Molecular Evolution and Protein Structure Variation of Dkk Family
Abstract
:1. Introduction
2. Results
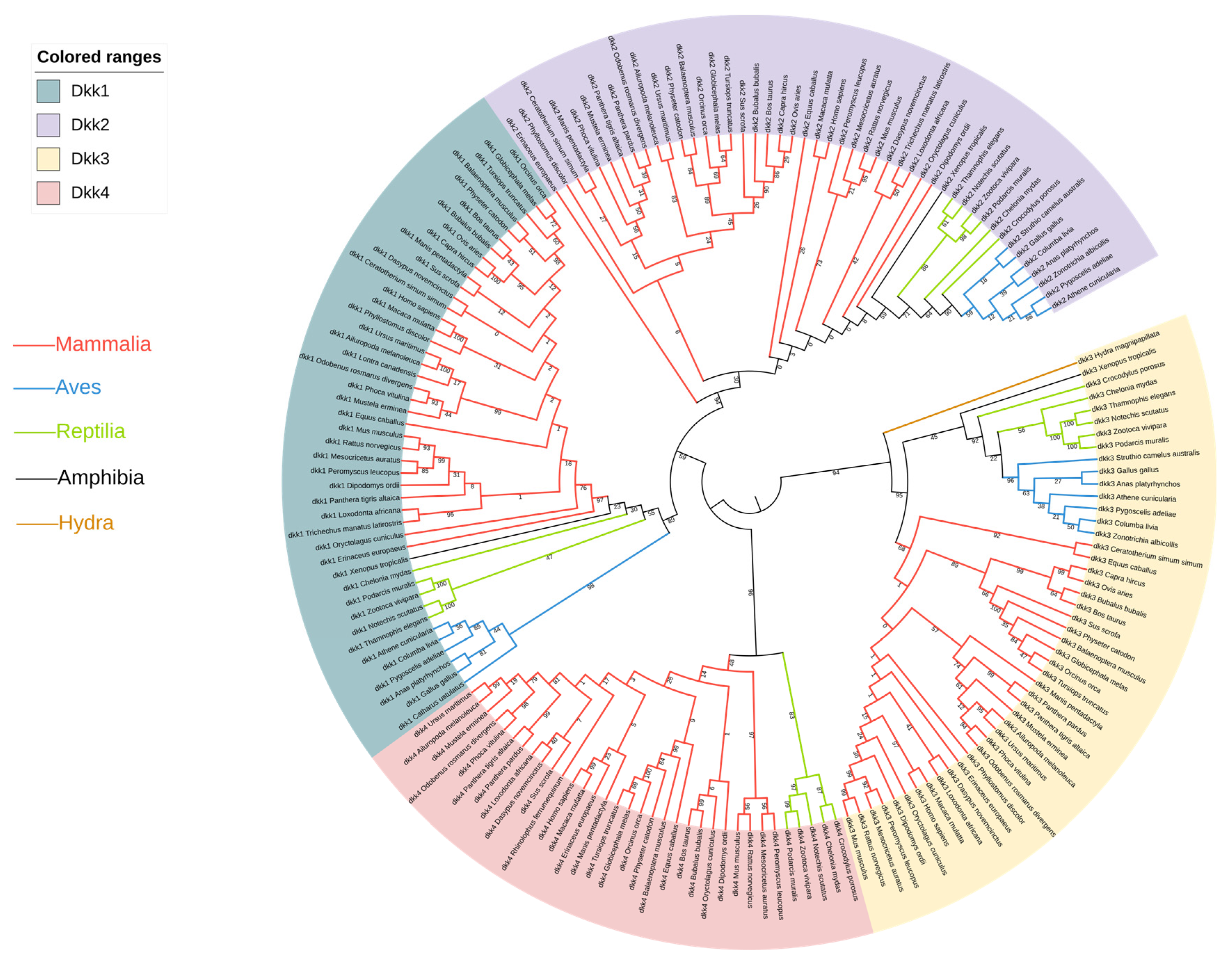
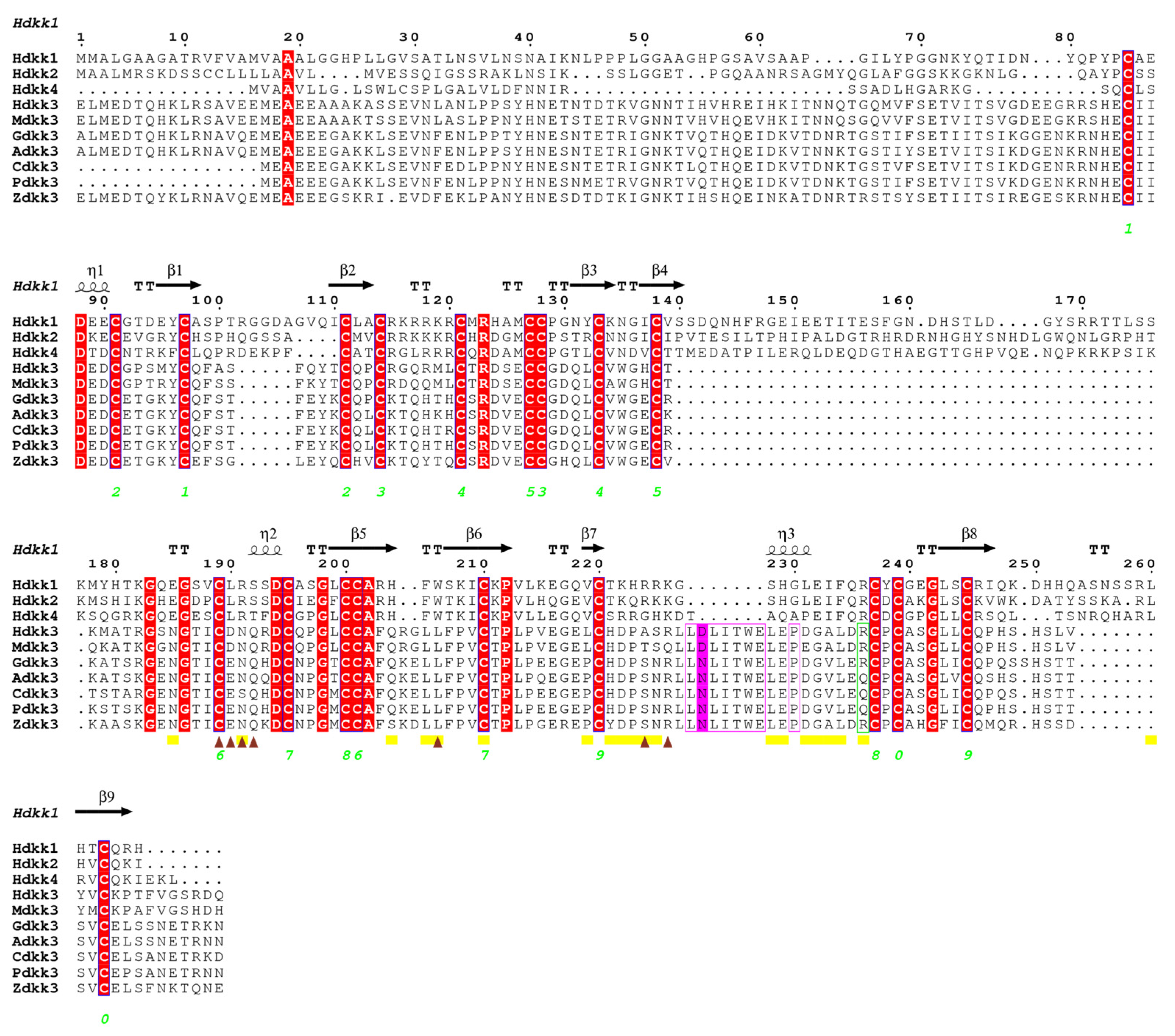
2.1. Sequence Alignment and Phylogenetic Analyses
2.2. Variation in Molecular Evolution Rate of Dkk
2.3. Dkks Functional Divergence Analysis Results
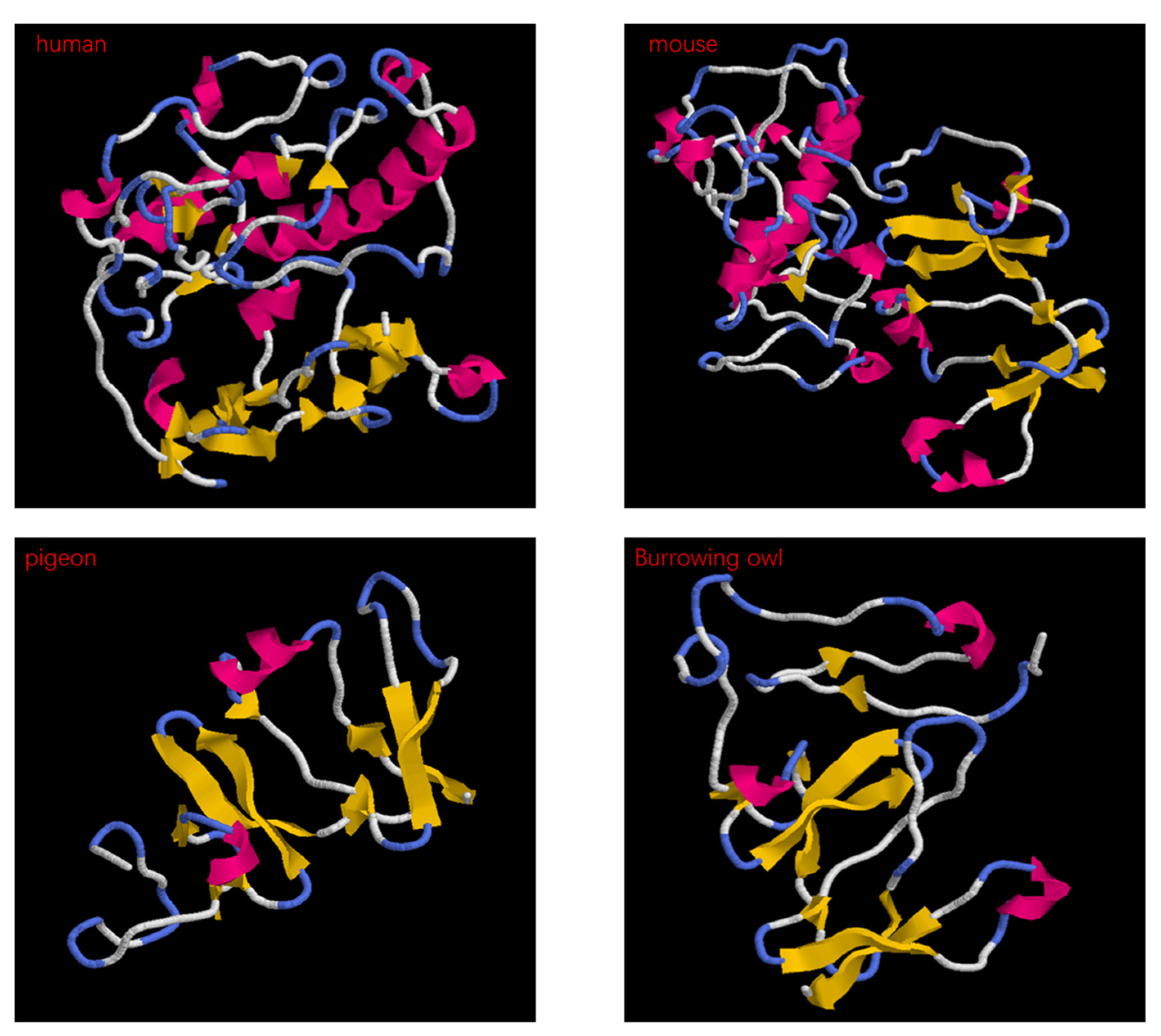
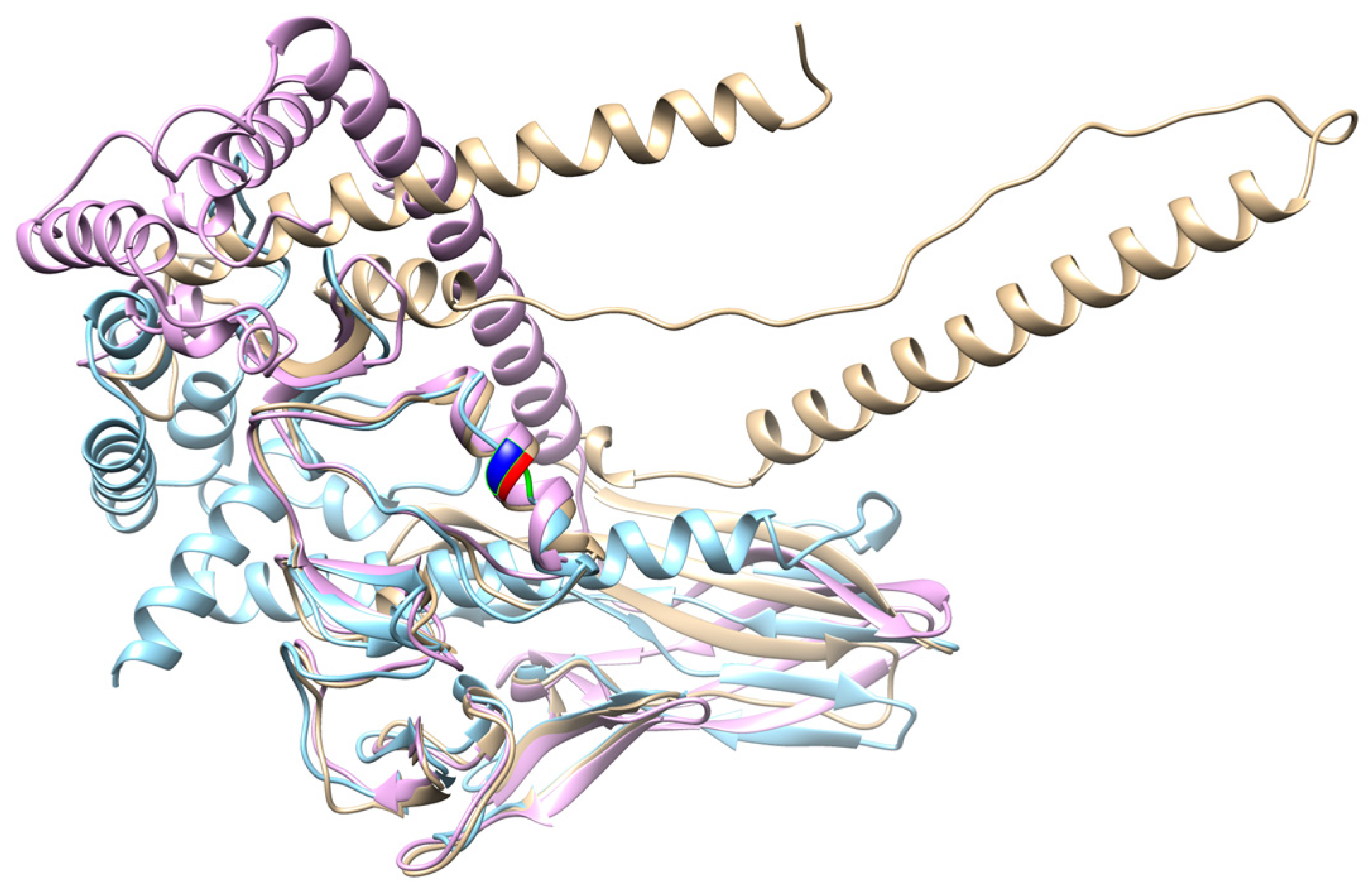
2.4. Dkk Protein Analysis Results
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sequence Acquisition
5.2. Nucleotide Sequence Analysis
5.3. Molecular Evolution Analysis
5.4. Protein Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glinka, A.; Wu, W.; Delius, H.; Monaghan, A.P.; Blumenstock, C.; Niehrs, C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 1998, 391, 357–362. [Google Scholar] [CrossRef]
- Krupnik, V.E.; Sharp, J.D.; Jiang, C.; Robison, K.; Chickering, T.W.; Amaravadi, L.; Brown, D.E.; Guyot, D.; Mays, G.; Leiby, K.; et al. Functional and structural diversity of the human Dickkopf gene family. Gene 1999, 238, 301–313. [Google Scholar] [CrossRef]
- Guder, C.; Pinho, S.; Nacak, T.G.; Schmidt, H.A.; Hobmayer, B.; Niehrs, C.; Holstein, T.W. An ancient Wnt-Dickkopf antagonism in Hydra. Development 2006, 133, 901–911. [Google Scholar] [CrossRef]
- Hino, K.; Satou, Y.; Yagi, K.; Satoh, N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. VI. Genes for Wnt, TGFbeta, Hedgehog and JAK/STAT signaling pathways. Dev. Genes Evol. 2003, 213, 264–272. [Google Scholar] [CrossRef]
- Yoshida, S.; Satoh, H.; Mitsuya, T.; Tate, G. Assignment1 of the human dickkopf (Xenopus) homolog 4 (DKK4) to chromosome 8p11.2→p11.1 by fluorescence in situ hybridization. Cytogenet. Cell Genet. 2001, 94, 88–89. [Google Scholar] [CrossRef]
- Patel, S.; Barkell, A.M.; Gupta, D.; Strong, S.L.; Bruton, S.; Muskett, F.W.; Addis, P.W.; Renshaw, P.S.; Slocombe, P.M.; Doyle, C.; et al. Structural and functional analysis of Dickkopf 4 (Dkk4): New insights into Dkk evolution and regulation of Wnt signaling by Dkk and Kremen proteins. J. Biol. Chem. 2018, 293, 12149–12166. [Google Scholar] [CrossRef]
- Song, Y.; Boncompagni, A.C.; Kim, S.S.; Gochnauer, H.R.; Zhang, Y.; Loots, G.G.; Wu, D.; Li, Y.; Xu, M.; Millar, S.E. Regional Control of Hairless versus Hair-Bearing Skin by Dkk2. Cell Rep. 2018, 25, 2981–2991. [Google Scholar] [CrossRef]
- Andl, T.; Reddy, S.T.; Gaddapara, T.; Millar, S.E. WNT signals are required for the initiation of hair follicle development. Dev. Cell 2002, 2, 643–653. [Google Scholar] [CrossRef]
- Chu, Q.; Cai, L.; Fu, Y.; Chen, X.; Yan, Z.; Lin, X.; Zhou, G.; Han, H.; Widelitz, R.B.; Chuong, C.M.; et al. Dkk2/Frzb in the dermal papillae regulates feather regeneration. Dev. Biol. 2014, 387, 167–178. [Google Scholar] [CrossRef]
- Hiramitsu, S.; Terauchi, M.; Kubota, T. The effects of Dickkopf-4 on the proliferation, differentiation, and apoptosis of osteoblasts. Endocrinology 2013, 154, 4618–4626. [Google Scholar] [CrossRef]
- Cui, C.Y.; Kunisada, M.; Piao, Y.; Childress, V.; Ko, M.S.; Schlessinger, D. Dkk4 and Eda regulate distinctive developmental mechanisms for subtypes of mouse hair. PLoS ONE 2010, 5, e10009. [Google Scholar] [CrossRef] [PubMed]
- Fedders, H.; Augustin, R.; Bosch, T.C. A Dickkopf-3-related gene is expressed in differentiating nematocytes in the basal metazoan Hydra. Dev. Genes Evol. 2004, 214, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 2006, 25, 7469–7481. [Google Scholar] [CrossRef] [PubMed]
- Veeck, J.; Dahl, E. Targeting the Wnt pathway in cancer: The emerging role of Dickkopf-3. Biochim. Biophys. Acta 2012, 1825, 18–28. [Google Scholar] [CrossRef]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Maderson, P.F.A. When? Why? and How? Some Speculations on the Evolution of the Vertebrate Integument. Am. Zool. 2015, 12, 159–171. [Google Scholar] [CrossRef]
- Yu, M.; Wu, P.; Widelitz, R.B.; Chuong, C.M. The morphogenesis of feathers. Nature 2002, 420, 308–312. [Google Scholar] [CrossRef]
- Winter, H.; Langbein, L.; Krawczak, M.; Cooper, D.N.; Jave-Suarez, L.F.; Rogers, M.A.; Praetzel, S.; Heidt, P.J.; Schweizer, J. Human type I hair keratin pseudogene phihHaA has functional orthologs in the chimpanzee and gorilla: Evidence for recent inactivation of the human gene after the Pan-Homo divergence. Hum. Genet. 2001, 108, 37–42. [Google Scholar] [CrossRef]
- Bhanot, P.; Brink, M.; Samos, C.H.; Hsieh, J.C.; Wang, Y.; Macke, J.P.; Andrew, D.; Nathans, J.; Nusse, R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature 1996, 382, 225–230. [Google Scholar] [CrossRef]
- Dann, C.E.; Hsieh, J.C.; Rattner, A.; Sharma, D.; Nathans, J.; Leahy, D.J. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 2001, 412, 86–90. [Google Scholar] [CrossRef]
- Lei, M.X.; Chuong, C.M.; Widelitz, R.B. Tuning Wnt signals for more or fewer hairs. J. Investig. Dermatol. 2013, 133, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Y.; Sennett, R.; Rezza, A.; Clavel, C.; Grisanti, L.; Zemla, R.; Najam, S.; Rendl, M. Wnt/β-catenin signaling in dermal condensates is required for hair follicle formation. Dev. Biol. 2014, 385, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Gu, X. Maximum-likelihood approach for gene family evolution under functional divergence. Mol. Biol. Evol. 2001, 18, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Fedi, P.; Bafico, A.; Nieto Soria, A.; Burgess, W.H.; Miki, T.; Bottaro, D.P.; Kraus, M.H.; Aaronson, S.A. Isolation and biochemical characterization of the human Dkk-1 homologue, a novel inhibitor of mammalian Wnt signaling. J. Biol. Chem. 1999, 274, 19465–19472. [Google Scholar] [CrossRef]
- Mao, B.; Wu, W.; Li, Y.; Hoppe, D.; Stannek, P.; Glinka, A.; Niehrs, C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature 2001, 411, 321–325. [Google Scholar] [CrossRef]
- Bafico, A.; Liu, G.; Yaniv, A.; Gazit, A.; Aaronson, S.A. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat. Cell Biol. 2001, 3, 683–686. [Google Scholar] [CrossRef]
- Caffrey, B.E.; Williams, T.A.; Jiang, X.; Toft, C.; Hokamp, K.; Fares, M.A. Proteome-wide analysis of functional divergence in bacteria: Exploring a host of ecological adaptations. PLoS ONE 2012, 7, e35659. [Google Scholar] [CrossRef]
- Cheng, Z.; Biechele, T.; Wei, Z.; Morrone, S.; Moon, R.T.; Wang, L.; Xu, W. Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat. Struct. Mol. Biol. 2011, 18, 1204–1210. [Google Scholar] [CrossRef]
- Ahn, V.E.; Chu, M.L.; Choi, H.J.; Tran, D.; Abo, A.; Weis, W.I. Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. Dev. Cell 2011, 21, 862–873. [Google Scholar] [CrossRef]
- Nakamura, T.; Nakamura, T.; Matsumoto, K. The functions and possible significance of Kremen as the gatekeeper of Wnt signalling in development and pathology. J. Cell Mol. Med. 2008, 12, 391–408. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, Y.; Li, X.; Chen, L.; Wang, H.; Wu, J.; Zheng, J.; Wu, D. Characterization of the Kremen-binding site on Dkk1 and elucidation of the role of Kremen in Dkk-mediated Wnt antagonism. J. Biol. Chem. 2008, 283, 23371–23375. [Google Scholar] [CrossRef]
- Mohammadpour, H.; Pourfathollah, A.A.; Nikougoftar Zarif, M.; Khalili, S. Key role of Dkk3 protein in inhibition of cancer cell proliferation: An in silico identification. J. Theor. Biol. 2016, 393, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Hoshino, T.; Kumon, H. Molecular simulation analysis of the structure complex of C2 domains of DKK family members and β-propeller domains of LRP5/6: Explaining why DKK3 does not bind to LRP5/6. Acta Med. Okayama 2014, 68, 63–78. [Google Scholar]
- David, A.; Razali, R.; Wass, M.N.; Sternberg, M.J. Protein-protein interaction sites are hot spots for disease-associated nonsynonymous SNPs. Hum. Mutat. 2012, 33, 359–363. [Google Scholar] [CrossRef]
- Poorebrahim, M.; Sadeghi, S.; Rahimi, H.; Karimipoor, M.; Azadmanesh, K.; Mazlomi, M.A.; Teimoori-Toolabi, L. Rational design of DKK3 structure-based small peptides as antagonists of Wnt signaling pathway and in silico evaluation of their efficiency. PLoS ONE 2017, 12, e0172217. [Google Scholar] [CrossRef]
- Barlow, D.J.; Thornton, J.M. Ion-pairs in proteins. J. Mol. Biol. 1983, 168, 867–885. [Google Scholar] [CrossRef]
- Kumar, S.; Nussinov, R. Close-range electrostatic interactions in proteins. Chembiochem 2002, 3, 604–617. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Xu, B.; Yang, Z. PAMLX: A graphical user interface for PAML. Mol. Biol. Evol. 2013, 30, 2723–2724. [Google Scholar] [CrossRef]
- Murphy, W.J.; Eizirik, E.; Johnson, W.E.; Zhang, Y.P.; Ryder, O.A.; O’Brien, S.J. Molecular phylogenetics and the origins of placental mammals. Nature 2001, 409, 614–618. [Google Scholar] [CrossRef]
- Liang, D.; Wu, R.; Geng, J.; Wang, C.; Zhang, P. A general scenario of HOX gene inventory variation among major sarcopterygian lineages. BMC Evol. Biol. 2011, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R.; Goldman, N.; Pedersen, A.M. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 2000, 155, 431–449. [Google Scholar] [CrossRef]
- Zhang, J.; Nielsen, R.; Yang, Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005, 22, 2472–2479. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Vander Velden, K. DIVERGE: Phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics 2002, 18, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Wu, P.; Baker, R.E.; Maini, P.K.; Alibardi, L.; Chuong, C.M. Reptile scale paradigm: Evo-Devo, pattern formation and regeneration. Int. J. Dev. Biol. 2009, 53, 813–826. [Google Scholar] [CrossRef] [PubMed]
| Gene | Foreground Branch | Possible Positive Selection Sites |
|---|---|---|
| Dkk1 | Aves | 17 H 0.997 **, 75 Y 0.998 **, 91 S 0.981 * |
| Aves and Reptilia | 7 H 0.979, 75 Y 1.000 ** | |
| Dkk2 | Lepidoptera (pangolin) | 27 V 0.998 ** |
| Dkk3 | Aves and Reptilia | 24 R 0.997 **, 41 R 0.998 **, 52 P 1.000 **, 58 V 0.999 ** |
| Dkk4 | Reptilia | 103 S 0.975 *, 105 K 0.970 *, 108 Q 0.961 * |
| Gene | Clusters | θI | MFE SE | Possible Positive Selection Sites (Qk > 0.90) |
|---|---|---|---|---|
| Dkk1 | A/B | −0.064754 | 0.145614 | |
| A/C | 0.143104 | 0.155194 | ||
| B/C | 0.068558 | 0.216823 | ||
| Dkk2 | A/B | 0.239265 | 0.140598 | |
| A/C | 0.403401 | 0.100621 | 171G | |
| B/C | 0.514811 | 0.144675 | ||
| Dkk3 | A/B | 0.427149 | 0.301757 | 367T, 377H, 410R, 414H, 416L, 423T, 443V, 450G, 495E |
| A/C | 0.630269 | 0.134886 | 399R, 407L, 411H | |
| B/C | 0.623537 | 0.348995 | 399R | |
| Dkk4 | A/B | 0.332477 | 0.090430 | 185Q |
| Gene | Clusters | θII | MFE SE | Possible Positive Selection Sites (Qk > 0.90) |
|---|---|---|---|---|
| Dkk1 | A/B | −0.038624 | 0.086041 | |
| A/C | 0.059911 | 0.081074 | 263Y, 268Q, 271S, 312S, 324G, 328S, 341N, 342S | |
| B/C | 0.010027 | 0.088840 | 252Y, 261K, 324G | |
| Dkk2 | A/B | 0.035892 | 0.052704 | 165H, 188H |
| A/C | 0.097338 | 0.043583 | 36A, 38L, 44S, 46G, 92Q, 97S, 98S, 114G, 161D, 165H, 166R, 170H, 174S, 176N, 177H, 188H, 212F, 266Y, 270A | |
| B/C | −0.020593 | 0.043545 | ||
| Dkk3 | A/B | 0.141160 | 0.144634 | 360G, 371V, 373G, 375L, 377H, 380A, 385D, 404S, 410P, 411H, 416L, 417V, 418Y, 421K, 422P, 423T, 443V, 444G, 446R, 447D, 450G, 472D, 477G, 484E, 487R, 488Q, 492D, 495E, 496R, 507P, 516G |
| A/C | 0.225194 | 0.105301 | 360G, 371V, 375L, 380A, 381S, 385D, 398D, 411H, 416L, 417V, 418Y, 421K, 422P, 423T, 443V, 444G, 447D, 448Q, 450G, 471P, 472D, 484E, 488Q, 496R, 507P, 516G | |
| B/C | −0.016133 | 0.132199 | 373G, 471P, 477G | |
| Dkk4 | A/B | −0.081373 | 0.135288 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, B.; Hu, S.; Yin, J.; Wu, J.; Guo, W. Molecular Evolution and Protein Structure Variation of Dkk Family. Genes 2023, 14, 1863. https://doi.org/10.3390/genes14101863
Wen B, Hu S, Yin J, Wu J, Guo W. Molecular Evolution and Protein Structure Variation of Dkk Family. Genes. 2023; 14(10):1863. https://doi.org/10.3390/genes14101863
Chicago/Turabian StyleWen, Binhong, Sile Hu, Jun Yin, Jianghong Wu, and Wenrui Guo. 2023. "Molecular Evolution and Protein Structure Variation of Dkk Family" Genes 14, no. 10: 1863. https://doi.org/10.3390/genes14101863
APA StyleWen, B., Hu, S., Yin, J., Wu, J., & Guo, W. (2023). Molecular Evolution and Protein Structure Variation of Dkk Family. Genes, 14(10), 1863. https://doi.org/10.3390/genes14101863





