; Luo, W.; et al. miR-429-3p/LPIN1 Axis Promotes Chicken Abdominal Fat Deposition via PPARgamma Pathway. Front. Cell Dev. Biol. 2020, 8, 595637. [Google Scholar] [CrossRef]
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef]
- Haeusler, R.A.; McGraw, T.E.; Accili, D. Biochemical and cellular properties of insulin receptor signalling. Nat. Rev. Mol. Cell Biol. 2018, 19, 31–44. [Google Scholar] [CrossRef]
- Besse-Patin, A.; Jeromson, S.; Levesque-Damphousse, P.; Secco, B.; Laplante, M.; Estall, J.L. PGC1A regulates the IRS1:IRS2 ratio during fasting to influence hepatic metabolism downstream of insulin. Proc. Natl. Acad. Sci. USA 2019, 116, 4285–4290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckstein, S.S.; Weigert, C.; Lehmann, R. Divergent Roles of IRS (Insulin Receptor Substrate) 1 and 2 in Liver and Skeletal Muscle. Curr. Med. Chem. 2017, 24, 1827–1852. [Google Scholar] [CrossRef] [PubMed]
- Man, X.F.; Hu, N.; Tan, S.W.; Tang, H.N.; Guo, Y.; Tang, C.Y.; Liu, Y.Q.; Tang, J.; Zhou, C.L.; Wang, F.; et al. Insulin receptor substrate-1 inhibits high-fat diet-induced obesity by browning of white adipose tissue through miR-503. FASEB J. 2020, 34, 12308–12323. [Google Scholar] [CrossRef]
- Kovacs, P.; Hanson, R.L.; Lee, Y.H.; Yang, X.; Kobes, S.; Permana, P.A.; Bogardus, C.; Baier, L.J. The role of insulin receptor substrate-1 gene (IRS1) in type 2 diabetes in Pima Indians. Diabetes 2003, 52, 3005–3009. [Google Scholar] [CrossRef] [Green Version]
- Alamdari-Palangi, V.; Amini, R.; Karami, H. MiRNA-7 enhances erlotinib sensitivity of glioblastoma cells by blocking the IRS-1 and IRS-2 expression. J. Pharm. Pharmacol. 2020, 72, 531–538. [Google Scholar] [CrossRef]
- Gymnopoulos, M.; Elsliger, M.A.; Vogt, P.K. Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl. Acad. Sci. USA 2007, 104, 5569–5574. [Google Scholar] [CrossRef] [Green Version]
- Mazloumi Gavgani, F.; Smith Arnesen, V.; Jacobsen, R.G.; Krakstad, C.; Hoivik, E.A.; Lewis, A.E. Class I Phosphoinositide 3-Kinase PIK3CA/p110alpha and PIK3CB/p110beta Isoforms in Endometrial Cancer. Int. J. Mol. Sci. 2018, 19, 3931. [Google Scholar] [CrossRef] [Green Version]
- Dibble, C.C.; Manning, B.D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 2013, 15, 555–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.D.; Han, C.C.; Wan, H.F.; He, F.; Xu, H.Y.; Wei, S.H.; Du, X.H.; Xu, F. Effects of inhibiting PI3K-Akt-mTOR pathway on lipid metabolism homeostasis in goose primary hepatocytes. Animal 2016, 10, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, T.; Rios, Z.; Mei, Q.; Lin, X.; Cao, S. Heat Shock Proteins and Cancer. Trends Pharmacol. Sci. 2017, 38, 226–256. [Google Scholar] [CrossRef] [PubMed]
- Nan, J.; Hu, X.; Guo, B.; Xu, M.; Yao, Y. Inhibition of endoplasmic reticulum stress alleviates triple-negative breast cancer cell viability, migration, and invasion by Syntenin/SOX4/Wnt/beta-catenin pathway via regulation of heat shock protein A4. Bioengineered 2022, 13, 10564–10577. [Google Scholar] [CrossRef]
- Chen, N.; Wang, J. Wnt/beta-Catenin Signaling and Obesity. Front. Physiol. 2018, 9, 792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cristancho, A.G.; Lazar, M.A. Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011, 12, 722–734. [Google Scholar] [CrossRef]
- Bagchi, D.P.; MacDougald, O.A. Wnt Signaling: From Mesenchymal Cell Fate to Lipogenesis and Other Mature Adipocyte Functions. Diabetes 2021, 70, 1419–1430. [Google Scholar] [CrossRef]
- Robichaud, S.; Fairman, G.; Vijithakumar, V.; Mak, E.; Cook, D.P.; Pelletier, A.R.; Huard, S.; Vanderhyden, B.C.; Figeys, D.; Lavallee-Adam, M.; et al. Identification of novel lipid droplet factors that regulate lipophagy and cholesterol efflux in macrophage foam cells. Autophagy 2021, 17, 3671–3689. [Google Scholar] [CrossRef]
- Huang, X.; Liu, G.; Guo, J.; Su, Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int. J. Biol. Sci. 2018, 14, 1483–1496. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Qian, D.W.; Jiang, S.; Shang, E.X.; Zhu, Z.H.; Duan, J.A. Scutellariae Radix and Coptidis Rhizoma Improve Glucose and Lipid Metabolism in T2DM Rats via Regulation of the Metabolic Profiling and MAPK/PI3K/Akt Signaling Pathway. Int. J. Mol. Sci. 2018, 19, 3634. [Google Scholar] [CrossRef]





| DEL | DEGs of Adjacent Protein Coding Genes | Distance (kb) | Spearman Correlation Coefficient | p-Value |
|---|---|---|---|---|
| MSTRG.10885.1 | DAPK3 | Coincide | 0.812767 | 0.001311 |
| MSTRG.13115.1 | LPIN1 | 55 | 0.603267 | 0.037831 |
| MSTRG.13120.1 | LPIN1 | 33.2 | 0.802451 | 0.001683 |
| MSTRG.17466.1 | DNAJB4 | 41.8 | 0.777584 | 0.00291 |
| FUBP1 | 4.5 | 0.679511 | 0.01507 | |
| NEXN | Coincide | 0.735553 | 0.006402 | |
| MSTRG.21882.1 | CNKSR2 | Coincide | 0.532917 | 0.074422 |
| KLHL34 | 45.7 | 0.691861 | 0.012674 | |
| MSTRG.383.1 | SYNCRIP | 29.2 | 0.313079 | 0.321749 |
| MSTRG.6525.3 | HERC4 | 13.8 | 0.751009 | 0.004874 |
| MSTRG.8414.1 | DUSP1 | 92.5 | 0.552691 | 0.062372 |
| Term | ID | p-Value | Gene |
|---|---|---|---|
| Insulin resistance | ssc04931 | 4.43 × 10−7 | RPS6KA1, CPT1B, PPP1R3C, IRS2, PIK3CA, PIK3CB, PRKAG2, PPARGC1A, PRKAA1, IRS1, SREBF1, PPARGC1B, G6PC3 |
| Insulin signaling pathway | ssc04910 | 1.96 × 10−5 | PPP1R3C, IRS2, PIK3CA, PIK3CB, PRKAG2, PPARGC1A, PRKAA1, IRS1, SORBS1, SREBF1, G6PC3, EIF4E |
| Adipocytokine signaling pathway | ssc04920 | 0.000106564 | CPT1B, IRS2, ACSL1, PRKAG2, PPARGC1A, PRKAA1, IRS1, G6PC3 |
| mTOR signaling pathway | ssc04150 | 0.000821208 | RPS6KA1, DDIT4, PIK3CA, PIK3CB, IRS1, PRKAA1, CLIP1, EIF4E, WNT5B, RICTOR |
| PI3K-Akt signaling pathway | ssc04151 | 0.000919504 | ERBB2, PDGFA, DDIT4, IL4R, PPP2R3C, PIK3CB, IRS1, PRKAA1, PIK3CA, VEGFA, ITGAV, COL1A1, EIF4E, HSP90AA1, MYC, G6PC3 |
| Non-alcoholic fatty liver disease (NAFLD) | ssc04932 | 0.002901411 | DDIT3, PIK3CA, EIF2AK3, PRKAG2, IRS1, PRKAA1, SREBF1, IRS2, PIK3CB |
| Regulation of lipolysis in adipocytes | ssc04923 | 0.005012828 | PIK3CA, PIK3CB, IRS1, IRS2, ABHD5 |
| Glycerolipid metabolism | ssc00561 | 0.006591252 | GPAM, LPIN1, LCLAT1, DGKD, DGKE |
| PPAR signaling pathway | ssc03320 | 0.016374973 | SORBS1, ACSL1, PLIN2, ACADM, CPT1B |
| Fatty acid metabolism | ssc01212 | 0.026319948 | FADS1, ACSL1, ACADM, CPT1B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Chen, S.; Yuan, J.; Shi, Y.; Wang, Y.; Xi, Y.; Qi, X.; Guo, Y.; Sheng, X.; Liu, J.; et al. Comprehensive Analysis of the lncRNA–miRNA–mRNA Regulatory Network for Intramuscular Fat in Pigs. Genes 2023, 14, 168. https://doi.org/10.3390/genes14010168
Zhao Y, Chen S, Yuan J, Shi Y, Wang Y, Xi Y, Qi X, Guo Y, Sheng X, Liu J, et al. Comprehensive Analysis of the lncRNA–miRNA–mRNA Regulatory Network for Intramuscular Fat in Pigs. Genes. 2023; 14(1):168. https://doi.org/10.3390/genes14010168
Chicago/Turabian StyleZhao, Yanhui, Shaokang Chen, Jiani Yuan, Yumei Shi, Yan Wang, Yufei Xi, Xiaolong Qi, Yong Guo, Xihui Sheng, Jianfeng Liu, and et al. 2023. "Comprehensive Analysis of the lncRNA–miRNA–mRNA Regulatory Network for Intramuscular Fat in Pigs" Genes 14, no. 1: 168. https://doi.org/10.3390/genes14010168







