Potential Involvement of LncRNAs in Cardiometabolic Diseases
Abstract
:1. Introduction
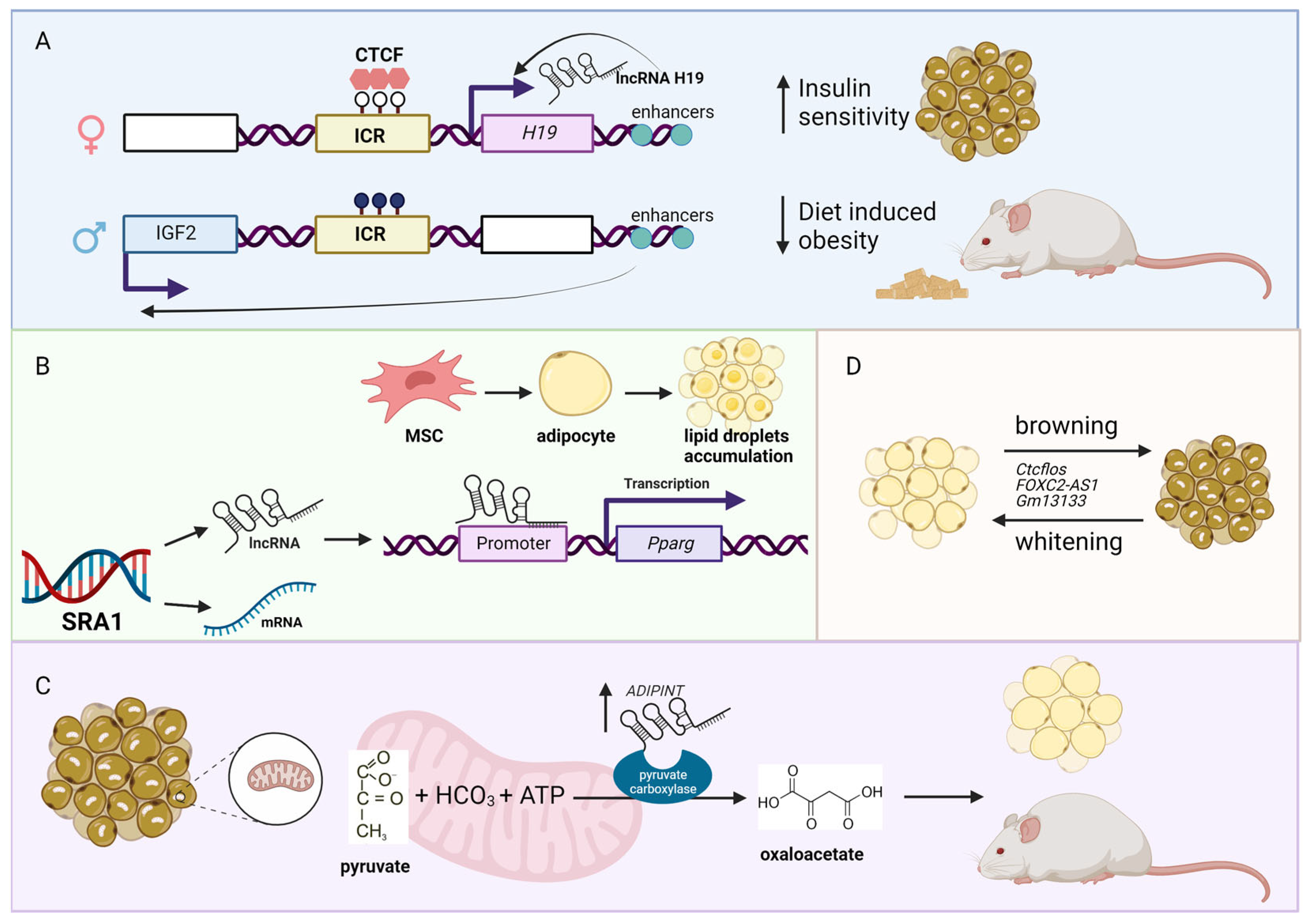
2. Obesity-Associated lncRNAs
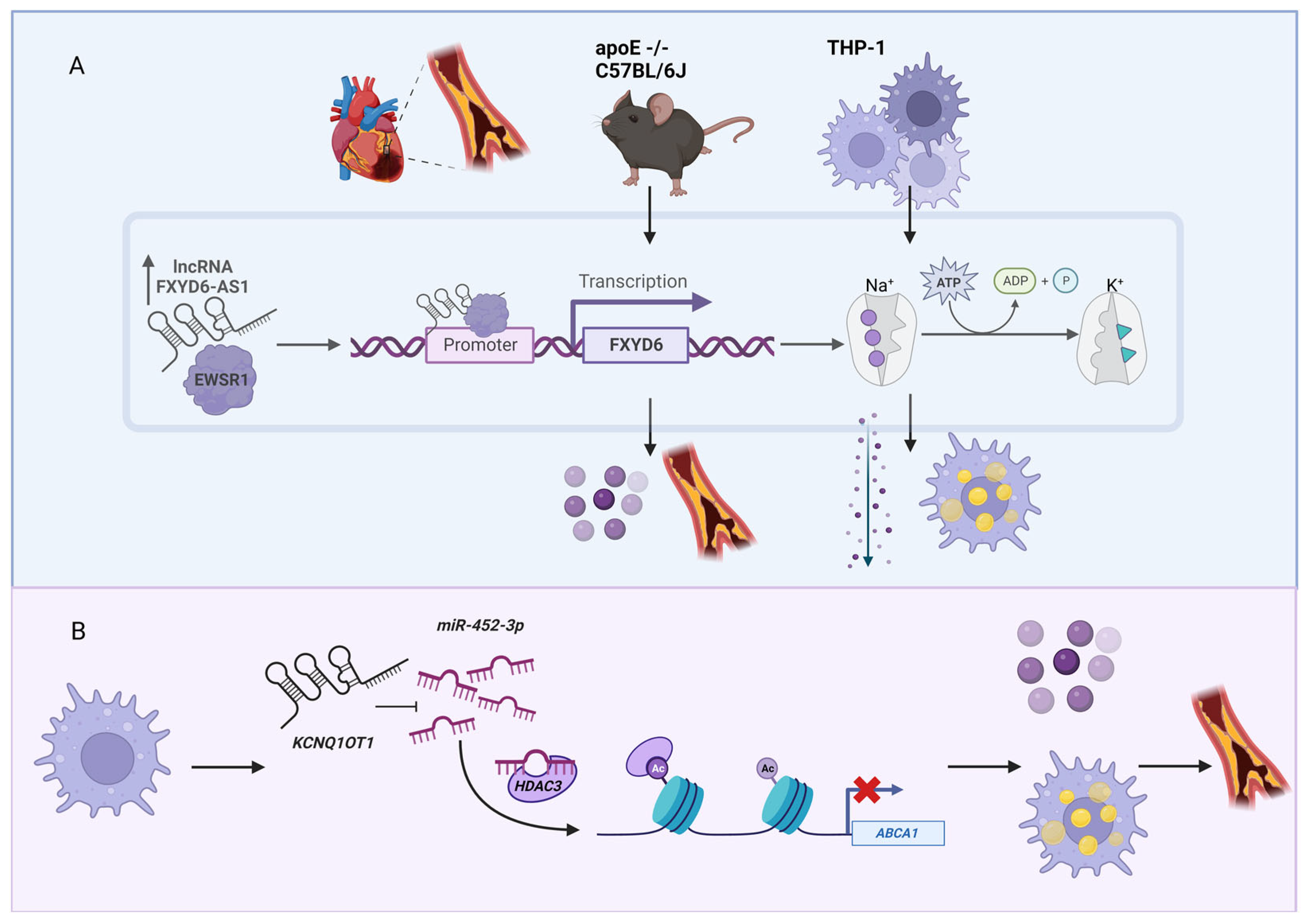
3. Cholesterol-Associated lncRNAs
4. Diabetes-Associated lncRNAs
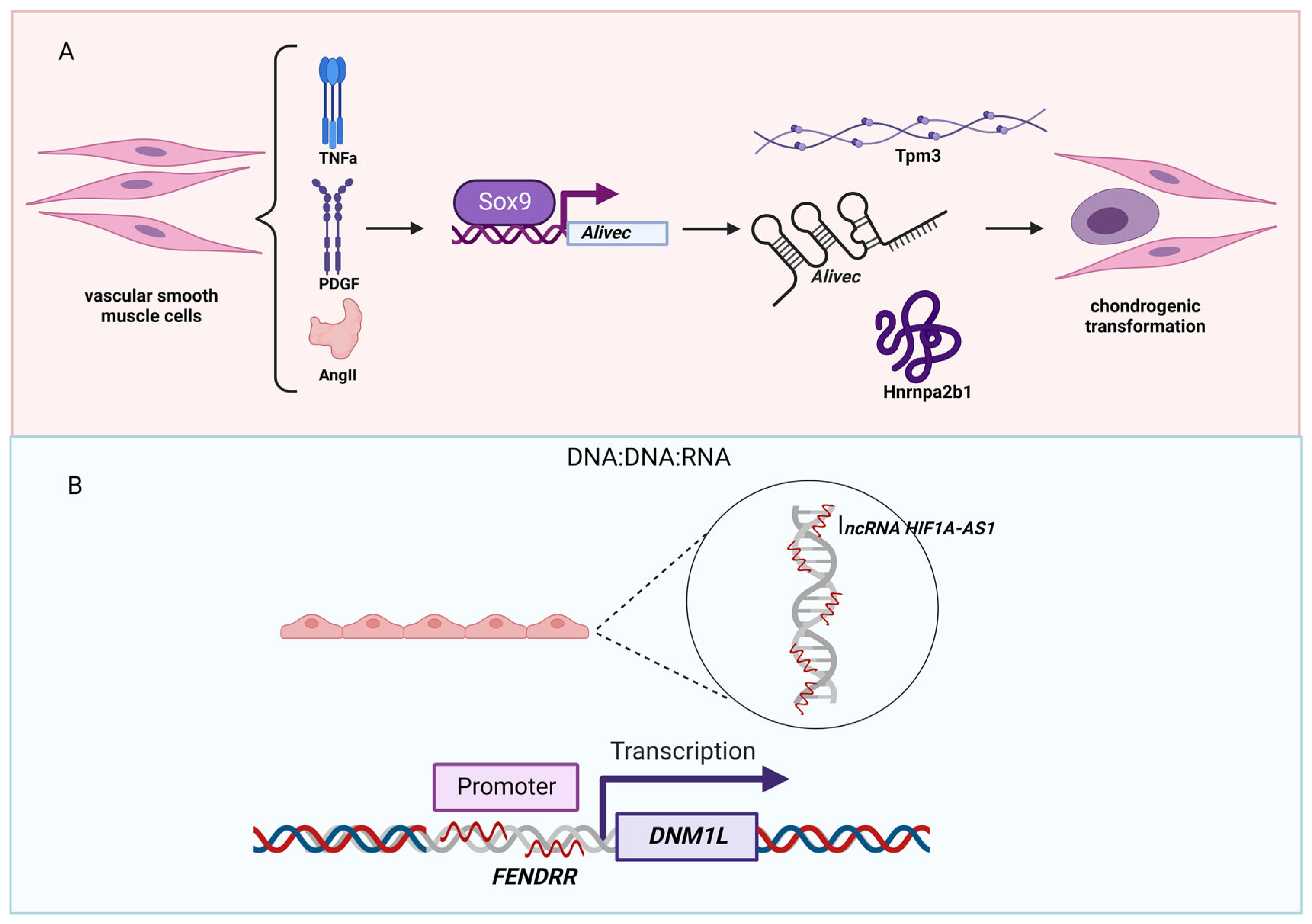
5. Hypertension-Associated lncRNAs
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, T.; Yi, J.; He, Y.; Zhang, J.; Li, X.; Ke, S.; Xia, L.; Liu, L. Associations of Dietary Fats with All-Cause Mortality and Cardiovascular Disease Mortality among Patients with Cardiometabolic Disease. Nutrients 2022, 14, 3608. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Menche, J. The regulatory network architecture of cardiometabolic diseases. Nat. Genet. 2022, 54, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Association, T.H. HjerteTal.dk. Available online: www.hjertetal.dk (accessed on 9 December 2022).
- Musunuru, K.; Hershberger, R.E.; Day, S.M.; Klinedinst, N.J.; Landstrom, A.P.; Parikh, V.N.; Prakash, S.; Semsarian, C.; Sturm, A.C.; American Heart Association Council on Genomic and Precision Medicine; et al. Genetic Testing for Inherited Cardiovascular Diseases: A Scientific Statement From the American Heart Association. Circ. Genom. Precis. Med. 2020, 13, e000067. [Google Scholar] [CrossRef]
- Kolber, M.R.; Scrimshaw, C. Family history of cardiovascular disease. Can. Fam. Physician 2014, 60, 1016. [Google Scholar] [PubMed]
- Arvanitis, M.; Tampakakis, E.; Zhang, Y.; Wang, W.; Auton, A.; 23andMe Research Team; Dutta, D.; Glavaris, S.; Keramati, A.; Chatterjee, N.; et al. Genome-wide association and multi-omic analyses reveal ACTN2 as a gene linked to heart failure. Nat. Commun. 2020, 11, 1122. [Google Scholar] [CrossRef] [Green Version]
- Benn, M.; Nordestgaard, B.G. From genome-wide association studies to Mendelian randomization: Novel opportunities for understanding cardiovascular disease causality, pathogenesis, prevention, and treatment. Cardiovasc. Res. 2018, 114, 1192–1208. [Google Scholar] [CrossRef] [PubMed]
- Kessler, T.; Vilne, B.; Schunkert, H. The impact of genome-wide association studies on the pathophysiology and therapy of cardiovascular disease. EMBO Mol. Med. 2016, 8, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Grow, M.A.; Pallaud, C.; Klitz, W.; Erlich, H.A.; Visvikis, S.; Chen, J.J.; Pullinger, C.R.; Malloy, M.J.; Siest, G.; et al. A multilocus genotyping assay for candidate markers of cardiovascular disease risk. Genome Res. 1999, 9, 936–949. [Google Scholar] [CrossRef] [Green Version]
- Aragam, K.G.; Jiang, T.; Goel, A.; Kanoni, S.; Wolford, B.N.; Atri, D.S.; Weeks, E.M.; Wang, M.; Hindy, G.; Zhou, W.; et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat. Genet. 2022, 54, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Adams, J.C. Physiological roles of non-coding RNAs. Am. J. Physiol. Cell Physiol. 2019, 317, C1–C2. [Google Scholar] [CrossRef]
- Nurnberg, S.T.; Zhang, H.; Hand, N.J.; Bauer, R.C.; Saleheen, D.; Reilly, M.P.; Rader, D.J. From Loci to Biology: Functional Genomics of Genome-Wide Association for Coronary Disease. Circ. Res. 2016, 118, 586–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirza, A.H.; Kaur, S.; Brorsson, C.A.; Pociot, F. Effects of GWAS-associated genetic variants on lncRNAs within IBD and T1D candidate loci. PLoS ONE 2014, 9, e105723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holdt, L.M.; Teupser, D. Long Noncoding RNA ANRIL: Lnc-ing Genetic Variation at the Chromosome 9p21 Locus to Molecular Mechanisms of Atherosclerosis. Front. Cardiovasc. Med. 2018, 5, 145. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Beutner, F.; Scholz, M.; Gielen, S.; Gabel, G.; Bergert, H.; Schuler, G.; Thiery, J.; Teupser, D. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arter. Thromb. Vasc. Biol. 2010, 30, 620–627. [Google Scholar] [CrossRef] [Green Version]
- Matarin, M.; Brown, W.M.; Singleton, A.; Hardy, J.A.; Meschia, J.F. Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chromosome 9p21. Stroke 2008, 39, 1586–1589. [Google Scholar] [CrossRef]
- Larson, M.G.; Atwood, L.D.; Benjamin, E.J.; Cupples, L.A.; D’Agostino, R.B., Sr.; Fox, C.S.; Govindaraju, D.R.; Guo, C.Y.; Heard-Costa, N.L.; Hwang, S.J.; et al. Framingham Heart Study 100K project: Genome-wide associations for cardiovascular disease outcomes. BMC Med. Genet. 2007, 8, S5. [Google Scholar] [CrossRef]
- Samani, N.J.; Erdmann, J.; Hall, A.S.; Hengstenberg, C.; Mangino, M.; Mayer, B.; Dixon, R.J.; Meitinger, T.; Braund, P.; Wichmann, H.E.; et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007, 357, 443–453. [Google Scholar] [CrossRef] [Green Version]
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature 2007, 447, 661–678. [Google Scholar] [CrossRef] [Green Version]
- McPherson, R.; Pertsemlidis, A.; Kavaslar, N.; Stewart, A.; Roberts, R.; Cox, D.R.; Hinds, D.A.; Pennacchio, L.A.; Tybjaerg-Hansen, A.; Folsom, A.R.; et al. A common allele on chromosome 9 associated with coronary heart disease. Science 2007, 316, 1488–1491. [Google Scholar] [CrossRef] [Green Version]
- Helgadottir, A.; Thorleifsson, G.; Manolescu, A.; Gretarsdottir, S.; Blondal, T.; Jonasdottir, A.; Jonasdottir, A.; Sigurdsson, A.; Baker, A.; Palsson, A.; et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007, 316, 1491–1493. [Google Scholar] [CrossRef]
- Fox, A.; Feng, W.; Asal, V. What is driving global obesity trends? Globalization or “modernization”? Glob. Health 2019, 15, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romieu, I.; Dossus, L.; Barquera, S.; Blottiere, H.M.; Franks, P.W.; Gunter, M.; Hwalla, N.; Hursting, S.D.; Leitzmann, M.; Margetts, B.; et al. Energy balance and obesity: What are the main drivers? Cancer Causes Control 2017, 28, 247–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shook, R.P.; Blair, S.N.; Duperly, J.; Hand, G.A.; Matsudo, S.M.; Slavin, J.L. What is Causing the Worldwide Rise in Body Weight? Eur. Endocrinol. 2014, 10, 136–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, E.; Dhaouadi, I.; Gaziano, I.; Oliverio, M.; Klemm, P.; Awazawa, M.; Mitterer, G.; Fernandez-Rebollo, E.; Pradas-Juni, M.; Wagner, W.; et al. LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat. Commun. 2018, 9, 3622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viereck, J.; Buhrke, A.; Foinquinos, A.; Chatterjee, S.; Kleeberger, J.A.; Xiao, K.; Janssen-Peters, H.; Batkai, S.; Ramanujam, D.; Kraft, T.; et al. Targeting muscle-enriched long non-coding RNA H19 reverses pathological cardiac hypertrophy. Eur. Heart J. 2020, 41, 3462–3474. [Google Scholar] [CrossRef]
- Hobuss, L.; Foinquinos, A.; Jung, M.; Kenneweg, F.; Xiao, K.; Wang, Y.; Zimmer, K.; Remke, J.; Just, A.; Nowak, J.; et al. Pleiotropic cardiac functions controlled by ischemia-induced lncRNA H19. J. Mol. Cell. Cardiol. 2020, 146, 43–59. [Google Scholar] [CrossRef]
- Liu, L.; An, X.; Li, Z.; Song, Y.; Li, L.; Zuo, S.; Liu, N.; Yang, G.; Wang, H.; Cheng, X.; et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 2016, 111, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Dong, B.; Chen, M.; Yao, C. LncRNA H19 suppresses pyroptosis of cardiomyocytes to attenuate myocardial infarction in a PBX3/CYP1B1-dependent manner. Mol. Cell. Biochem. 2021, 476, 1387–1400. [Google Scholar] [CrossRef]
- Yengo, L.; Sidorenko, J.; Kemper, K.E.; Zheng, Z.; Wood, A.R.; Weedon, M.N.; Frayling, T.M.; Hirschhorn, J.; Yang, J.; Visscher, P.M.; et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700,000 individuals of European ancestry. Hum. Mol. Genet. 2018, 27, 3641–3649. [Google Scholar] [CrossRef]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef]
- Manco, M.; Crudele, A.; Mosca, A.; Caccamo, R.; Braghini, M.R.; De Vito, R.; Alterio, A.; Pizzolante, F.; De Peppo, F.; Alisi, A. LncOb rs10487505 variant is associated with leptin levels in pediatric non-alcoholic fatty liver disease. Pediatr. Res. 2022, 92, 1737–1743. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Nguyen, T.T.L.; Gao, H.; Huang, H.; Kim, D.C.; Sharp, B.; Ye, Z.; Lee, J.H.; Coombes, B.J.; Ordog, T.; et al. TCF7L2 lncRNA: A link between bipolar disorder and body mass index through glucocorticoid signaling. Mol. Psychiatry 2021, 26, 7454–7464. [Google Scholar] [CrossRef] [PubMed]
- Delacretaz, A.; Preisig, M.; Vandenberghe, F.; Saigi Morgui, N.; Quteineh, L.; Choong, E.; Gholam-Rezaee, M.; Kutalik, Z.; Magistretti, P.; Aubry, J.M.; et al. Influence of MCHR2 and MCHR2-AS1 Genetic Polymorphisms on Body Mass Index in Psychiatric Patients and In Population-Based Subjects with Present or Past Atypical Depression. PLoS ONE 2015, 10, e0139155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yim, N.H.; Cha, M.H.; Kim, M.S. Hypermethylation of the TSPOAP1-AS1 Promoter May Be Associated with Obesity in Overweight/Obese Korean Subjects. Int. J. Mol. Sci. 2020, 21, 3307. [Google Scholar] [CrossRef]
- Sam, S.; Mazzone, T. Adipose tissue changes in obesity and the impact on metabolic function. Transl. Res. 2014, 164, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Attie, A.D.; Scherer, P.E. Adipocyte metabolism and obesity. J. Lipid Res. 2009, 50, S395–S399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Sheng, L.; Miao, H.; Saunders, T.L.; MacDougald, O.A.; Koenig, R.J.; Xu, B. SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J. Biol. Chem. 2014, 289, 13000–13009. [Google Scholar] [CrossRef] [Green Version]
- Lanz, R.B.; McKenna, N.J.; Onate, S.A.; Albrecht, U.; Wong, J.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 1999, 97, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Kerr, A.G.; Wang, Z.; Wang, N.; Kwok, K.H.M.; Jalkanen, J.; Ludzki, A.; Lecoutre, S.; Langin, D.; Bergo, M.O.; Dahlman, I.; et al. The long noncoding RNA ADIPINT regulates human adipocyte metabolism via pyruvate carboxylase. Nat. Commun. 2022, 13, 2958. [Google Scholar] [CrossRef]
- Alvarez-Dominguez, J.R.; Winther, S.; Hansen, J.B.; Lodish, H.F.; Knoll, M. An adipose lncRAP2-Igf2bp2 complex enhances adipogenesis and energy expenditure by stabilizing target mRNAs. iScience 2022, 25, 103680. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, J.; Zhou, X.; Hu, S.; Ge, L.; Sun, J.; Li, P.; Long, K.; Jin, L.; Tang, Q.; et al. Comprehensive Analysis of mRNA and lncRNA Transcriptomes Reveals the Differentially Hypoxic Response of Preadipocytes During Adipogenesis. Front. Genet. 2020, 11, 845. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Lim, Y.C.; Chia, S.Y.; Walet, A.C.E.; Xu, S.; Lo, K.A.; Zhao, Y.; Zhu, D.; Shan, Z.; Chen, Q.; et al. De novo reconstruction of human adipose transcriptome reveals conserved lncRNAs as regulators of brown adipogenesis. Nat. Commun. 2018, 9, 1329. [Google Scholar] [CrossRef] [Green Version]
- Lo, K.A.; Huang, S.; Walet, A.C.E.; Zhang, Z.C.; Leow, M.K.; Liu, M.; Sun, L. Adipocyte Long-Noncoding RNA Transcriptome Analysis of Obese Mice Identified Lnc-Leptin, Which Regulates Leptin. Diabetes 2018, 67, 1045–1056. [Google Scholar] [CrossRef] [Green Version]
- Bai, Z.; Chai, X.R.; Yoon, M.J.; Kim, H.J.; Lo, K.A.; Zhang, Z.C.; Xu, D.; Siang, D.T.C.; Walet, A.C.E.; Xu, S.H.; et al. Dynamic transcriptome changes during adipose tissue energy expenditure reveal critical roles for long noncoding RNA regulators. PLoS Biol. 2017, 15, e2002176. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Goff, L.A.; Trapnell, C.; Alexander, R.; Lo, K.A.; Hacisuleyman, E.; Sauvageau, M.; Tazon-Vega, B.; Kelley, D.R.; Hendrickson, D.G.; et al. Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3387–3392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saely, C.H.; Geiger, K.; Drexel, H. Brown versus white adipose tissue: A mini-review. Gerontology 2012, 58, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Samuelson, I.; Vidal-Puig, A. Studying Brown Adipose Tissue in a Human in vitro Context. Front. Endocrinol. 2020, 11, 629. [Google Scholar] [CrossRef]
- Wu, C.; Fang, S.; Zhang, H.; Li, X.; Du, Y.; Zhang, Y.; Lin, X.; Wang, L.; Ma, X.; Xue, Y.; et al. Long noncoding RNA XIST regulates brown preadipocytes differentiation and combats high-fat diet induced obesity by targeting C/EBPalpha. Mol. Med. 2022, 28, 6. [Google Scholar] [CrossRef]
- Herz, C.T.; Kiefer, F.W. Adipose tissue browning in mice and humans. J. Endocrinol. 2019, 241, R97–R109. [Google Scholar] [CrossRef]
- Gupta, A.; Shamsi, F.; Altemose, N.; Dorlhiac, G.F.; Cypess, A.M.; White, A.P.; Yosef, N.; Patti, M.E.; Tseng, Y.H.; Streets, A. Characterization of transcript enrichment and detection bias in single-nucleus RNA-seq for mapping of distinct human adipocyte lineages. Genome Res. 2022, 32, 242–257. [Google Scholar] [CrossRef]
- Wang, Y.; Hua, S.; Cui, X.; Cao, Y.; Wen, J.; Chi, X.; Ji, C.; Pang, L.; You, L. The Effect of FOXC2-AS1 on White Adipocyte Browning and the Possible Regulatory Mechanism. Front. Endocrinol. 2020, 11, 565483. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Zhou, Y.; Cui, X.; Wang, X.; Sun, Y.; Gao, Y.; Wang, X.; Wen, J.; Xie, K.; Tang, R.; et al. GM13133 is a negative regulator in mouse white adipocytes differentiation and drives the characteristics of brown adipocytes. J. Cell. Physiol. 2018, 233, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Malakar, A.K.; Choudhury, D.; Halder, B.; Paul, P.; Uddin, A.; Chakraborty, S. A review on coronary artery disease, its risk factors, and therapeutics. J. Cell. Physiol. 2019, 234, 16812–16823. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.C.; Gerhardt, T.E.; Kwon, E. Risk factors for coronary artery disease. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Zareba, L.; Fitas, A.; Wolska, M.; Junger, E.; Eyileten, C.; Wicik, Z.; De Rosa, S.; Siller-Matula, J.M.; Postula, M. MicroRNAs and Long Noncoding RNAs in Coronary Artery Disease: New and Potential Therapeutic Targets. Cardiol. Clin. 2020, 38, 601–617. [Google Scholar] [CrossRef] [PubMed]
- Cardona-Monzonis, A.; Garcia-Gimenez, J.L.; Mena-Molla, S.; Pareja-Galeano, H.; de la Guia-Galipienso, F.; Lippi, G.; Pallardo, F.V.; Sanchis-Gomar, F. Non-coding RNAs and Coronary Artery Disease. Adv. Exp. Med. Biol. 2020, 1229, 273–285. [Google Scholar] [CrossRef]
- Dong, X.H.; Lu, Z.F.; Kang, C.M.; Li, X.H.; Haworth, K.E.; Ma, X.; Lu, J.B.; Liu, X.H.; Fang, F.C.; Wang, C.S.; et al. The Long Noncoding RNA RP11-728F11.4 Promotes Atherosclerosis. Arter. Thromb. Vasc. Biol. 2021, 41, 1191–1204. [Google Scholar] [CrossRef]
- Camargo, F.N.; Matos, S.L.; Araujo, L.C.C.; Carvalho, C.R.O.; Amaral, A.G.; Camporez, J.P. Western Diet-Fed ApoE Knockout Male Mice as an Experimental Model of Non-Alcoholic Steatohepatitis. Curr. Issues Mol. Biol. 2022, 44, 4692–4703. [Google Scholar] [CrossRef]
- Kampschulte, M.; Stockl, C.; Langheinrich, A.C.; Althohn, U.; Bohle, R.M.; Krombach, G.A.; Stieger, P.; Churin, Y.; Kremer, S.; Dierkes, C.; et al. Western diet in ApoE-LDLR double-deficient mouse model of atherosclerosis leads to hepatic steatosis, fibrosis, and tumorigenesis. Lab. Invest. 2014, 94, 1273–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.H.; Deng, W.Y.; Chen, J.J.; Xu, X.D.; Liu, X.X.; Chen, L.; Shi, M.W.; Liu, Q.X.; Tao, M.; Ren, K. LncRNA kcnq1ot1 promotes lipid accumulation and accelerates atherosclerosis via functioning as a ceRNA through the miR-452-3p/HDAC3/ABCA1 axis. Cell Death Dis. 2020, 11, 1043. [Google Scholar] [CrossRef]
- Wang, B.; Su, Z.; Wan, L.; He, T. Relationship between long non-coding RNA polymorphism and the risk of coronary artery disease: A protocol for systematic review and meta-analysis. Medicine 2021, 100, e25146. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Gholipour, M.; Taheri, M. The Emerging Role of Long Non-coding RNAs and Circular RNAs in Coronary Artery Disease. Front. Cardiovasc. Med. 2021, 8, 632393. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Y.; Huang, J.; Liu, Y.; Zhu, Y. Association of genetic variants in lncRNA GAS5/miR-21/mTOR axis with risk and prognosis of coronary artery disease among a Chinese population. J. Clin. Lab. Anal. 2020, 34, e23430. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, M.; Li, J.; Liang, B.; Chen, Z.; Yang, J.; Guo, X.; Huang, S.; Gu, L.; Su, L. LncRNA H19 rs4929984 Variant is Associated with Coronary Artery Disease Susceptibility in Han Chinese Female Population. Biochem. Genet. 2021, 59, 1359–1380. [Google Scholar] [CrossRef]
- Hu, W.N.; Ding, H.X.; Xu, Q.; Zhang, X.Y.; Yang, D.T.; Jin, Y.Z. Relationship between Long Noncoding RNA H19 Polymorphisms and Risk of Coronary Artery Disease in a Chinese Population: A Case-Control Study. Dis. Markers 2020, 2020, 9839612. [Google Scholar] [CrossRef]
- Gao, W.; Zhu, M.; Wang, H.; Zhao, S.; Zhao, D.; Yang, Y.; Wang, Z.M.; Wang, F.; Yang, Z.J.; Lu, X.; et al. Association of polymorphisms in long non-coding RNA H19 with coronary artery disease risk in a Chinese population. Mutat. Res. 2015, 772, 15–22. [Google Scholar] [CrossRef]
- Kim, I.J.; Lee, J.Y.; Park, H.W.; Park, H.S.; Ko, E.J.; Sung, J.H.; Kim, N.K. Association between HOTAIR lncRNA Polymorphisms and Coronary Artery Disease Susceptibility. J. Pers. Med. 2021, 11, 375. [Google Scholar] [CrossRef]
- Song, N.; Luo, J.Y.; Zhao, Q.; Zhang, J.Y.; Liu, F.; Li, X.M.; Yang, Y.N. MALAT1 gene rs600231 polymorphism positively associated with acute coronary syndrome in Chinese population: A case-control study. Cardiovasc. Diagn. Ther. 2021, 11, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Ding, H.; Ouyang, A.; Zhang, X.; Xu, Q.; Han, Y.; Zhang, X.; Jin, Y. LncRNA MALAT1 gene polymorphisms in coronary artery disease: A case-control study in a Chinese population. Biosci. Rep. 2019, 39, BSR20182213. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Li, Y.; Peng, Y.; Tang, J.; Li, H. Association of polymorphisms in MALAT1 with risk of coronary atherosclerotic heart disease in a Chinese population. Lipids Health Dis. 2018, 17, 75. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.S.; Cheng, J.; Cai, M.Y.; Yang, X.L.; Liu, X.G.; Zheng, B.Y.; Xiong, X.D. Association of lincRNA-p21 Haplotype with Coronary Artery Disease in a Chinese Han Population. Dis. Markers 2016, 2016, 9109743. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeFronzo, R.A.; Ferrannini, E.; Groop, L.; Henry, R.R.; Herman, W.H.; Holst, J.J.; Hu, F.B.; Kahn, C.R.; Raz, I.; Shulman, G.I.; et al. Type 2 diabetes mellitus. Nat. Rev. Dis. Prim. 2015, 1, 15019. [Google Scholar] [CrossRef]
- Olokoba, A.B.; Obateru, O.A.; Olokoba, L.B. Type 2 diabetes mellitus: A review of current trends. Oman Med. J. 2012, 27, 269–273. [Google Scholar] [CrossRef]
- Khan, M.A.B.; Hashim, M.J.; King, J.K.; Govender, R.D.; Mustafa, H.; Al Kaabi, J. Epidemiology of Type 2 Diabetes—Global Burden of Disease and Forecasted Trends. J. Epidemiol. Glob. Health 2020, 10, 107–111. [Google Scholar] [CrossRef] [Green Version]
- Schnurr, T.M.; Jakupovic, H.; Carrasquilla, G.D.; Angquist, L.; Grarup, N.; Sorensen, T.I.A.; Tjonneland, A.; Overvad, K.; Pedersen, O.; Hansen, T.; et al. Obesity, unfavourable lifestyle and genetic risk of type 2 diabetes: A case-cohort study. Diabetologia 2020, 63, 1324–1332. [Google Scholar] [CrossRef]
- Kyrou, I.; Tsigos, C.; Mavrogianni, C.; Cardon, G.; Van Stappen, V.; Latomme, J.; Kivela, J.; Wikstrom, K.; Tsochev, K.; Nanasi, A.; et al. Sociodemographic and lifestyle-related risk factors for identifying vulnerable groups for type 2 diabetes: A narrative review with emphasis on data from Europe. BMC Endocr. Disord. 2020, 20, 134. [Google Scholar] [CrossRef] [Green Version]
- Leong, A.; Porneala, B.; Dupuis, J.; Florez, J.C.; Meigs, J.B. Type 2 Diabetes Genetic Predisposition, Obesity, and All-Cause Mortality Risk in the U.S.: A Multiethnic Analysis. Diabetes Care 2016, 39, 539–546. [Google Scholar] [CrossRef] [Green Version]
- Martin-Timon, I.; Sevillano-Collantes, C.; Segura-Galindo, A.; Del Canizo-Gomez, F.J. Type 2 diabetes and cardiovascular disease: Have all risk factors the same strength? World J. Diabetes 2014, 5, 444–470. [Google Scholar] [CrossRef]
- Sun, D.; Zhou, T.; Heianza, Y.; Li, X.; Fan, M.; Fonseca, V.A.; Qi, L. Type 2 Diabetes and Hypertension. Circ. Res. 2019, 124, 930–937. [Google Scholar] [CrossRef]
- Duh, E.J.; Sun, J.K.; Stitt, A.W. Diabetic retinopathy: Current understanding, mechanisms, and treatment strategies. JCI Insight 2017, 2, e93751. [Google Scholar] [CrossRef] [PubMed]
- McGill, J.B.; Haller, H.; Roy-Chaudhury, P.; Cherrington, A.; Wada, T.; Wanner, C.; Ji, L.; Rossing, P. Making an impact on kidney disease in people with type 2 diabetes: The importance of screening for albuminuria. BMJ Open Diabetes Res. Care 2022, 10, e002806. [Google Scholar] [CrossRef] [PubMed]
- Chi, T.; Lin, J.; Wang, M.; Zhao, Y.; Liao, Z.; Wei, P. Non-Coding RNA as Biomarkers for Type 2 Diabetes Development and Clinical Management. Front. Endocrinol. 2021, 12, 630032. [Google Scholar] [CrossRef] [PubMed]
- Formichi, C.; Nigi, L.; Grieco, G.E.; Maccora, C.; Fignani, D.; Brusco, N.; Licata, G.; Sebastiani, G.; Dotta, F. Non-Coding RNAs: Novel Players in Insulin Resistance and Related Diseases. Int. J. Mol. Sci. 2021, 22, 7716. [Google Scholar] [CrossRef]
- Dieter, C.; Lemos, N.E.; Correa, N.R.F.; Assmann, T.S.; Crispim, D. The Impact of lncRNAs in Diabetes Mellitus: A Systematic Review and In Silico Analyses. Front. Endocrinol. 2021, 12, 602597. [Google Scholar] [CrossRef]
- Chen, Y.; He, Y.; Zhou, H. The potential role of lncRNAs in diabetes and diabetic microvascular complications. Endocr. J. 2020, 67, 659–668. [Google Scholar] [CrossRef]
- Hosen, M.R.; Militello, G.; Weirick, T.; Ponomareva, Y.; Dassanayaka, S.; Moore, J.B.; Doring, C.; Wysoczynski, M.; Jones, S.P.; Dimmeler, S.; et al. Airn Regulates Igf2bp2 Translation in Cardiomyocytes. Circ. Res. 2018, 122, 1347–1353. [Google Scholar] [CrossRef]
- Peng, T.; Liu, M.; Hu, L.; Guo, D.; Wang, D.; Qi, B.; Ren, G.; Hu, C.; Zhang, F.; Chun, H.J.; et al. LncRNA Airn alleviates diabetic cardiac fibrosis by inhibiting activation of cardiac fibroblasts via a m6A-IMP2-p53 axis. Biol. Direct 2022, 17, 32. [Google Scholar] [CrossRef]
- Miller, H.E.; Ilieva, M.; Bishop, A.J.R.; Uchida, S. Current Status of Epitranscriptomic Marks Affecting lncRNA Structures and Functions. Noncoding RNA 2022, 8, 23. [Google Scholar] [CrossRef]
- Leon, B.M.; Maddox, T.M. Diabetes and cardiovascular disease: Epidemiology, biological mechanisms, treatment recommendations and future research. World J. Diabetes 2015, 6, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Xu, W.; Zhang, W.; Wang, W.; Liu, T.; Zhou, X. LncRNA DCRF regulates cardiomyocyte autophagy by targeting miR-551b-5p in diabetic cardiomyopathy. Theranostics 2019, 9, 4558–4566. [Google Scholar] [CrossRef]
- Furman, B.L. Streptozotocin-Induced Diabetic Models in Mice and Rats. Curr. Protoc. Pharmacol. 2015, 70, 5.47.1–5.47.20. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Lin, Y.; Shen, H.; Yu, S.; Xu, J. LncRNA TUG1 exacerbates myocardial fibrosis in diabetic cardiomyopathy by modulating the microRNA-145a-5p/Cfl2 axis. J. Cardiovasc. Pharmacol. 2022; Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tian, C.; Gao, Z.; Zhang, B.; Zhao, L. Research status and trends of the diabetic cardiomyopathy in the past 10 years (2012-2021): A bibliometric analysis. Front. Cardiovasc. Med. 2022, 9, 1018841. [Google Scholar] [CrossRef]
- Feng, B.; Liu, J.; Wang, E.; Su, Z.; Chakrabarti, S. Endothelial derived miRNA-9 mediated cardiac fibrosis in diabetes and its regulation by ZFAS1. PLoS ONE 2022, 17, e0276076. [Google Scholar] [CrossRef]
- Xiao, W.; Zheng, D.; Chen, X.; Yu, B.; Deng, K.; Ma, J.; Wen, X.; Hu, Y.; Hou, J. Long non-coding RNA MIAT is involved in the regulation of pyroptosis in diabetic cardiomyopathy via targeting miR-214-3p. iScience 2021, 24, 103518. [Google Scholar] [CrossRef]
- Ni, T.; Huang, X.; Pan, S.; Lu, Z. Inhibition of the long non-coding RNA ZFAS1 attenuates ferroptosis by sponging miR-150-5p and activates CCND2 against diabetic cardiomyopathy. J. Cell. Mol. Med. 2021, 25, 9995–10007. [Google Scholar] [CrossRef]
- Zhou, J.; Ji, X.; Zhang, F.; Yan, K.; Huang, P.; Huang, C. Bioinformatics analysis for identifying micro-RNAs, long noncoding RNAs, transcription factors, and immune genes regulatory networks in diabetic cardiomyopathy using an integrated bioinformatics analysis. Inflamm. Res. 2022, 71, 847–858. [Google Scholar] [CrossRef]
- Rai, A.K.; Lee, B.; Gomez, R.; Rajendran, D.; Khan, M.; Garikipati, V.N.S. Current Status and Potential Therapeutic Strategies for Using Non-coding RNA to Treat Diabetic Cardiomyopathy. Front. Physiol. 2020, 11, 612722. [Google Scholar] [CrossRef]
- Chen, K.; Ma, Y.; Wu, S.; Zhuang, Y.; Liu, X.; Lv, L.; Zhang, G. Construction and analysis of a lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveals functional lncRNAs in diabetic cardiomyopathy. Mol. Med. Rep. 2019, 20, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Whelton, P.K.; Carey, R.M.; Mancia, G.; Kreutz, R.; Bundy, J.D.; Williams, B. Harmonization of the American College of Cardiology/American Heart Association and European Society of Cardiology/European Society of Hypertension Blood Pressure/Hypertension Guidelines: Comparisons, Reflections, and Recommendations. J. Am. Coll. Cardiol. 2022, 80, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
- Samara, V.A.; Das, S.; Reddy, M.A.; Tanwar, V.S.; Stapleton, K.; Leung, A.; Abdollahi, M.; Ganguly, R.; Lanting, L.; Natarajan, R. Angiotensin II-Induced Long Non-Coding RNA Alivec Regulates Chondrogenesis in Vascular Smooth Muscle Cells. Cells 2021, 10, 2696. [Google Scholar] [CrossRef]
- Hassoun, P.M. Pulmonary Arterial Hypertension. N. Engl. J. Med. 2021, 385, 2361–2376. [Google Scholar] [CrossRef]
- Mathew, R. Pulmonary hypertension and metabolic syndrome: Possible connection, PPARgamma and Caveolin-1. World J. Cardiol. 2014, 6, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.J.; Vorla, M.; Kalra, D.K. Molecular Pathways in Pulmonary Arterial Hypertension. Int. J. Mol. Sci. 2022, 23, 10001. [Google Scholar] [CrossRef]
- Maron, B.A.; Loscalzo, J. Pulmonary hypertension: Pathophysiology and signaling pathways. Handb. Exp. Pharmacol. 2013, 218, 31–58. [Google Scholar] [CrossRef]
- Bernardi, N.; Bianconi, E.; Vecchi, A.; Ameri, P. Noncoding RNAs in Pulmonary Arterial Hypertension: Current Knowledge and Translational Perspectives. Heart Fail. Clin. 2023, 19, 137–152. [Google Scholar] [CrossRef]
- Deng, L.; Han, X.; Wang, Z.; Nie, X.; Bian, J. The Landscape of Noncoding RNA in Pulmonary Hypertension. Biomolecules 2022, 12, 796. [Google Scholar] [CrossRef]
- Shirazi-Tehrani, E.; Chamasemani, A.; Firouzabadi, N.; Mousaei, M. ncRNAs and polyphenols: New therapeutic strategies for hypertension. RNA Biol. 2022, 19, 575–587. [Google Scholar] [CrossRef]
- Zang, H.; Zhang, Q.; Li, X. Non-Coding RNA Networks in Pulmonary Hypertension. Front. Genet. 2021, 12, 703860. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Long, Y.; Kwoh, C.K. Deep learning based DNA:RNA triplex forming potential prediction. BMC Bioinform. 2020, 21, 522. [Google Scholar] [CrossRef] [PubMed]
- Antonov, I.; Medvedeva, Y.A. Purine-rich low complexity regions are potential RNA binding hubs in the human genome. F1000Research 2018, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Leisegang, M.S.; Bains, J.K.; Seredinski, S.; Oo, J.A.; Krause, N.M.; Kuo, C.C.; Gunther, S.; Senturk Cetin, N.; Warwick, T.; Cao, C.; et al. HIF1alpha-AS1 is a DNA:DNA:RNA triplex-forming lncRNA interacting with the HUSH complex. Nat. Commun. 2022, 13, 6563. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Wahrisch, S.; Beisaw, A.; Macura, K.; Blass, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, Q.; He, S.; Bai, J.; Ma, C.; Zhang, L.; Guan, X.; Yuan, H.; Li, Y.; Zhu, X.; et al. LncRNA FENDRR with m6A RNA methylation regulates hypoxia-induced pulmonary artery endothelial cell pyroptosis by mediating DRP1 DNA methylation. Mol. Med. 2022, 28, 126. [Google Scholar] [CrossRef]
- Ilieva, M.; Uchida, S. Perspectives of LncRNAs for therapy. Cell Biol. Toxicol. 2022, 38, 915–917. [Google Scholar] [CrossRef]
- Badowski, C.; He, B.; Garmire, L.X. Blood-derived lncRNAs as biomarkers for cancer diagnosis: The Good, the Bad and the Beauty. NPJ Precis. Oncol. 2022, 6, 40. [Google Scholar] [CrossRef]
- Lu, C.; Wei, D.; Zhang, Y.; Wang, P.; Zhang, W. Long Non-Coding RNAs as Potential Diagnostic and Prognostic Biomarkers in Breast Cancer: Progress and Prospects. Front. Oncol. 2021, 11, 710538. [Google Scholar] [CrossRef]
- Pant, T.; Dhanasekaran, A.; Zhao, M.; Thorp, E.B.; Forbess, J.M.; Bosnjak, Z.J.; Benjamin, I.J.; Ge, Z.D. Identification and analysis of circulating long non-coding RNAs with high significance in diabetic cardiomyopathy. Sci. Rep. 2021, 11, 2571. [Google Scholar] [CrossRef]
- Ismail, N.; Abdullah, N.; Abdul Murad, N.A.; Jamal, R.; Sulaiman, S.A. Long Non-Coding RNAs (lncRNAs) in Cardiovascular Disease Complication of Type 2 Diabetes. Diagnostics 2021, 11, 145. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ilieva, M.; Uchida, S. Potential Involvement of LncRNAs in Cardiometabolic Diseases. Genes 2023, 14, 213. https://doi.org/10.3390/genes14010213
Ilieva M, Uchida S. Potential Involvement of LncRNAs in Cardiometabolic Diseases. Genes. 2023; 14(1):213. https://doi.org/10.3390/genes14010213
Chicago/Turabian StyleIlieva, Mirolyuba, and Shizuka Uchida. 2023. "Potential Involvement of LncRNAs in Cardiometabolic Diseases" Genes 14, no. 1: 213. https://doi.org/10.3390/genes14010213
APA StyleIlieva, M., & Uchida, S. (2023). Potential Involvement of LncRNAs in Cardiometabolic Diseases. Genes, 14(1), 213. https://doi.org/10.3390/genes14010213






