Functional Assessment of a New PBX1 Variant in a 46,XY Fetus with Severe Syndromic Difference of Sexual Development through CRISPR-Cas9 Gene Editing
Abstract
1. Introduction
2. Patient and Methods
2.1. Editorial Policies and Ethical Considerations
2.2. Patient
2.3. Cytogenetic Analyses
2.4. Exome Sequencing and Bioinformatics Pipeline
2.5. Cell Culture
2.6. Cell DNA Extraction, PCR and Sanger Sequencing
2.7. RNA Extraction, Reverse Transcription and Quantitative PCR
2.8. Protein Extraction
2.9. Western Blot
2.10. Methylen Blue Viability Assays
2.11. siRNA Transfection
2.12. Cloning
2.13. CRISPR-KD
2.14. Site-Directed Mutagenesis on PBX1 Plasmid
2.15. Proliferation Studies
2.16. RNA-seq
3. Results
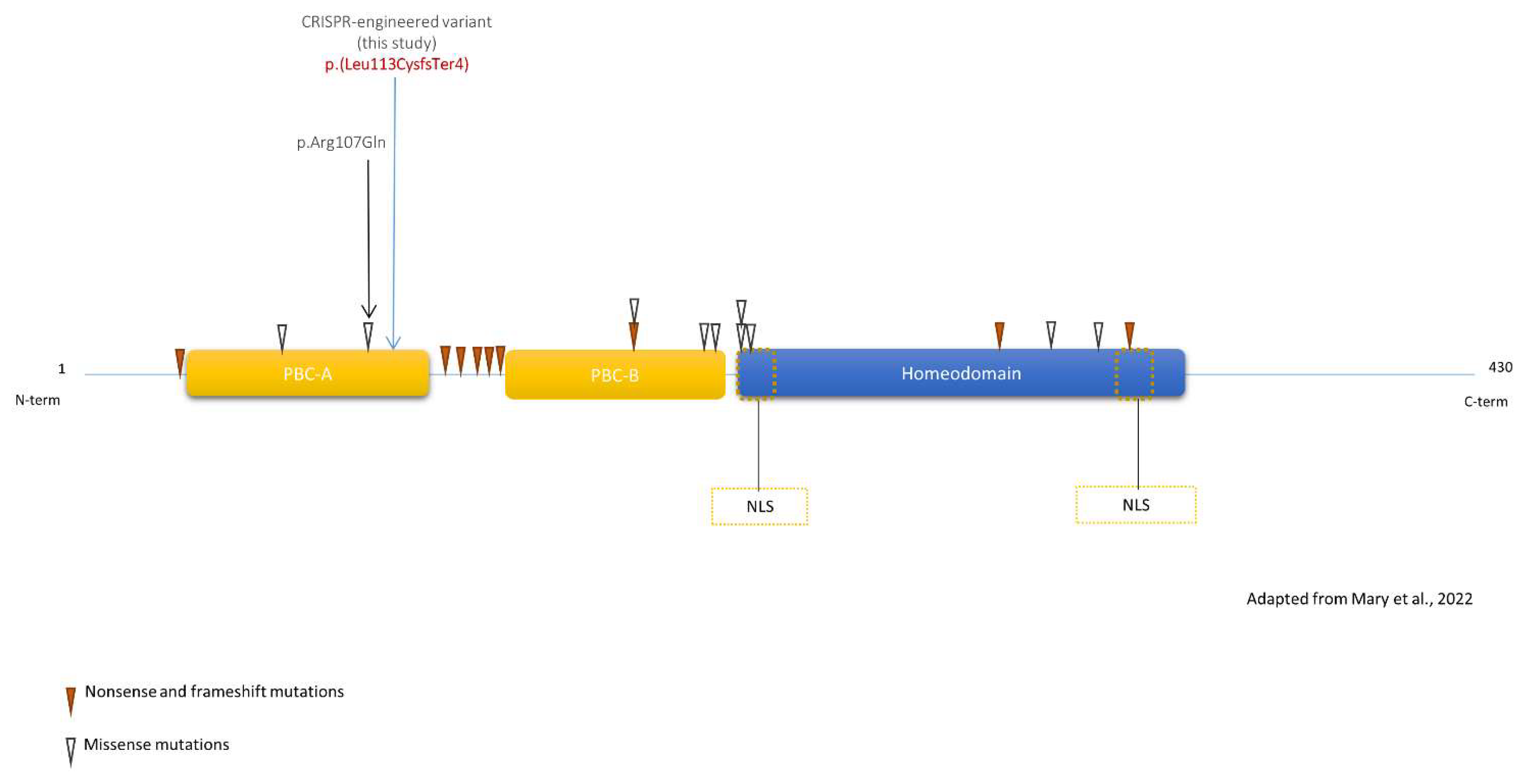
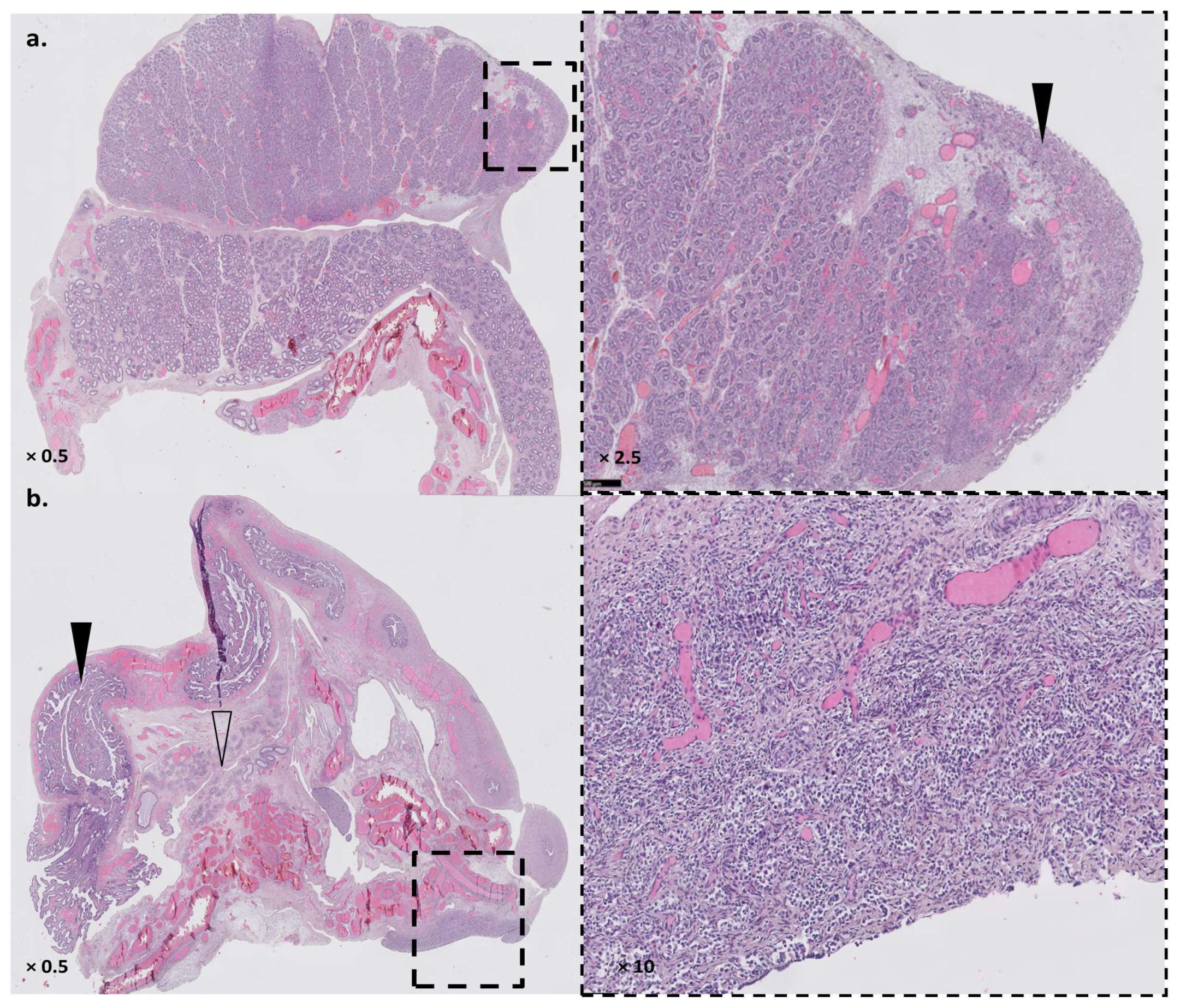
3.1. Patient
3.2. Generation of a Cellular Model Knocked-Down (KD) for PBX1, with Impaired Proliferation and Support Adhesion
3.3. Rescue Experiments by Transfection with PBX1a-WT or PBX1a 320G>A Plasmids
3.3.1. PBX1 Overexpression Enhances Cell Proliferation
3.3.2. Overexpression of PBX1a Slightly Rescues KD2 Normal Adhesion
3.3.3. Targeted RT-qPCR Suggests MEIS1 as a Potential Disrupted Gene
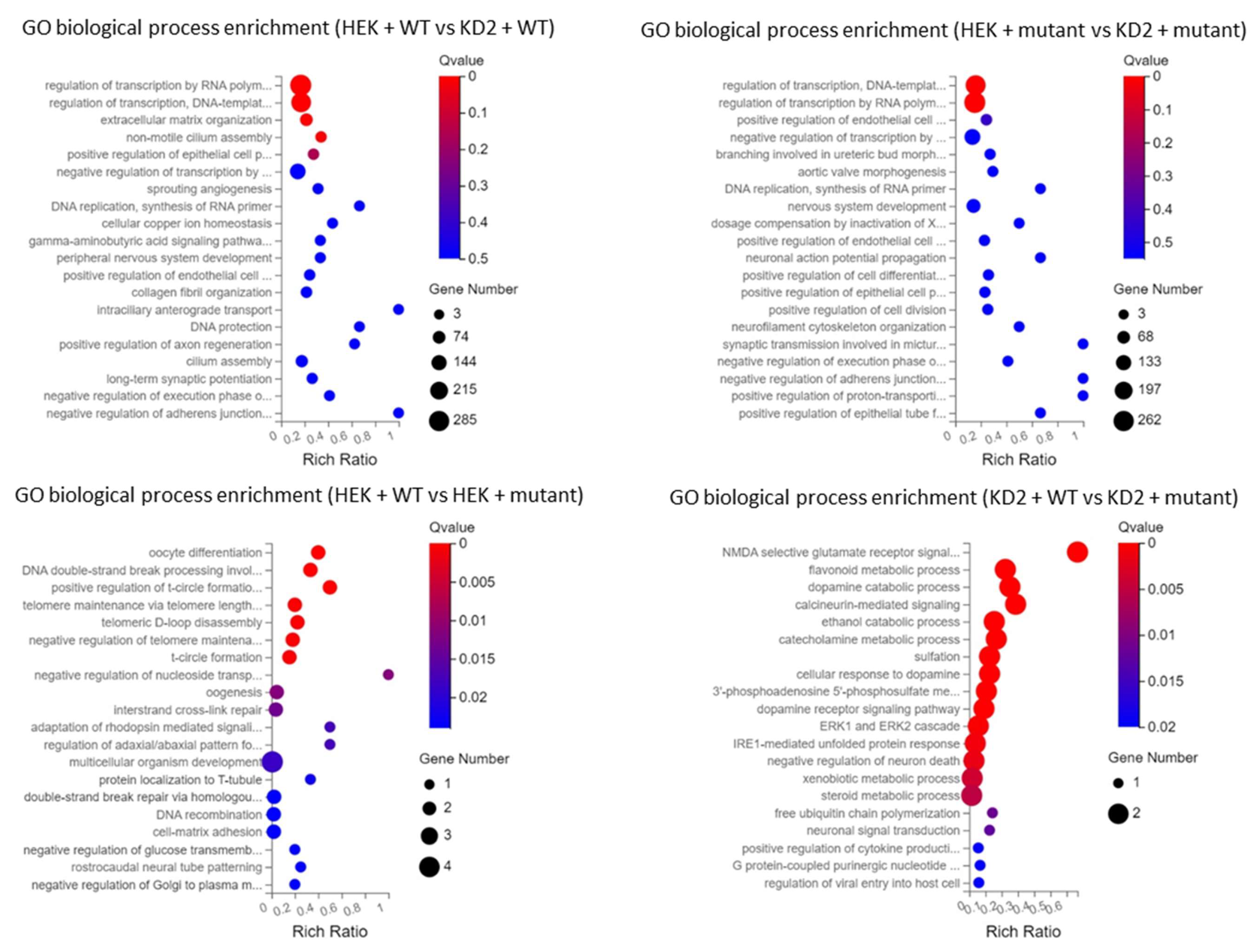
3.4. RNA-seq
3.4.1. Both Cell Lines Expressed Multiple PBX1 Transcripts
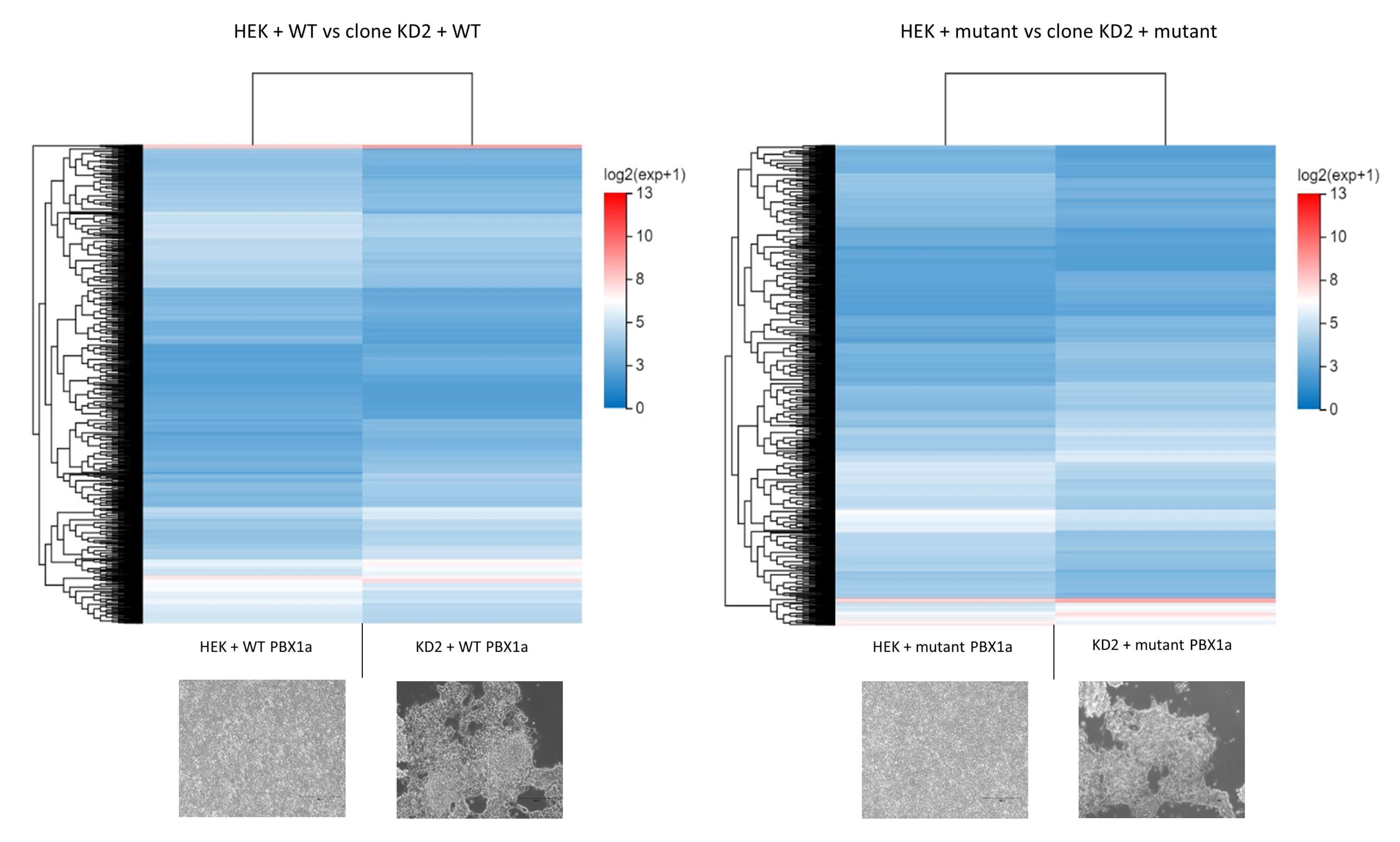
3.4.2. Gene Expression between WT HEK and KD2 Cell Lines Remains Different despite Being Transfected with the Same Plasmid
3.4.3. DEGs between WT and Mutant PBX1a Transfections Depend on the Cell Line
3.4.4. Transfection with Mutant PBX1a May Alter Skipped Exon Splicing Events
4. Discussion
4.1. PBX1 Is a DSD Gene
4.2. Partial PBX1 Loss of Function (LoF) Impairs Cell Proliferation
4.3. Transfection with PBX1-Coding Plasmids Does Not Fully Rescue a PBX1-KD Phenotype
4.4. PBX1 Transfection Gives Fluctuant Results in HEK Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hughes, I.A.; Houk, C.; Ahmed, S.F.; Lee, P.A.; LWPES/ESPE Consensus Group. Consensus Statement on Management of Intersex Disorders. Arch. Dis. Child. 2006, 91, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.A.; Nordenström, A.; Houk, C.P.; Ahmed, S.F.; Auchus, R.; Baratz, A.; Baratz Dalke, K.; Liao, L.-M.; Lin-Su, K.; Looijenga, L.H.J.; et al. Global Disorders of Sex Development Update since 2006: Perceptions, Approach and Care. Horm. Res. Paediatr. 2016, 85, 158–180. [Google Scholar] [CrossRef] [PubMed]
- Okashita, N.; Tachibana, M. Transcriptional Regulation of the Y-Linked Mammalian Testis-Determining Gene SRY. Sex. Dev. 2021, 15, 351–359. [Google Scholar] [CrossRef]
- Baetens, D.; Verdin, H.; De Baere, E.; Cools, M. Update on the Genetics of Differences of Sex Development (DSD). Best Pract. Res. Clin. Endocrinol. Metab. 2019, 33, 101271. [Google Scholar] [CrossRef]
- McElreavey, K.; Bashamboo, A. Monogenic Forms of DSD: An Update. Horm. Res. Paediatr. 2021. [CrossRef]
- Globa, E.; Zelinska, N.; Shcherbak, Y.; Bignon-Topalovic, J.; Bashamboo, A.; McElreavey, K. Disorders of Sex Development in a Large Ukrainian Cohort: Clinical Diversity and Genetic Findings. Front. Endocrinol. 2022, 13, 810782. [Google Scholar] [CrossRef]
- Mary, L.; Leclerc, D.; Gilot, D.; Belaud-Rotureau, M.-A.; Jaillard, S. The TALE Never Ends: A Comprehensive Overview of the Role of PBX1, a TALE Transcription Factor, in Human Developmental Defects. Hum. Mutat. 2022, 43, 1125–1148. [Google Scholar] [CrossRef]
- Selleri, L.; Zappavigna, V.; Ferretti, E. “Building a Perfect Body”: Control of Vertebrate Organogenesis by PBX-Dependent Regulatory Networks. Genes Dev. 2019, 33, 258–275. [Google Scholar] [CrossRef]
- Heidet, L.; Morinière, V.; Henry, C.; De Tomasi, L.; Reilly, M.L.; Humbert, C.; Alibeu, O.; Fourrage, C.; Bole-Feysot, C.; Nitschké, P.; et al. Targeted Exome Sequencing Identifies PBX1 as Involved in Monogenic Congenital Anomalies of the Kidney and Urinary Tract. J. Am. Soc. Nephrol. 2017, 28, 2901–2914. [Google Scholar] [CrossRef]
- Le Tanno, P.; Breton, J.; Bidart, M.; Satre, V.; Harbuz, R.; Ray, P.F.; Bosson, C.; Dieterich, K.; Jaillard, S.; Odent, S.; et al. PBX1 Haploinsufficiency Leads to Syndromic Congenital Anomalies of the Kidney and Urinary Tract (CAKUT) in Humans. J. Med. Genet. 2017, 54, 502–510. [Google Scholar] [CrossRef]
- Schnabel, C.A.; Selleri, L.; Cleary, M.L. Pbx1 Is Essential for Adrenal Development and Urogenital Differentiation. Genesis 2003, 37, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, C.A.; Godin, R.E.; Cleary, M.L. Pbx1 Regulates Nephrogenesis and Ureteric Branching in the Developing Kidney. Dev. Biol. 2003, 254, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Li, Y.; Wang, M.; Li, H.; Su, T.; Li, Y.; Wang, S. Associations of Polymorphisms in WNT9B and PBX1 with Mayer-Rokitansky-Küster-Hauser Syndrome in Chinese Han. PLoS ONE 2015, 10, e0130202. [Google Scholar] [CrossRef]
- Chen, N.; Zhao, S.; Jolly, A.; Wang, L.; Pan, H.; Yuan, J.; Chen, S.; Koch, A.; Ma, C.; Tian, W.; et al. Perturbations of Genes Essential for Müllerian Duct and Wölffian Duct Development in Mayer-Rokitansky-Küster-Hauser Syndrome. Am. J. Hum. Genet. 2021, 108, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Slavotinek, A.; Risolino, M.; Losa, M.; Cho, M.T.; Monaghan, K.G.; Schneidman-Duhovny, D.; Parisotto, S.; Herkert, J.C.; Stegmann, A.P.A.; Miller, K.; et al. De Novo, Deleterious Sequence Variants That Alter the Transcriptional Activity of the Homeoprotein PBX1 Are Associated with Intellectual Disability and Pleiotropic Developmental Defects. Hum. Mol. Genet. 2017, 26, 4849–4860. [Google Scholar] [CrossRef] [PubMed]
- Eozenou, C.; Bashamboo, A.; Bignon-Topalovic, J.; Merel, T.; Zwermann, O.; Lourenco, D.; Lottmann, H.; Lichtenauer, U.; Rojo, S.; Beuschlein, F.; et al. The TALE Homeodomain of PBX1 Is Involved in Human Primary Testis-Determination. Hum. Mutat. 2019, 40, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained Genome Targeting with Near-PAMless Engineered CRISPR-Cas9 Variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef]
- Engler, C.; Marillonnet, S. Golden Gate Cloning. Methods Mol. Biol. 2014, 1116, 119–131. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce Acceleration-Supported Software for Integrated Quality Control and Preprocessing of High-Throughput Sequencing Data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R Package for Identifying Differentially Expressed Genes from RNA-Seq Data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Shen, S.; Park, J.W.; Lu, Z.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. RMATS: Robust and Flexible Detection of Differential Alternative Splicing from Replicate RNA-Seq Data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef]
- Benelli, M.; Pescucci, C.; Marseglia, G.; Severgnini, M.; Torricelli, F.; Magi, A. Discovering Chimeric Transcripts in Paired-End RNA-Seq Data by Using EricScript. Bioinformatics 2012, 28, 3232–3239. [Google Scholar] [CrossRef]
- Thiaville, M.M.; Stoeck, A.; Chen, L.; Wu, R.-C.; Magnani, L.; Oidtman, J.; Shih, I.-M.; Lupien, M.; Wang, T.-L. Identification of PBX1 Target Genes in Cancer Cells by Global Mapping of PBX1 Binding Sites. PLoS ONE 2012, 7, e36054. [Google Scholar] [CrossRef]
- Villaescusa, J.C.; Li, B.; Toledo, E.M.; Rivetti di Val Cervo, P.; Yang, S.; Stott, S.R.; Kaiser, K.; Islam, S.; Gyllborg, D.; Laguna-Goya, R.; et al. A PBX1 Transcriptional Network Controls Dopaminergic Neuron Development and Is Impaired in Parkinson’s Disease. EMBO J. 2016, 35, 1963–1978. [Google Scholar] [CrossRef]
- Arts, P.; Garland, J.; Byrne, A.B.; Hardy, T.S.E.; Babic, M.; Feng, J.; Wang, P.; Ha, T.; King-Smith, S.L.; Schreiber, A.W.; et al. Paternal Mosaicism for a Novel PBX1 Mutation Associated with Recurrent Perinatal Death: Phenotypic Expansion of the PBX1-Related Syndrome. Am. J. Med. Genet. Part A 2020, 182, 1273–1277. [Google Scholar] [CrossRef]
- Blasi, F.; Bruckmann, C.; Penkov, D.; Dardaei, L. A Tale of TALE, PREP1, PBX1, and MEIS1: Interconnections and Competition in Cancer. Bioessays 2017, 39, 1600245. [Google Scholar] [CrossRef]
- He, C.; Wang, Z.; Zhang, L.; Yang, L.; Li, J.; Chen, X.; Zhang, J.; Chang, Q.; Yu, Y.; Liu, B.; et al. A Hydrophobic Residue in the TALE Homeodomain of PBX1 Promotes Epithelial-to-Mesenchymal Transition of Gastric Carcinoma. Oncotarget 2017, 8, 46818–46833. [Google Scholar] [CrossRef] [PubMed]
- Stankunas, K.; Shang, C.; Twu, K.Y.; Kao, S.-C.; Jenkins, N.A.; Copeland, N.G.; Sanyal, M.; Selleri, L.; Cleary, M.L.; Chang, C.-P. Pbx/Meis Deficiencies Demonstrate Multigenetic Origins of Congenital Heart Disease. Circ. Res. 2008, 103, 702–709. [Google Scholar] [CrossRef]
- Fernandez, L.C.; Errico, M.C.; Bottero, L.; Penkov, D.; Resnati, M.; Blasi, F.; Caré, A. Oncogenic HoxB7 Requires TALE Cofactors and Is Inactivated by a Dominant-Negative Pbx1 Mutant in a Cell-Specific Manner. Cancer Lett. 2008, 266, 144–155. [Google Scholar] [CrossRef]
- Kyei-Barffour, I.; Margetts, M.; Vash-Margita, A.; Pelosi, E. The Embryological Landscape of Mayer-Rokitansky-Kuster-Hauser Syndrome: Genetics and Environmental Factors. Yale J. Biol. Med. 2021, 94, 657–672. [Google Scholar] [PubMed]
- Hatzistergos, K.E.; Williams, A.R.; Dykxhoorn, D.; Bellio, M.A.; Yu, W.; Hare, J.M. Tumor Suppressors RB1 and CDKN2a Cooperatively Regulate Cell-Cycle Progression and Differentiation During Cardiomyocyte Development and Repair. Circ. Res. 2019, 124, 1184–1197. [Google Scholar] [CrossRef]
- Galvin, K.M.; Donovan, M.J.; Lynch, C.A.; Meyer, R.I.; Paul, R.J.; Lorenz, J.N.; Fairchild-Huntress, V.; Dixon, K.L.; Dunmore, J.H.; Gimbrone, M.A.; et al. A Role for Smad6 in Development and Homeostasis of the Cardiovascular System. Nat. Genet. 2000, 24, 171–174. [Google Scholar] [CrossRef]
- Davis, B.N.; Hilyard, A.C.; Lagna, G.; Hata, A. SMAD Proteins Control DROSHA-Mediated MicroRNA Maturation. Nature 2008, 454, 56–61. [Google Scholar] [CrossRef]
- Portnoi, M.-F.; Dumargne, M.-C.; Rojo, S.; Witchel, S.F.; Duncan, A.J.; Eozenou, C.; Bignon-Topalovic, J.; Yatsenko, S.A.; Rajkovic, A.; Reyes-Mugica, M.; et al. Mutations Involving the SRY-Related Gene SOX8 Are Associated with a Spectrum of Human Reproductive Anomalies. Hum. Mol. Genet. 2018, 27, 1228–1240. [Google Scholar] [CrossRef]
- Soares, L.M.M.; Zanier, K.; Mackereth, C.; Sattler, M.; Valcárcel, J. Intron Removal Requires Proofreading of U2AF/3’ Splice Site Recognition by DEK. Science 2006, 312, 1961–1965. [Google Scholar] [CrossRef]
- Zhao, Y.; Cai, W.; Hua, Y.; Yang, X.; Zhou, J. The Biological and Clinical Consequences of RNA Splicing Factor U2AF1 Mutation in Myeloid Malignancies. Cancers 2022, 14, 4406. [Google Scholar] [CrossRef]
- Svendsen, J.M.; Smogorzewska, A.; Sowa, M.E.; O’Connell, B.C.; Gygi, S.P.; Elledge, S.J.; Harper, J.W. Mammalian BTBD12/SLX4 Assembles a Holliday Junction Resolvase and Is Required for DNA Repair. Cell 2009, 138, 63–77. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Ji, B.; Sun, D. TRIM34 Localizes to the Mitochondria and Mediates Apoptosis through the Mitochondrial Pathway in HEK293T Cells. Heliyon 2020, 6, e03115. [Google Scholar] [CrossRef] [PubMed]
- Rosa-Mercado, N.A.; Steitz, J.A. Who Let the DoGs out? - Biogenesis of Stress-Induced Readthrough Transcripts. Trends Biochem. Sci. 2022, 47, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.; Morse, S.; Ararat, M.; Graham, F.L. Preferential Transformation of Human Neuronal Cells by Human Adenoviruses and the Origin of HEK 293 Cells. FASEB J. 2002, 16, 869–871. [Google Scholar] [CrossRef]
- Stepanenko, A.A.; Dmitrenko, V.V. HEK293 in Cell Biology and Cancer Research: Phenotype, Karyotype, Tumorigenicity, and Stress-Induced Genome-Phenotype Evolution. Gene 2015, 569, 182–190. [Google Scholar] [CrossRef]




| PBX1 Isoform | WT HEK + WT TPM | WT HEK + Mutant TPM | Clone KD2 + WT TPM | Clone KD2 + Mutant TPM |
|---|---|---|---|---|
| NM_001204961 = PBX1b | 1.60 | 2.56 | 0.22 | 1.10 |
| NM_001204963 | NA | NA | NA | NA |
| NM_001353130 | 0 | 0.31 | 0 | 0.16 |
| NM_001353131 = PBX1b | 1.91 | 1.79 | 0 | 1.07 |
| NM_002585= XM_005245229 = PBX1a | 1208.94 | 976.54 | 447.82 | 408.52 |
| XM_005245228 | NA | NA | NA | NA |
| XM_011509590 | 0 | 0 | 0 | 0.25 |
| XM_011509591 | 0 | 0 | 0.46 | 0 |
| XM_011509592 | 1.47 | 0.18 | 0.31 | 0 |
| XM_017001395 (PBX1a ?) | 2665.33 | 2435.52 | 1155.85 | 887.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mary, L.; Leclerc, D.; Labalme, A.; Bellaud, P.; Mazaud-Guittot, S.; Dréano, S.; Evrard, B.; Bigand, A.; Cauchoix, A.; Loget, P.; et al. Functional Assessment of a New PBX1 Variant in a 46,XY Fetus with Severe Syndromic Difference of Sexual Development through CRISPR-Cas9 Gene Editing. Genes 2023, 14, 273. https://doi.org/10.3390/genes14020273
Mary L, Leclerc D, Labalme A, Bellaud P, Mazaud-Guittot S, Dréano S, Evrard B, Bigand A, Cauchoix A, Loget P, et al. Functional Assessment of a New PBX1 Variant in a 46,XY Fetus with Severe Syndromic Difference of Sexual Development through CRISPR-Cas9 Gene Editing. Genes. 2023; 14(2):273. https://doi.org/10.3390/genes14020273
Chicago/Turabian StyleMary, Laura, Delphine Leclerc, Audrey Labalme, Pascale Bellaud, Séverine Mazaud-Guittot, Stéphane Dréano, Bertrand Evrard, Antoine Bigand, Aurélie Cauchoix, Philippe Loget, and et al. 2023. "Functional Assessment of a New PBX1 Variant in a 46,XY Fetus with Severe Syndromic Difference of Sexual Development through CRISPR-Cas9 Gene Editing" Genes 14, no. 2: 273. https://doi.org/10.3390/genes14020273
APA StyleMary, L., Leclerc, D., Labalme, A., Bellaud, P., Mazaud-Guittot, S., Dréano, S., Evrard, B., Bigand, A., Cauchoix, A., Loget, P., Lokchine, A., Cluzeau, L., Gilot, D., Belaud-Rotureau, M.-A., & Jaillard, S. (2023). Functional Assessment of a New PBX1 Variant in a 46,XY Fetus with Severe Syndromic Difference of Sexual Development through CRISPR-Cas9 Gene Editing. Genes, 14(2), 273. https://doi.org/10.3390/genes14020273







