Adaptive Evolution of Rhizobial Symbiosis beyond Horizontal Gene Transfer: From Genome Innovation to Regulation Reconstruction
Abstract
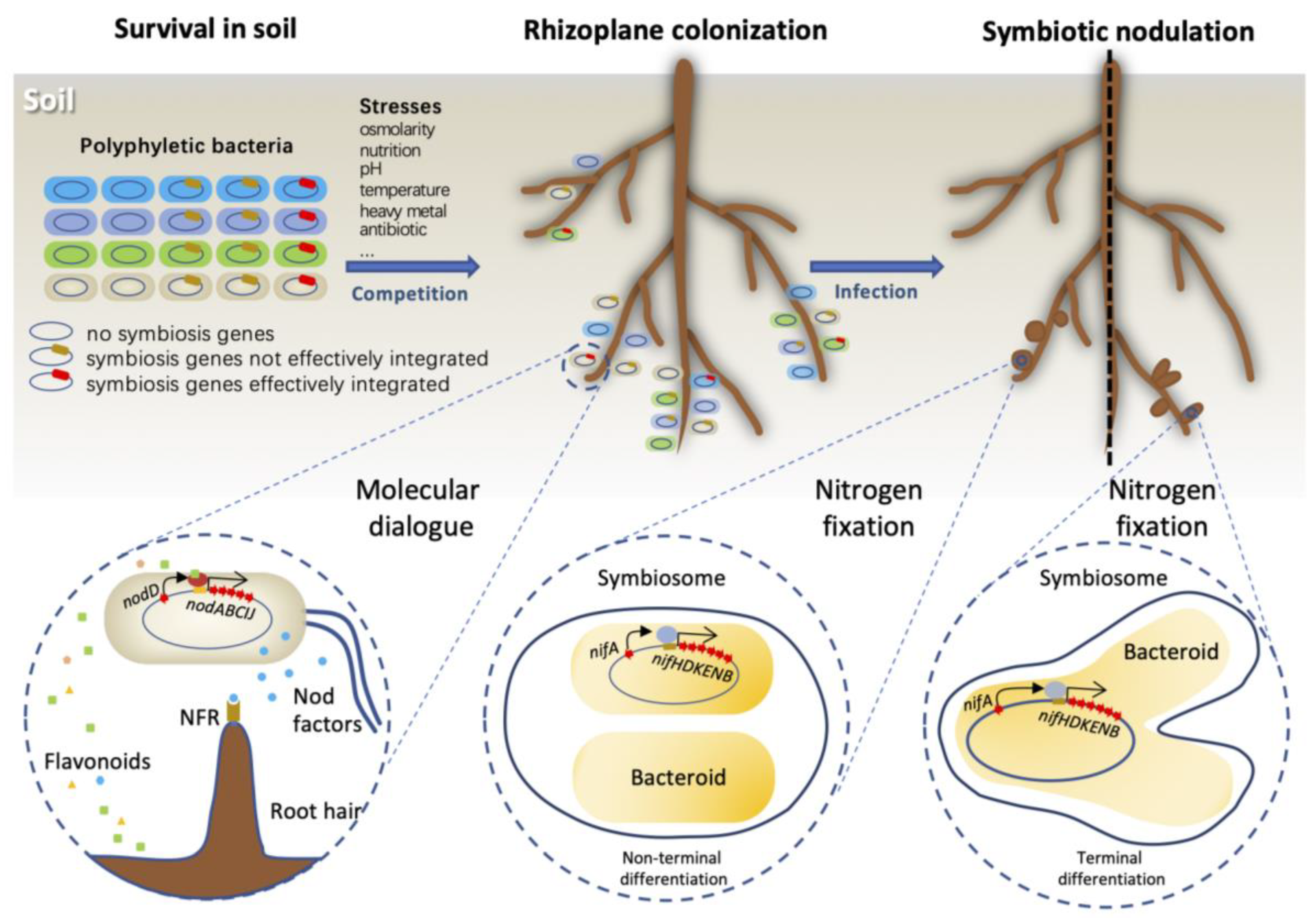
1. Introduction
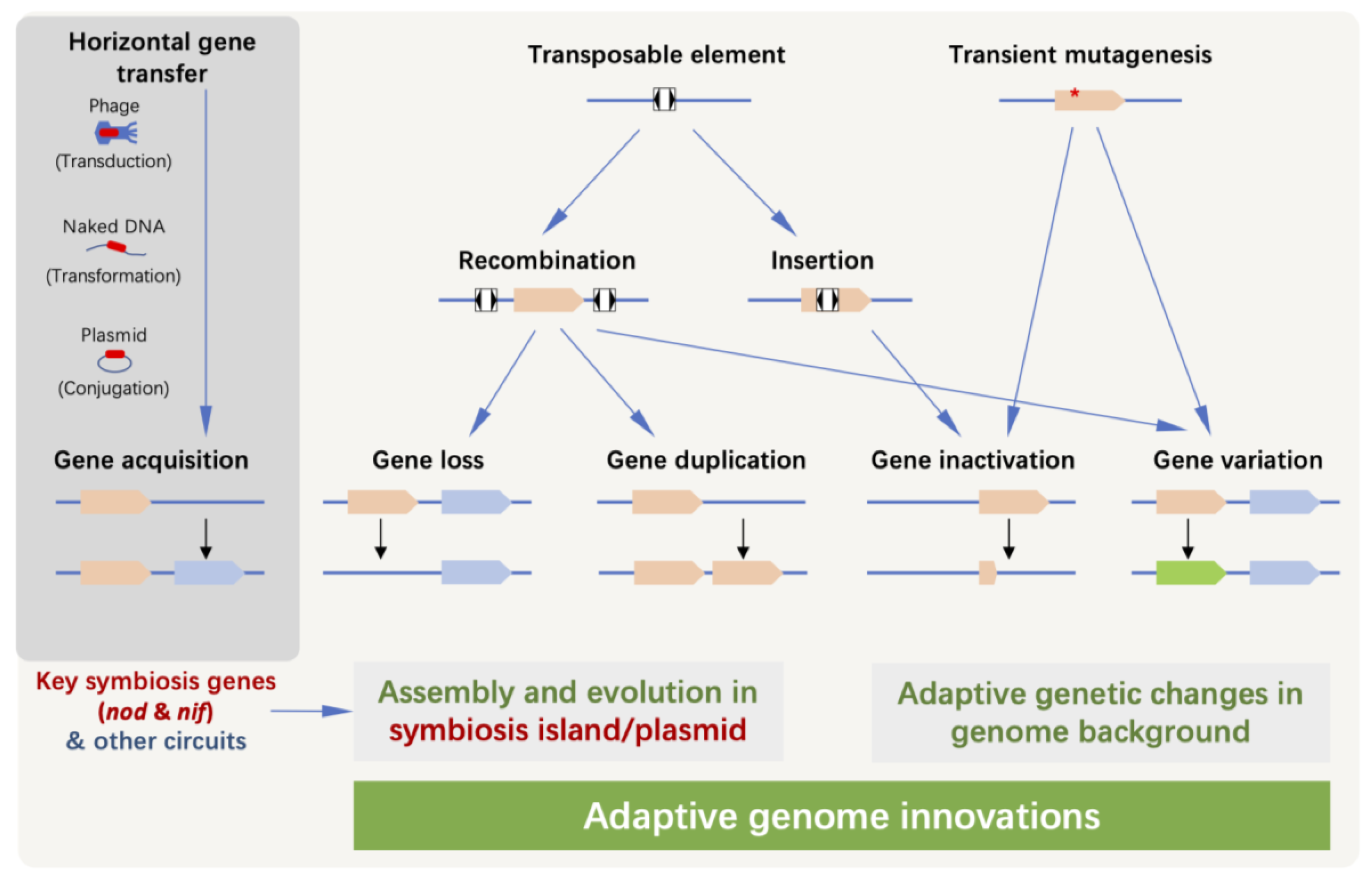
2. Genome Innovations after Receiving Key Symbiosis Genes
2.1. Continuous Evolution of Key Symbiosis Genes
2.2. Horizontal Transfer of Genes beyond Key Symbiosis Genes
2.3. Gene Inactivation, Gene Loss and Genome Rearrangement in Symbiosis Plasmid/Islands
3. Reconstruction of Regulatory Networks
3.1. Recruitment of Indigenous Functions to Support Symbiosis
3.2. Integration of Key Symbiosis Circuits with Recipient Regulation Network
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| nod | nodulation |
| nif | nitrogen fixation |
| NFR | nodulation factor receptor |
| NCR | nodule-specific cysteine-rich |
| IRLC | inverted repeat lacking clade |
| SNF | symbiotic nitrogen fixation |
| HGT | horizontal gene transfer |
| NF | nodulation factor |
| T1SS | type I secretion system |
| T3SS | type III secretion system |
| T4SS | type IV secretion system |
| T6SS | type VI secretion system |
| NO | nitric oxide |
| IS | insertion sequence |
| TE | transposable element |
| EPS | exopolysaccharide |
| APS | arabinose-containing polysaccharide |
| IAA | indole-3-acetic acid |
| ChIP-seq | chromatin immunoprecipitation sequencing |
| Sf | Sinorhizobium fredii |
| Sm | Sinorhizobium meliloti |
| Rt | Rhizobium tropici |
| Ml | Mesorhizobium loti |
| Bd | Bradyrhizobium diazoefficiens |
References
- de Lajudie, P.M.; Andrews, M.; Ardley, J.; Eardly, B.; Jumas-bilak, E.; Kuzmanovic, N.; Lassalle, F.; Lindström, K.; Mhamdi, R.; Martínez-Romero, E.; et al. Minimal Standards for the Description of New Genera and Species of Rhizobia and Agrobacteria. Int. J. Syst. Evol. Microbiol. 2019, 69, 1852–1863. [Google Scholar] [CrossRef]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From Saprophytes to Endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 Years of Genetic Discoveries in Legume Nodulation and Symbiotic Nitrogen Fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Reznikoff, W.S. Mobile DNA in Obligate Intracellular Bacteria. Nat. Rev. Microbiol. 2005, 3, 688–699. [Google Scholar] [CrossRef]
- Coba de la Peña, T.; Fedorova, E.; Pueyo, J.J.; Mercedes Lucas, M. The Symbiosome: Legume and Rhizobia Co-Evolution toward a Nitrogen-Fixing Organelle? Front. Plant Sci. 2018, 8, 2229. [Google Scholar] [CrossRef]
- Mergaert, P.; Uchiumi, T.; Alunni, B.; Evanno, G.; Cheron, A.; Catrice, O.; Mausset, A.-E.E.; Barloy-Hubler, F.; Galibert, F.; Kondorosi, A.; et al. Eukaryotic Control on Bacterial Cell Cycle and Differentiation in the Rhizobium-Legume Symbiosis. Proc. Natl. Acad. Sci. USA 2006, 103, 5230–5235. [Google Scholar] [CrossRef]
- Van de Velde, W.; Zehirov, G.; Szatmari, A.; Debreczeny, M.; Ishihara, H.; Kevei, Z.; Farkas, A.; Mikulass, K.; Nagy, A.; Tiricz, H.; et al. Plant Peptides Govern Terminal Differentiation of Bacteria in Symbiosis. Science 2010, 327, 1122–1126. [Google Scholar] [CrossRef]
- Montiel, J.; Downie, J.A.; Farkas, A.; Bihari, P.; Herczeg, R.; Bálint, B.; Mergaert, P.; Kereszt, A.; Kondorosi, É. Morphotype of Bacteroids in Different Legumes Correlates with the Number and Type of Symbiotic NCR Peptides. Proc. Natl. Acad. Sci. USA 2017, 114, 5041–5046. [Google Scholar] [CrossRef]
- Czernic, P.; Gully, D.; Cartieaux, F.; Moulin, L.; Guefrachi, I.; Patrel, D.; Pierre, O.; Fardoux, J.; Chaintreuil, C.; Nguyen, P.; et al. Convergent Evolution of Endosymbiont Differentiation in Dalbergioid and Inverted Repeat-Lacking Clade Legumes Mediated by Nodule-Specific Cysteine-Rich Peptides. Plant Physiol. 2015, 169, 1254–1265. [Google Scholar] [CrossRef]
- Oono, R.; Schmitt, I.; Sprent, J.I.; Denison, R.F. Multiple Evolutionary Origins of Legume Traits Leading to Extreme Rhizobial Differentiation. New Phytol. 2010, 187, 508–520. [Google Scholar] [CrossRef]
- Dixon, R.; Kahn, D. Genetic Regulation of Biological Nitrogen Fixation. Nat. Rev. Microbiol. 2004, 2, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Becana, M.; Klucas, R.V. Oxidation and Reduction of Leghemoglobin in Root Nodules of Leguminous Plants. Plant Physiol. 1992, 98, 1217–1221. [Google Scholar] [CrossRef] [PubMed]
- Ott, T.; van Dongen, J.T.; Günther, C.; Krusell, L.; Desbrosses, G.; Vigeolas, H.; Bock, V.; Czechowski, T.; Geigenberger, P.; Udvardi, M.K. Symbiotic Leghemoglobins are Crucial for Nitrogen Fixation in Legume Root Nodules but not for General Plant Growth and Development. Curr. Biol. 2005, 15, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Rubio, M.C.; Xin, X.; Zhang, B.; Fan, Q.; Wang, Q.; Ning, G.; Becana, M.; Duanmu, D. CRISPR/Cas9 Knockout of Leghemoglobin Genes in Lotus Japonicus Uncovers Their Synergistic Roles in Symbiotic Nitrogen Fixation. New Phytol. 2019, 224, 818–832. [Google Scholar] [CrossRef]
- Herridge, D.F.; Peoples, M.B.; Boddey, R.M. Global Inputs of Biological Nitrogen Fixation in Agricultural Systems. Plant Soil 2008, 311, 1–18. [Google Scholar] [CrossRef]
- Masson-Boivin, C.; Giraud, E.; Perret, X.; Batut, J. Establishing Nitrogen-Fixing Symbiosis with Legumes: How Many Rhizobium Recipes? Trends Microbiol. 2009, 17, 458–466. [Google Scholar] [CrossRef]
- Remigi, P.; Zhu, J.; Young, J.P.W.; Masson-Boivin, C. Symbiosis within Symbiosis: Evolving Nitrogen-Fixing Legume Symbionts. Trends Microbiol. 2016, 24, 63–75. [Google Scholar] [CrossRef]
- Galibert, F.; Finan, T.M.; Long, S.R.; Puhler, A.; Abola, P.; Ampe, F.; Barloy-Hubler, F.; Barnett, M.J.; Becker, A.; Boistard, P.; et al. The Composite Genome of the Legume Symbiont Sinorhizobium meliloti. Science 2001, 293, 668–672. [Google Scholar] [CrossRef]
- Kaneko, T.; Nakamura, Y.; Sato, S.; Minamisawa, K.; Uchiumi, T.; Sasamoto, S.; Watanabe, A.; Idesawa, K.; Iriguchi, M.; Kawashima, K.; et al. Complete Genomic Sequence of Nitrogen-Fixing Symbiotic Bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002, 9, 189–197. [Google Scholar] [CrossRef]
- Giraud, E.; Moulin, L.; Vallenet, D.; Barbe, V.; Cytryn, E.; Avarre, J.C.; Jaubert, M.; Simon, D.; Cartieaux, F.; Prin, Y.; et al. Legumes Symbioses: Absence of Nod Genes in Photosynthetic Bradyrhizobia. Science 2007, 316, 1307–1312. [Google Scholar] [CrossRef]
- Okazaki, S.; Tittabutr, P.; Teulet, A.; Thouin, J.; Fardoux, J.; Chaintreuil, C.; Gully, D.; Arrighi, J.-F.; Furuta, N. Rhizobium—Legume Symbiosis in the Absence of Nod Factors: Two Possible Scenarios with or without the T3SS. ISME J. 2015, 10, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Kaneko, T.; Sato, S.; Saeki, K. Hijacking of Leguminous Nodulation Signaling by the Rhizobial Type III Secretion System. Proc. Natl. Acad. Sci. USA 2013, 110, 17131–17136. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Oter, R.; Nakano, R.T.; Dombrowski, N.; Ma, K.W.; McHardy, A.C.; Schulze-Lefert, P. Modular Traits of the Rhizobiales Root Microbiota and Their Evolutionary Relationship with Symbiotic Rhizobia. Cell Host Microbe 2018, 24, 155–167. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone Taxa as Drivers of Microbiome Structure and Functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Dennis, P.G.; Paungfoo-Lonhienne, C.; Weber, L.; Brackin, R.; Ragan, M.A.; Schmidt, S.; Hugenholtz, P. Evolutionary Conservation of a Core Root Microbiome across Plant Phyla along a Tropical Soil Chronosequence. Nat. Commun. 2017, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Bontemps, C.; Elliott, G.N.; Simon, M.F.; Dos Reis Junior, F.B.; Gross, E.; Lawton, R.C.; Neto, N.E.; de Fatima Loureiro, M.; De Faria, S.M.; Sprent, J.I.; et al. Burkholderia Species Are Ancient Symbionts of Legumes. Mol. Ecol. 2010, 19, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Sachs, J.L.; Skophammer, R.G.; Regus, J.U. Evolutionary Transitions in Bacterial Symbiosis. Proc. Natl. Acad. Sci. USA 2011, 108, 10800–10807. [Google Scholar] [CrossRef]
- Wang, S.; Meade, A.; Lam, H.-M.; Luo, H. Evolutionary Timeline and Genomic Plasticity Underlying the Lifestyle Diversity in Rhizobiales. mSystems 2020, 5, e00438-20. [Google Scholar] [CrossRef]
- Martinez-Romero, E. Coevolution in Rhizobium-Legume Symbiosis? DNA Cell Biol. 2009, 28, 361–370. [Google Scholar] [CrossRef]
- Hirsch, A.M.; Wilson, K.J.; Jones, J.D.G. Rhizobium meliloti Nodulation Genes Allow Agrobacterium tumefaciens and Escherichia coli to Form Pseudonodules on Alfalfa. J. Bacteriol. 1984, 158, 1133–1143. [Google Scholar] [CrossRef]
- Marchetti, M.; Capela, D.; Glew, M.; Cruveiller, S.; Chane-Woon-Ming, B.; Gris, C.; Timmers, T.; Poinsot, V.; Gilbert, L.B.; Heeb, P.; et al. Experimental Evolution of a Plant Pathogen into a Legume Symbiont. PLoS Biol. 2010, 8, e1000280. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.H.; Gris, C.; Cruveiller, S.; Pouzet, C.; Tasse, L.; Leru, A.; Maillard, A.; Médigue, C.; Batut, J.; Masson-Boivin, C.; et al. Experimental Evolution of Nodule Intracellular Infection in Legume Symbionts. ISME J. 2013, 7, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Capela, D.; Marchetti, M.; Clérissi, C.; Perrier, A.; Guetta, D.; Gris, C.; Valls, M.; Jauneau, A.; Cruveiller, S.; Rocha, E.P.C.; et al. Recruitment of a Lineage-Specific Virulence Regulatory Pathway Promotes Intracellular Infection by a Plant Pathogen Experimentally Evolved into a Legume Symbiont. Mol. Biol. Evol. 2017, 34, 2503–2521. [Google Scholar] [CrossRef] [PubMed]
- Hill, Y.; Colombi, E.; Bonello, E.; Haskett, T.; Ramsay, J.; O’Hara, G.; Terpolilli, J. Evolution of Diverse Effective N2-Fixing Microsymbionts of Cicer arietinum Following Horizontal Transfer of the Mesorhizobium ciceri CC1192 Symbiosis Integrative and Conjugative Element. Appl. Environ. Microbiol. 2021, 87, e02558-20. [Google Scholar] [CrossRef]
- Sullivan, J.T.; Patrick, H.N.; Lowther, W.L.; Scott, D.B.; Ronson, C.W. Nodulating Strains of Rhizobium loti Arise through Chromosomal Symbiotic Gene Transfer in the Environment. Proc. Natl. Acad. Sci. USA 1995, 92, 8985–8989. [Google Scholar] [CrossRef]
- Sullivan, J.T.; Ronson, C.W. Evolution of Rhizobia by Acquisition of a 500-kb Symbiosis Island That Integrates into a Phe-tRNA Gene. Proc. Natl. Acad. Sci. USA 1998, 95, 5145–5149. [Google Scholar] [CrossRef]
- Ling, J.; Wang, H.; Wu, P.; Li, T.; Tang, Y.; Naseer, N.; Zheng, H.; Masson-boivin, C.; Zhong, Z.; Zhu, J. Plant Nodulation Inducers Enhance Horizontal Gene Transfer of Azorhizobium caulinodans Symbiosis Island. Proc. Natl. Acad. Sci. USA 2016, 113, 13875–13880. [Google Scholar] [CrossRef]
- Barcellos, F.G.; Menna, P.; Batista, J.S.D.; Hungria, M.; da Silva Batista, J.S. Evidence of Horizontal Transfer of Symbiotic Genes from a Bradyrhizobium japonicum Inoculant Strain to Indigenous Diazotrophs Sinorhizobium (Ensifer) fredii and Bradyrhizobium elkanii in a Brazilian Savannah Soil. Appl. Environ. Microbiol. 2007, 73, 2635–2643. [Google Scholar] [CrossRef]
- Brockwell, J.; Bottomley, P.J. Recent Advances in Inoculant Technology and Prospects for the Future. Soil Biol. Biochem. 1995, 27, 683–697. [Google Scholar] [CrossRef]
- Liu, L.X.; Li, Q.Q.; Zhang, Y.Z.; Hu, Y.; Jiao, J.; Guo, H.J.; Zhang, X.X.; Zhang, B.; Chen, W.X.; Tian, C.F. The Nitrate-Reduction Gene Cluster Components Exert Lineage-Dependent Contributions to Optimization of Sinorhizobium Symbiosis with Soybeans. Environ. Microbiol. 2017, 19, 4926–4938. [Google Scholar] [CrossRef]
- Peoples, M.B.; Brockwell, J.; Hunt, J.R.; Swan, A.D.; Watson, L.; Hayes, R.C.; Li, G.D.; Hackney, B.; Nuttall, J.G.; Davies, S.L.; et al. Factors Affecting the Potential Contributions of N2 Fixation by Legumes in Australian Pasture Systems. Crop Pasture Sci. 2012, 63, 759. [Google Scholar] [CrossRef]
- Svensson, E.I.; Berger, D. The Role of Mutation Bias in Adaptive Evolution. Trends Ecol. Evol. 2019, 34, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, D.; Barton, N.H.; Charlesworth, B. The Sources of Adaptive Variation. Proc. R. Soc. B-Biol. Sci. 2017, 284, 20162864. [Google Scholar] [CrossRef] [PubMed]
- Begon, M.; Townsend, C.R. Ecology: From Individuals to Ecosystems, 5th ed.; Wiley: Hoboken, NJ, USA, 2021; ISBN 978-1-119-27931-0. [Google Scholar]
- Barton, N.H.; Etheridge, A.M.; Véber, A. The Infinitesimal Model: Definition, Derivation, and Implications. Theor. Popul. Biol. 2017, 118, 50–73. [Google Scholar] [CrossRef]
- Golicz, A.A.; Bayer, P.E.; Bhalla, P.L.; Batley, J.; Edwards, D. Pangenomics Comes of Age: From Bacteria to Plant and Animal Applications. Trends Genet. 2020, 36, 132–145. [Google Scholar] [CrossRef] [PubMed]
- McInerney, J.O.; McNally, A.; O’Connell, M.J. Why Prokaryotes Have Pangenomes. Nat. Microbiol. 2017, 2, 17040. [Google Scholar] [CrossRef]
- Persson, T.; Battenberg, K.; Demina, I.V.; Vigil-Stenman, T.; Vanden Heuvel, B.; Pujic, P.; Facciotti, M.T.; Wilbanks, E.G.; O’Brien, A.; Fournier, P.; et al. Candidatus Frankia Datiscae Dg1, the Actinobacterial Microsymbiont of Datisca glomerata, Expresses the Canonical nod Genes nodABC in Symbiosis with Its Host Plant. PLoS ONE 2015, 10, e0127630. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Wibberg, D.; Battenberg, K.; Blom, J.; Vanden Heuvel, B.; Berry, A.M.; Kalinowski, J.; Pawlowski, K. An Assemblage of Frankia Cluster II Strains from California Contains the Canonical nod Genes and Also the Sulfotransferase Gene nodH. BMC Genom. 2016, 17, 796. [Google Scholar] [CrossRef]
- Ktari, A.; Nouioui, I.; Furnholm, T.; Swanson, E.; Ghodhbane-Gtari, F.; Tisa, L.S.; Gtari, M. Permanent Draft Genome Sequence of Frankia sp. NRRL B-16219 Reveals the Presence of Canonical nod Genes, Which Are Highly Homologous to Those Detected in Candidatus Frankia Dg1 Genome. Stand. Genom. Sci. 2017, 12, 51. [Google Scholar] [CrossRef]
- Aoki, S.; Ito, M.; Iwasaki, W. From β- to α-Proteobacteria: The Origin and Evolution of Rhizobial Nodulation Genes nodIJ. Mol. Biol. Evol. 2013, 30, 2494–2508. [Google Scholar] [CrossRef]
- Tian, C.F.; Zhou, Y.J.; Zhang, Y.M.; Li, Q.Q.; Zhang, Y.Z.; Li, D.F.; Wang, S.; Wang, J.; Gilbert, L.B.; Li, Y.R.; et al. Comparative Genomics of Rhizobia Nodulating Soybean Suggests Extensive Recruitment of Lineage-Specific Genes in Adaptations. Proc. Natl. Acad. Sci. USA 2012, 109, 8629–8634. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.F.; Young, J.P.W. Evolution of Symbiosis Genes: Vertical and Horizontal Gene Transfer. In Ecology and Evolution of Rhizobia: Principles and Applications; Wang, E.T., Tian, C.F., Chen, W.F., Young, J.P.W., Chen, W.X., Eds.; Springer: Singapore, 2019; pp. 145–152. ISBN 978-981-32-9554-4. [Google Scholar] [CrossRef]
- Perret, X.; Staehelin, C.; Broughton, W.J. Molecular Basis of Symbiotic Promiscuity. Microbiol. Mol. Biol. Rev. 2000, 64, 180–201. [Google Scholar] [CrossRef] [PubMed]
- Roche, P.; Maillet, F.; Plazanet, C.; Debellé, F.; Ferro, M.; Truchet, G.; Promé, J.-C.; Denarié, J. The Common nodABC Genes of Rhizobium meliloti Are Host-Range Determinants. Proc. Natl. Acad. Sci. USA 1996, 93, 15305–15310. [Google Scholar] [CrossRef]
- Kamst, E.; Pilling, J.; Raamsdonk, L.M.; Lugtenberg, B.J.; Spaink, H.P. Rhizobium Nodulation Protein NodC Is an Important Determinant of Chitin Oligosaccharide Chain Length in Nod Factor Biosynthesis. J. Bacteriol. 1997, 179, 2103–2108. [Google Scholar] [CrossRef] [PubMed]
- Barran, L.R.; Bromfield, E.S.P.; Brown, D.C.W. Identification and Cloning of the Bacterial Nodulation Specificity Gene in the Sinorhizobium meliloti-Medicago laciniata Symbiosis. Can. J. Microbiol. 2002, 48, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Rogel, M.A.; Ormeño-Orrillo, E.; Martinez Romero, E. Symbiovars in Rhizobia Reflect Bacterial Adaptation to Legumes. Syst. Appl. Microbiol. 2011, 34, 96–104. [Google Scholar] [CrossRef]
- Wang, E.T.; Chen, W.F.; Tian, C.F.; Young, J.P.W.; Chen, W.X. Ecology and Evolution of Rhizobia: Principles and Applications; Springer: Singapore, 2019; ISBN 978-981-32-9554-4. [Google Scholar] [CrossRef]
- Moron, B.; Soria-Diaz, M.E.; Ault, J.; Verroios, G.; Noreen, S.; Rodriguez-Navarro, D.N.; Gil-Serrano, A.; Thomas-oates, J.; Megias, M.; Sousa, C.; et al. Low pH Changes the Profile of Nodulation Factors Produced by Rhizobium tropici CIAT899. Chem. Biol. 2005, 12, 1029–1040. [Google Scholar] [CrossRef]
- Ormeño-Orrillo, E.; Menna, P.; Almeida, L.G.P.; Ollero, F.J.; Nicolás, M.F.; Pains Rodrigues, E.; Shigueyoshi Nakatani, A.; Silva Batista, J.S.; Oliveira Chueire, L.M.; Souza, R.C.; et al. Genomic Basis of Broad Host Range and Environmental Adaptability of Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 Which Are Used in Inoculants for Common Bean (Phaseolus vulgaris L.). BMC Genom. 2012, 13, 735. [Google Scholar] [CrossRef]
- Mulligan, J.T.; Long, S.R. Induction of Rhizobium meliloti nodC Expression by Plant Exudate Requires nodD. Proc. Natl. Acad. Sci. USA 1985, 82, 6609–6613. [Google Scholar] [CrossRef]
- Hu, H.; Liu, S.; Yang, Y.; Chang, W.; Hong, G. In Rhizobium leguminosarum, NodD Represses Its Own Transcription by Competing with RNA Polymerase for Binding Sites. Nucleic Acids Res. 2000, 28, 2784–2793. [Google Scholar] [CrossRef]
- Györgypal, Z.; Kondorosi, É.; Kondorosi, A. Diverse Signal Sensitivity of NodD Protein Homologs from Narrow and Broad Host Range Rhizobia. Mol. Plant Microbe Interact. 1991, 4, 356–364. [Google Scholar] [CrossRef]
- Machado, D.; Krishnan, H.B. nodD Alleles of Sinorhizobium fredii USDA191 Differentially Influence Soybean Nodulation, nodC Expression, and Production of Exopolysaccharides. Curr. Microbiol. 2003, 47, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, H.B.; Kuo, C.I.; Pueppke, S.G. Elaboration of Flavonoid-Induced Proteins by the Nitrogen-Fixing Soybean Symbiont Rhizobium fredii Is Regulated by Both nodD1 and nodD2, and Is Dependent on the Cultivar-Specificity Locus, nolXWBTUV. Microbiology 1995, 141, 2245–2251. [Google Scholar] [CrossRef]
- del Cerro, P.; Rolla-Santos, A.A.P.; Gomes, D.F.; Marks, B.B.; Espuny, M.d.R.; Rodríguez-Carvajal, M.Á.; Soria-Díaz, M.E.; Nakatani, A.S.; Hungria, M.; Ollero, F.J.; et al. Opening the “black Box” of nodD3, nodD4 and nodD5 Genes of Rhizobium tropici Strain CIAT 899. BMC Genom. 2015, 16, 864. [Google Scholar] [CrossRef]
- Lestrange, K.K.; Bender, G.L.; Djordjevic, M.A.; Rolfe, B.G.; Redmond, J.W. The Rhizobium Strain NGR234 nodD1 Gene-Product Responds to Activation by the Simple Phenolic-Compounds Vanillin and Isovanillin Present in Wheat Seedling Extracts. Mol. Plant Microbe Interact. 1990, 3, 214–220. [Google Scholar] [CrossRef]
- Jiao, Y.S.; Liu, Y.H.; Yan, H.; Wang, E.T.; Tian, C.F.; Chen, W.X.; Guo, B.L.; Chen, W.F. Rhizobial Diversity and Nodulation Characteristics of the Extremely Promiscuous Legume Sophora flavescens. Mol. Plant Microbe Interact. 2015, 28, 1338–1352. [Google Scholar] [CrossRef]
- Guo, H.J.; Wang, E.T.; Zhang, X.X.; Li, Q.Q.; Zhang, Y.M.; Tian, C.F.; Chen, W.X. Replicon-Dependent Differentiation of Symbiosis-Related Genes in Sinorhizobium Strains Nodulating Glycine max. Appl. Environ. Microbiol. 2014, 80, 1245–1255. [Google Scholar] [CrossRef]
- Zhang, X.X.; Guo, H.J.; Wang, R.; Sui, X.H.; Zhang, Y.M.; Wang, E.T.; Tian, C.F.; Chen, W.X. Genetic Divergence of Bradyrhizobium Nodulating Soybeans as Revealed by Multilocus Sequence Analysis of Genes inside and Outside the Symbiosis Island. Appl. Environ. Microbiol. 2014, 80, 3181–3190. [Google Scholar] [CrossRef]
- Pueppke, S.G.; Broughton, W.J. Rhizobium sp. Strain NGR234 and R. fredii USDA257 Share Exceptionally Broad, Nested Host Ranges. Mol. Plant Microbe Interact. 1999, 12, 293–318. [Google Scholar] [CrossRef]
- Songwattana, P.; Tittabutr, P.; Wongdee, J.; Teamtisong, K.; Wulandari, D.; Teulet, A.; Fardoux, J.; Boonkerd, N.; Giraud, E.; Teaumroong, N. Symbiotic Properties of a Chimeric Nod-independent Photosynthetic Bradyrhizobium Strain Obtained by Conjugative Transfer of a Symbiotic Plasmid. Environ. Microbiol. 2019, 21, 3442–3454. [Google Scholar] [CrossRef]
- Pi, H.W.; Lin, J.J.; Chen, C.A.; Wang, P.H.; Chiang, Y.R.; Huang, C.C.; Young, C.C.; Li, W.H. Origin and Evolution of Nitrogen Fixation in Prokaryotes. Mol. Biol. Evol. 2022, 39, msac181. [Google Scholar] [CrossRef]
- Okubo, T.; Piromyou, P.; Tittabutr, P.; Teaumroong, N.; Minamisawa, K. Origin and Evolution of Nitrogen Fixation Genes on Symbiosis Islands and Plasmid in Bradyrhizobium. Microbes Environ. 2016, 31, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.F.; Young, J.P.W. Genomics and Evolution of Rhizobia. In Ecology and Evolution of Rhizobia: Principles and Applications; Wang, E.T., Tian, C.F., Chen, W.F., Young, J.P.W., Chen, W.X., Eds.; Springer: Singapore, 2019; pp. 103–119. ISBN 978-981-32-9554-4. [Google Scholar] [CrossRef]
- Cui, W.; Zhang, B.; Zhao, R.; Liu, L.; Jiao, J.; Zhang, Z.; Tian, C.-F. Lineage-Specific Rewiring of Core Pathways Predating Innovation of Legume Nodules Shapes Symbiotic Efficiency. mSystems 2021, 6, e01299-20. [Google Scholar] [CrossRef] [PubMed]
- Cavassim, M.I.A.; Moeskjær, S.; Moslemi, C.; Fields, B.; Bachmann, A.; Vilhjálmsson, B.J.; Schierup, M.H.; Young, J.P.W.; Andersen, S.U. Symbiosis Genes Show a Unique Pattern of Introgression and Selection within a Rhizobium leguminosarum Species Complex. Microb. Genom. 2020, 6, e000351. [Google Scholar] [CrossRef] [PubMed]
- Perez Carrascal, O.M.; VanInsberghe, D.; Juarez, S.; Polz, M.F.; Vinuesa, P.; Gonzalez, V. Population Genomics of the Symbiotic Plasmids of Sympatric Nitrogen-Fixing Rhizobium Species Associated with Phaseolus vulgaris. Environ. Microbiol. 2016, 18, 2660–2676. [Google Scholar] [CrossRef] [PubMed]
- Epstein, B.; Branca, A.; Mudge, J.; Bharti, A.K.; Briskine, R.; Farmer, A.D.; Sugawara, M.; Young, N.D.; Sadowsky, M.J.; Tiffin, P. Population Genomics of the Facultatively Mutualistic Bacteria Sinorhizobium meliloti and S. medicae. PLoS Genet. 2012, 8, e1002868. [Google Scholar] [CrossRef] [PubMed]
- Ledermann, R.; Schulte, C.C.M.; Poole, P.S. How Rhizobia Adapt to the Nodule Environment. J. Bacteriol. 2021, 203, e00539-20. [Google Scholar] [CrossRef]
- Preisig, O.; Zufferey, R.; Hennecke, H. The Bradyrhizobium japonicum fixGHIS Genes Are Required for the Formation of the High-Affinity cbb3-Type Cytochrome Oxidase. Arch. Microbiol. 1996, 165, 297–305. [Google Scholar] [CrossRef]
- Staehelin, C.; Krishnan, H.B. Nodulation Outer Proteins: Double-Edged Swords of Symbiotic Rhizobia. Biochem. J. 2015, 470, 263–274. [Google Scholar] [CrossRef]
- Zhao, R.; Liu, L.X.; Zhang, Y.Z.; Jiao, J.; Cui, W.J.; Zhang, B.; Wang, X.L.; Li, M.L.; Chen, Y.; Xiong, Z.Q.; et al. Adaptive Evolution of Rhizobial Symbiotic Compatibility Mediated by Co-Evolved Insertion Sequences. ISME J. 2018, 12, 101–111. [Google Scholar] [CrossRef]
- Sugawara, M.; Takahashi, S.; Umehara, Y.; Iwano, H.; Tsurumaru, H.; Odake, H.; Suzuki, Y.; Kondo, H.; Konno, Y.; Yamakawa, T.; et al. Variation in Bradyrhizobial NopP Effector Determines Symbiotic Incompatibility with Rj2-Soybeans via Effector-Triggered Immunity. Nat. Commun. 2018, 9, 3139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, M.; Sun, Y.; Zhao, P.; Liu, C.; Qing, K.; Hu, X.; Zhong, Z.; Cheng, J.; Wang, H.; et al. Glycine max NNL1 Restricts Symbiotic Compatibility with Widely Distributed Bradyrhizobia via Root Hair Infection. Nat. Plants 2021, 7, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Hubber, A.; Vergunst, A.C.; Sullivan, J.T.; Hooykaas, P.J.J.; Ronson, C.W. Symbiotic Phenotypes and Translocated Effector Proteins of the Mesorhizobium loti Strain R7A VirB/D4 Type IV Secretion System. Mol. Microbiol. 2004, 54, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Paço, A.; Da-Silva, J.R.; Eliziário, F.; Brígido, C.; Oliveira, S.; Alexandre, A. traG Gene Is Conserved across Mesorhizobium spp. Able to Nodulate the Same Host Plant and Expressed in Response to Root Exudates. Biomed Res. Int. 2019, 2019, 3715271. [Google Scholar] [CrossRef]
- Sullivan, J.T.; Trzebiatowski, J.R.; Cruickshank, R.W.; Gouzy, J.; Brown, S.D.; Elliot, R.M.; Fleetwood, D.J.; McCallum, N.G.; Rossbach, U.; Stuart, G.S.; et al. Comparative Sequence Analysis of the Symbiosis Island of Mesorhizobium loti Strain R7A. J. Bacteriol. 2002, 184, 3086–3095. [Google Scholar] [CrossRef]
- Hubber, A.M.; Sullivan, J.T.; Ronson, C.W. Symbiosis-Induced Cascade Regulation of the Mesorhizobium loti R7A VirB/D4 Type IV Secretion System. Mol. Plant-Microbe Interact. 2007, 20, 255–261. [Google Scholar] [CrossRef]
- Yang, L.L.; Jiang, Z.; Li, Y.; Wang, E.T.; Zhi, X.Y. Plasmids Related to the Symbiotic Nitrogen Fixation Are Not Only Cooperated Functionally but Also May Have Evolved over a Time Span in Family Rhizobiaceae. Genome. Biol. Evol. 2020, 12, 2002–2014. [Google Scholar] [CrossRef]
- Nelson, M.; Guhlin, J.; Epstein, B.; Tiffin, P.; Sadowsky, M.J. The Complete Replicons of 16 Ensifer meliloti Strains Offer Insights into Intra-and Inter-Replicon Gene Transfer, Transposon-Associated Loci, and Repeat Elements. Microb. Genom. 2018, 4, e000174. [Google Scholar] [CrossRef]
- Jozefkowicz, C.; Brambilla, S.; Frare, R.; Stritzler, M.; Piccinetti, C.; Puente, M.; Berini, C.A.; Pérez, P.R.; Soto, G.; Ayub, N. Stable Symbiotic Nitrogen Fixation under Water-Deficit Field Conditions by a Stress-Tolerant Alfalfa Microsymbiont and Its Complete Genome Sequence. J. Biotechnol. 2017, 263, 52–54. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, B.; Jiao, J.; Dai, S.-Q.; Chen, W.-X.; Tian, C.-F. Modulation of Symbiotic Compatibility by Rhizobial Zinc Starvation Machinery. mBio 2020, 11, e03193-19. [Google Scholar] [CrossRef]
- Price, P.A.; Tanner, H.R.; Dillon, B.A.; Shabab, M.; Walker, G.C.; Griffitts, J.S. Rhizobial Peptidase HrrP Cleaves Host-Encoded Signaling Peptides and Mediates Symbiotic Compatibility. Proc. Natl. Acad. Sci. USA 2015, 112, 15244–15249. [Google Scholar] [CrossRef] [PubMed]
- Crook, M.B.; Lindsay, D.P.; Biggs, M.B.; Bentley, J.S.; Price, J.C.; Clement, S.C.; Clement, M.J.; Long, S.R.; Griffitts, J.S. Rhizobial Plasmids That Cause Impaired Symbiotic Nitrogen Fixation and Enhanced Host Invasion. Mol. Plant-Microbe Interact. 2012, 25, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Arashida, H.; Odake, H.; Sugawara, M.; Noda, R.; Kakizaki, K.; Ohkubo, S.; Mitsui, H.; Sato, S.; Minamisawa, K. Evolution of Rhizobial Symbiosis Islands through Insertion Sequence-Mediated Deletion and Duplication. ISME J. 2022, 16, 112–121. [Google Scholar] [CrossRef]
- Remigi, P.; Capela, D.; Clerissi, C.; Tasse, L.; Torchet, R.; Bouchez, O.; Batut, J.; Cruveiller, S.; Rocha, E.P.C.; Masson-Boivin, C. Transient Hypermutagenesis Accelerates the Evolution of Legume Endosymbionts Following Horizontal Gene Transfer. PLoS Biol. 2014, 12, e1001942. [Google Scholar] [CrossRef]
- Tang, M.; Bouchez, O.; Masson-boivin, C.; Capela, D. Modulation of Quorum Sensing as an Adaptation to Nodule Cell Infection during Experimental Evolution of Legume. mBio 2020, 11, e03129-19. [Google Scholar] [CrossRef]
- Doin de Moura, G.G.; Remigi, P.; Masson-boivin, C.; Capela, D. Experimental Evolution of Legume Symbionts: What Have We Learnt? Genes 2020, 11, 339. [Google Scholar] [CrossRef]
- Biemont, C. A Brief History of the Status of Transposable Elements: From Junk DNA to Major Players in Evolution. Genetics 2010, 1093, 1085–1093. [Google Scholar] [CrossRef]
- Niu, X.M.; Xu, Y.C.; Li, Z.W.; Bian, Y.T.; Hou, X.H.; Chen, J.F.; Zou, Y.P.; Jiang, J.; Wu, Q.; Ge, S.; et al. Transposable Elements Drive Rapid Phenotypic Variation in Capsella rubella. Proc. Natl. Acad. Sci. USA 2019, 116, 6908–6913. [Google Scholar] [CrossRef]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The Impact of Insertion Sequences on Bacterial Genome Plasticity and Adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial Insertion Sequences: Their Genomic Impact and Diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef]
- Freiberg, C.; Fellay, R.; Bairoch, A.; Broughton, W.J.; Rosenthal, A.; Perret, X. Molecular Basis of Symbiosis between Rhizobium and Legumes. Nature 1997, 387, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Barnett, M.J.; Fisher, R.F.; Jones, T.; Komp, C.; Abola, A.P.; Barloy-Hubler, F.; Bowser, L.; Capela, D.; Galibert, F.; Gouzy, J.; et al. Nucleotide Sequence and Predicted Functions of the Entire Sinorhizobium meliloti pSymA Megaplasmid. Proc. Natl. Acad. Sci. USA 2001, 98, 9883–9888. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Nakamura, Y.; Sato, S.; Asamizu, E.; Kato, T.; Sasamoto, S.; Watanabe, A.; Idesawa, K.; Ishikawa, A.; Kawashima, K.; et al. Complete Genome Structure of the Nitrogen-Fixing Symbiotic Bacterium Mesorhizobium loti. DNA Res. 2000, 7, 331–338. [Google Scholar] [CrossRef]
- Barros-Carvalho, G.A.; Hungria, M.; Lopes, F.M.; Van Sluys, M.A. Brazilian-Adapted Soybean Bradyrhizobium Strains Uncover IS Elements with Potential Impact on Biological Nitrogen Fixation. FEMS Microbiol. Lett. 2019, 366, fnz046. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Wang, S.; Liao, T.; Luo, H. Evolutionary Origin and Ecological Implication of a Unique nif Island in Free-Living Bradyrhizobium Lineages. ISME J. 2021, 15, 3195–3206. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Shi, W.; Zhang, B.; Xu, Y.; Jiao, J. Intracellular Common Gardens Reveal Niche Differentiation in Transposable Element Community during Bacterial Adaptive Evolution. ISME J. 2022. [Google Scholar] [CrossRef]
- Jiao, J.; Zhang, B.; Li, M.-L.; Zhang, Z.; Tian, C.-F. The Zinc-Finger Bearing Xenogeneic Silencer MucR in α-Proteobacteria Balances Adaptation and Regulatory Integrity. ISME J. 2022, 16, 738–749. [Google Scholar] [CrossRef]
- Shi, W.-T.; Zhang, B.; Li, M.-L.; Liu, K.-H.; Jiao, J.; Tian, C.-F. The Convergent Xenogeneic Silencer MucR Predisposes α-Proteobacteria to Integrate AT-Rich Symbiosis Genes. Nucleic Acids Res. 2022, 50, 8580–8598. [Google Scholar] [CrossRef]
- Jiao, J.; Tian, C.-F. Ancestral Zinc-Finger Bearing Protein MucR in Alpha-Proteobacteria: A Novel Xenogeneic Silencer? Comput. Struct. Biotechnol. J. 2020, 18, 3623–3631. [Google Scholar] [CrossRef]
- Jiao, J.; Ni, M.; Zhang, B.; Zhang, Z.; Young, J.P.W.; Chan, T.-F.; Chen, W.X.; Lam, H.-M.; Tian, C.F. Coordinated Regulation of Core and Accessory Genes in the Multipartite Genome of Sinorhizobium fredii. PLoS Genet. 2018, 14, e1007428. [Google Scholar] [CrossRef]
- Wheatley, R.M.; Ford, B.L.; Li, L.; Aroney, S.T.N.; Knights, H.E.; Ledermann, R.; East, A.K.; Ramachandran, V.K.; Poole, P.S. Lifestyle Adaptations of Rhizobium from Rhizosphere to Symbiosis. Proc. Natl. Acad. Sci. USA 2020, 117, 23823–23834. [Google Scholar] [CrossRef] [PubMed]
- DiCenzo, G.C.; Checcucci, A.; Bazzicalupo, M.; Mengoni, A.; Viti, C.; Dziewit, L.; Finan, T.M.; Galardini, M.; Fondi, M. Metabolic Modelling Reveals the Specialization of Secondary Replicons for Niche Adaptation in Sinorhizobium meliloti. Nat. Commun. 2016, 7, 12219. [Google Scholar] [CrossRef] [PubMed]
- Galardini, M.; Pini, F.; Bazzicalupo, M.; Biondi, E.G.; Mengoni, A. Replicon-Dependent Bacterial Genome Evolution: The Case of Sinorhizobium meliloti. Genome Biol. Evol. 2013, 5, 542–558. [Google Scholar] [CrossRef]
- Ramachandran, V.K.; East, A.K.; Karunakaran, R.; Downie, J.A.; Poole, P.S. Adaptation of Rhizobium leguminosarum to Pea, Alfalfa and Sugar Beet Rhizospheres Investigated by Comparative Transcriptomics. Genome Biol. 2011, 12, R106. [Google Scholar] [CrossRef]
- Vercruysse, M.; Fauvart, M.; Beullens, S.; Braeken, K.; Cloots, L.; Engelen, K.; Marchal, K.; Michiels, J. A Comparative Transcriptome Analysis of Rhizobium etli Bacteroids: Specific Gene Expression During Symbiotic Nongrowth. Mol. Plant Microbe Interact. 2011, 24, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Capela, D.; Filipe, C.; Bobilk, C.; Batut, J.; Bruand, C.; Bobik, C.; Plantes-microorganismes, L.I.; Inra-cnrs, U.M.R. Sinorhizobium meliloti Differentiation during Symbiosis with Alfalfa: A Transcriptomic Dissection. Mol. Plant Microbe Interact. 2006, 19, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tian, C.F.; Chen, W.F.; Wang, L.; Sui, X.H.; Chen, W.X. High-Resolution Transcriptomic Analyses of Sinorhizobium sp. NGR234 Bacteroids in Determinate Nodules of Vigna unguiculata and Indeterminate Nodules of Leucaena leucocephala. PLoS ONE 2013, 8, e70531. [Google Scholar] [CrossRef]
- Green, R.T.; East, A.K.; Karunakaran, R.; Downie, J.A.; Poole, P.S. Transcriptomic Analysis of Rhizobium leguminosarum Bacteroids in Determinate and Indeterminate Nodules. Microb. Genom. 2019, 5, e000254. [Google Scholar] [CrossRef]
- Schulte, C.C.M.; Borah, K.; Wheatley, R.M.; Terpolilli, J.J.; Saalbach, G.; Crang, N.; de Groot, D.H.; Ratcliffe, R.G.; Kruger, N.J.; Papachristodoulou, A.; et al. Metabolic Control of Nitrogen Fixation in Rhizobium-Legume Symbioses. Sci. Adv. 2021, 7, eabh2433. [Google Scholar] [CrossRef]
- Aroney, S.T.N.; Poole, P.S.; Sánchez-Cañizares, C. Rhizobial Chemotaxis and Motility Systems at Work in the Soil. Front. Plant Sci. 2021, 12, 1856. [Google Scholar] [CrossRef]
- Acosta-jurado, S.; Fuentes-romero, F.; Ruiz-sainz, J.E.; Janczarek, M.; Vinardell, J.M. Rhizobial Exopolysaccharides: Genetic Regulation of Their Synthesis and Relevance in Symbiosis with Legumes. Int. J. Mol. Sci. 2021, 22, 6233. [Google Scholar] [CrossRef] [PubMed]
- Janczarek, M.; Rachwał, K.; Marzec, A.; Grzadziel, J.; Palusińska-Szysz, M. Signal Molecules and Cell-Surface Components Involved in Early Stages of the Legume-Rhizobium Interactions. Appl. Soil Ecol. 2015, 85, 94–113. [Google Scholar] [CrossRef]
- Kawaharada, Y.; Kelly, S.; Nielsen, M.W.; Hjuler, C.T.; Gysel, K.; Muszyński, A.; Carlson, R.W.; Thygesen, M.B.; Sandal, N.; Asmussen, M.H.; et al. Receptor-Mediated Exopolysaccharide Perception Controls Bacterial Infection. Nature 2015, 523, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Kawaharada, Y.; Nielsen, M.W.; Kelly, S.; James, E.K.; Andersen, K.R.; Madsen, L.H.; Heckmann, A.B.; Radutoiu, S.; Rasmussen, S.R.; Fu, W.; et al. Differential Regulation of the Epr3 Receptor Coordinates Membrane-Restricted Rhizobial Colonization of Root Nodule Primordia. Nat. Commun. 2017, 8, 14534. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.F.F.; Penterman, J.; Shabab, M.; Chen, E.J.; Walker, C. Important Late-Stage Symbiotic Role of the Sinorhizobium meliloti Exopolysaccharide Succinoglycan. J. Bacteriol. 2018, 200, e00665-17. [Google Scholar] [CrossRef]
- Li, D.; Li, Z.; Wu, J.; Tang, Z.; Xie, F.; Chen, D.; Lin, H.; Li, Y. Analysis of Outer Membrane Vesicles Indicates That Glycerophospholipid Metabolism Contributes to Early Symbiosis Between Sinorhizobium fredii HH103 and Soybean. Mol. Plant-Microbe Interact. 2022, 35, 311–322. [Google Scholar] [CrossRef]
- Yang, M.H.; Sun, K.J.; Zhou, L.; Yang, R.F.; Zhong, Z.T.; Zhu, J. Functional Analysis of Three AHL Autoinducer Synthase Genes in Mesorhizobium loti Reveals the Important Role of Quorum Sensing in Symbiotic Nodulation. Can. J. Microbiol. 2009, 55, 210–214. [Google Scholar] [CrossRef]
- Marketon, M.M.; Gronquist, M.R.; Eberhard, A.; Gonza, J.E. Characterization of the Sinorhizobium meliloti sinR/sinI Locus and the Production of Novel N-Acyl Homoserine Lactones. J. Bacteriol. 2002, 184, 5686–5695. [Google Scholar] [CrossRef]
- Gurich, N.; Gonzalez, J.E. Role of Quorum Sensing in Sinorhizobium meliloti-Alfalfa Symbiosis. J. Bacteriol. 2009, 191, 4372–4382. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Alías-Villegas, C.; Almozara, A.; Espuny, M.R.; Vinardell, J.M.; Pérez-Montaño, F. Deciphering the Symbiotic Significance of Quorum Sensing Systems of Sinorhizobium fredii HH103. Microorganisms 2020, 8, 68. [Google Scholar] [CrossRef]
- Calatrava-Morales, N.; McIntosh, M.; Soto, M.J. Regulation Mediated by N-Acyl Homoserine Lactone Quorum Sensing Signals in the Rhizobium-Legume Symbiosis. Genes 2018, 9, 263. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Medina, C.; Vinardell, J.M.; Ollero, F.J.; López-Baena, F.J. The Rhizobial Type 3 Secretion System: The Dr. Jekyll and Mr. Hyde in the Rhizobium–Legume Symbiosis. Int. J. Mol. Sci. 2022, 23, 11089. [Google Scholar] [CrossRef] [PubMed]
- Ratu, S.T.N.; Amelia, L.; Okazaki, S. Type III Effector Provides a Novel Symbiotic Pathway in Legume-Rhizobia Symbiosis. Biosci. Biotechnol. Biochem. 2023, 87, 28–37. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, B.F.S.; Castellane, T.C.L.; Tighilt, L.; Lemos, E.G. de M.; Rey, L. Rhizobial Exopolysaccharides and Type VI Secretion Systems: A Promising Way to Improve Nitrogen Acquisition by Legumes. Front. Agron. 2021, 3, e661468. [Google Scholar] [CrossRef]
- Salinero-Lanzarote, A.; Pacheco-Moreno, A.; Domingo-Serrano, L.; Durán, D.; Ormeño-Orrillo, E.; Martínez-Romero, E.; Albareda, M.; Palacios, J.M.; Rey, L. The Type VI Secretion System of Rhizobium etli Mim1 Has a Positive Effect in Symbiosis. FEMS Microbiol. Ecol. 2019, 95, efiz054. [Google Scholar] [CrossRef]
- Fauvart, M.; Michiels, J. Rhizobial Secreted Proteins as Determinants of Host Specificity in the Rhizobium-Legume Symbiosis. FEMS Microbiol. Lett. 2008, 285, 1–9. [Google Scholar] [CrossRef]
- Finnie, C.; Hartley, N.M.; Findlay, K.C.; Downie, J.A. The Rhizobium leguminosarum prsDE Genes Are Required for Secretion of Several Proteins, Some of Which Influence Nodulation, Symbiotic Nitrogen Fixation and Exopolysaccharide Modification. Mol. Microbiol. 1997, 25, 135–146. [Google Scholar] [CrossRef]
- Liu, J.; Wang, T.; Qin, Q.; Yu, X.; Yang, S.; Dinkins, R.D.; Kuczmog, A.; Putnoky, P.; Muszyński, A.; Griffitts, J.S.; et al. Paired Medicago Receptors Mediate Broad-Spectrum Resistance to Nodulation by Sinorhizobium meliloti Carrying a Species-Specific Gene. Proc. Natl. Acad. Sci. USA 2022, 119, e2214703119. [Google Scholar] [CrossRef]
- Yurgel, S.N.; Kahn, M.L. Dicarboxylate Transport by Rhizobia. FEMS Microbiol. Rev. 2004, 28, 489–501. [Google Scholar] [CrossRef]
- Wang, C.; Saldanha, M.; Sheng, X.; Shelswell, K.J.; Walsh, K.T.; Sobral, B.W.S.; Charles, T.C. Roles of Poly-3-Hydroxybutyrate (PHB) and Glycogen in Symbiosis of Sinorhizobium meliloti with Medicago sp. Microbiology 2007, 153, 388–398. [Google Scholar] [CrossRef]
- Mandon, K.; Michel-reydellet, N.; Encarnacio, S.; Kaminski, P.A.; Leija, A.; Cevallos, M.A.; Elmerich, C.; Mora, J. Poly-β-Hydroxybutyrate Turnover in Azorhizobium caulinodans Is Required for Growth and Affects nifA Expression. J. Bacteriol. 1998, 180, 5070–5076. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-W.; Li, Y.; Hu, Y.; Chen, W.-X.; Tian, C.-F. Coordinated Regulation of the Size and Number of Polyhydroxybutyrate Granules by Core and Accessory Phasins in the Facultative Microsymbiont Sinorhizobium fredii NGR234. Appl. Environ. Microbiol. 2019, 85, e00717-19. [Google Scholar] [CrossRef] [PubMed]
- Lodwig, E.M.; Hosie, A.H.; Bourdes, A.; Findlay, K.; Allaway, D.; Karunakaran, R.; Downie, J.A.; Poole, P.S. Amino-Acid Cycling Drives Nitrogen Fixation in the Legume-Rhizobium Symbiosis. Nature 2003, 422, 722–726. [Google Scholar] [CrossRef]
- Prell, J.; White, J.P.; Bourdes, A.; Bunnewell, S.; Bongaerts, R.J.; Poole, P.S. Legumes Regulate Rhizobium Bacteroid Development and Persistence by the Supply of Branched-Chain Amino Acids. Proc. Natl. Acad. Sci. USA 2009, 106, 12477–12482. [Google Scholar] [CrossRef]
- Prell, J.; Bourdes, A.; Kumar, S.; Lodwig, E.; Hosie, A.; Kinghorn, S.; White, J.; Poole, P. Role of Symbiotic Auxotrophy in the Rhizobium-Legume Symbioses. PLoS ONE 2010, 5, e13933. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiao, J.; Liu, L.X.; Sun, Y.W.; Chen, W.; Sui, X.; Chen, W.; Tian, C.F. Evidence for Phosphate Starvation of Rhizobia without Terminal Differentiation in Legume Nodules. Mol. Plant-Microbe Interact. 2018, 31, 1060–1068. [Google Scholar] [CrossRef]
- Bardin, S.D.; Finan, T.M. Regulation of Phosphate Assimilation in Rhizobium (Sinorhizobium) meliloti. Genetics 1998, 148, 1689–1700. [Google Scholar] [CrossRef]
- Yuan, Z.C.; Zaheer, R.; Finan, T.M. Regulation and Properties of PstSCAB, a High-Affinity, High-Velocity Phosphate Transport System of Sinorhizobium meliloti. J. Bacteriol. 2006, 188, 1089–1102. [Google Scholar] [CrossRef]
- Feng, X.-Y.; Tian, Y.; Cui, W.-J.; Li, Y.-Z.; Wang, D.; Liu, Y.; Jiao, J.; Chen, W.-X.; Tian, C.-F. The PTSNtr-KdpDE-KdpFABC Pathway Contributes to Low Potassium Stress Adaptation and Competitive Nodulation of Sinorhizobium fredii. mBio 2022, 13, e03721-21. [Google Scholar] [CrossRef]
- Dominguez-Ferreras, A.; Munoz, S.; Olivares, J.; Soto, M.J.; Sanjuan, J.; Domínguez-ferreras, A.; Mun, S.; Sanjua, J. Role of Potassium Uptake Systems in Sinorhizobium meliloti Osmoadaptation and Symbiotic Performance. J. Bacteriol. 2009, 191, 2133–2143. [Google Scholar] [CrossRef]
- Cheng, G.; Karunakaran, R.; East, A.K.; Poole, P.S. Multiplicity of Sulfate and Molybdate Transporters and Their Role in Nitrogen Fixation in Rhizobium leguminosarum bv. viciae Rlv3841. Mol. Plant-Microbe Interact. 2016, 29, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-H.; Zhang, B.; Yang, B.-S.; Shi, W.-T.; Li, Y.-F.; Wang, Y.; Zhang, P.; Jiao, J.; Tian, C.-F. Rhizobiales-Specific RirA Represses a Naturally “Synthetic” Foreign Siderophore Gene Cluster to Maintain Sinorhizobium-Legume Mutualism. mBio 2022, 13, e02900-21. [Google Scholar] [CrossRef] [PubMed]
- Sankari, S.; Babu, V.M.P.; Bian, K.; Alhhazmi, A.; Andorfer, M.C.; Avalos, D.M.; Smith, T.A.; Yoon, K.; Drennan, C.L.; Yaffe, M.B.; et al. A Haem-Sequestering Plant Peptide Promotes Iron Uptake in Symbiotic Bacteria. Nat. Microbiol. 2022, 7, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Hood, G.; Ramachandran, V.K.; East, A.; Downie, J.A.; Poole, P.S. Manganese Transport Is Essential for N2-Fixation by Rhizobium leguminosarum in Bacteroids from Galegoid but Not Phaseoloid Nodules. Environ. Microbiol. 2017, 19, 2715–2726. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, B.; Le Scornet, A.; Sauviac, L.; Rémy, A.; Bruand, C.; Meilhoc, E. The Nitrate Assimilatory Pathway in Sinorhizobium meliloti: Contribution to NO Production. Front. Microbiol. 2019, 10, 1526. [Google Scholar] [CrossRef] [PubMed]
- Cam, Y.; Pierre, O.; Boncompagni, E.; Herouart, D.; Meilhoc, E.; Bruand, C. Nitric Oxide (NO) a Key Player in the Senescence of Medicago truncatula Root Nodules. New Phytol. 2012, 196, 548–560. [Google Scholar] [CrossRef]
- Bobik, C.; Meilhoc, E.; Batut, J. FixJ: A Major Regulator of the Oxygen Limitation Response and Late Symbiotic Functions of Sinorhizobium meliloti. J. Bacteriol. 2006, 188, 4890–4902. [Google Scholar] [CrossRef]
- Bauer, E.; Kaspar, T.; Fischer, H.M.; Hennecke, H. Expression of the fixR-nifA Operon in Bradyrhizobium japonicum Depends on a New Response Regulator, RegR. J. Bacteriol. 1998, 180, 3853–3863. [Google Scholar] [CrossRef]
- Rutten, P.J.; Steel, H.; Hood, G.A.; Ramachandran, V.K.; McMurtry, L.; Geddes, B.; Papachristodoulou, A.; Poole, P.S. Multiple Sensors Provide Spatiotemporal Oxygen Regulation of Gene Expression in a Rhizobium-Legume Symbiosis. PLoS Genet. 2021, 17, e1009099. [Google Scholar] [CrossRef]
- De Nisco, N.J.; Abo, R.P.; Wu, C.M.; Penterman, J.; Walker, G.C. Global Analysis of Cell Cycle Gene Expression of the Legume Symbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 2014, 111, 3217–3224. [Google Scholar] [CrossRef]
- Gibson, K.E.; Campbell, G.R.; Lloret, J.; Walker, G.C. CbrA Is a Stationary-Phase Regulator of Cell Surface Physiology and Legume Symbiosis in Sinorhizobium meliloti. J. Bacteriol. 2006, 188, 4508–4521. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; De Nisco, N.J.; Chien, P.; Simmons, L.A.; Walker, G.C. Sinorhizobium meliloti CpdR1 Is Critical for Co-Ordinating Cell Cycle Progression and the Symbiotic Chronic Infection. Mol. Microbiol. 2009, 73, 586–600. [Google Scholar] [CrossRef] [PubMed]
- diCenzo, G.C.; Zamani, M.; Ludwig, H.N.; Finan, T.M. Heterologous Complementation Reveals a Specialized Activity for BacA in the Medicago-Sinorhizobium meliloti Symbiosis. Mol. Plant-Microbe Interact. 2017, 30, 312–324. [Google Scholar] [CrossRef]
- Marlow, V.L.; Haag, A.F.; Kobayashi, H.; Fletcher, V.; Scocchi, M.; Walker, G.C.; Ferguson, G.P. Essential Role for the BacA Protein in the Uptake of a Truncated Eukaryotic Peptide in Sinorhizobium meliloti. J. Bacteriol. 2009, 191, 1519–1527. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, R.; Haag, A.F.; East, A.K.; Ramachandran, V.K.; Prell, J.; James, E.K.; Scocchi, M.; Ferguson, G.P.; Poole, P.S. BacA Is Essential for Bacteroid Development in Nodules of Galegoid, but Not Phaseoloid, Legumes. J. Bacteriol. 2010, 192, 2920–2928. [Google Scholar] [CrossRef] [PubMed]
- Haag, A.F.; Baloban, M.; Sani, M.; Kerscher, B.; Pierre, O.; Farkas, A.; Longhi, R.; Boncompagni, E.; Hérouart, D.; Dall’Angelo, S.; et al. Protection of Sinorhizobium against Host Cysteine-Rich Antimicrobial Peptides Is Critical for Symbiosis. PLoS Biol. 2011, 9, e1001169. [Google Scholar] [CrossRef]
- Nicoud, Q.; Barrière, Q.; Busset, N.; Dendene, S.; Travin, D.; Bourge, M.; Le Bars, R.; Boulogne, C.; Lecroël, M.; Jenei, S.; et al. Sinorhizobium meliloti Functions Required for Resistance to Antimicrobial NCR Peptides and Bacteroid Differentiation. mBio 2021, 12, e00895-21. [Google Scholar] [CrossRef]
- Ren, B.; Wang, X.; Duan, J.; Ma, J. Rhizobial tRNA-Derived Small RNAs Are Signal Molecules Regulating Plant Nodulation. Science 2019, 365, 919–922. [Google Scholar] [CrossRef]
- García-Tomsig, N.I.; Robledo, M.; diCenzo, G.C.; Mengoni, A.; Millán, V.; Peregrina, A.; Uceta, A.; Jiménez-Zurdo, J.I. Pervasive RNA Regulation of Metabolism Enhances the Root Colonization Ability of Nitrogen-Fixing Symbiotic α-Rhizobia. mBio 2022, 13, e03576-21. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cañizares, C.; Prell, J.; Pini, F.; Rutten, P.; Kraxner, K.; Wynands, B.; Karunakaran, R.; Poole, P.S. Global Control of Bacterial Nitrogen and Carbon Metabolism by a PTSNtr-Regulated Switch. Proc. Natl. Acad. Sci. USA 2020, 117, 10234–10245. [Google Scholar] [CrossRef]
- Li, Y.Z.; Wang, D.; Feng, X.Y.; Jiao, J.; Chen, W.X.; Tian, C.F. Genetic Analysis Reveals the Essential Role of Nitrogen Phosphotransferase System Components in Sinorhizobium fredii CCBAU 45436 Symbioses with Soybean and Pigeonpea Plants. Appl. Environ. Microbiol. 2016, 82, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- David, M.; Daveran, M.L.; Batut, J.; Dedieu, A.; Domergue, O.; Ghai, J.; Hertig, C.; Boistard, P.; Kahn, D. Cascade Regulation of nif Gene Expression in Rhizobium meliloti. Cell 1988, 54, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, P.A.; Elmerich, C. Involvement of fixLJ in the Regulation of Nitrogen Fixation in Azorhizobium caulinodans. Mol. Microbiol. 1991, 5, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, P.A.; Mandon, K.; Arigoni, F.; Desnoues, N.; Elmerich, C. Regulation of Nitrogen Fixation in Azorhizobium caulinodans: Identification of a fixK-like Gene, a Positive Regulator of nifA. Mol. Microbiol. 1991, 5, 1983–1991. [Google Scholar] [CrossRef]
- Emmerich, R.; Hennecke, H.; Fischer, H.M. Evidence for a Functional Similarity between the Two-Component Regulatory Systems RegSR, ActSR, and RegBA (PrrBA) in α-Proteobacteria. Arch. Microbiol. 2000, 174, 307–313. [Google Scholar] [CrossRef]
- Lindemann, A.; Moser, A.; Pessi, G.; Hauser, F.; Friberg, M.; Hennecke, H.; Fischer, H.M. New Target Genes Controlled by the Bradyrhizobium japonicum Two-Component Regulatory System RegSR. J. Bacteriol. 2007, 189, 8928–8943. [Google Scholar] [CrossRef]
- Anthamatten, D.; Scherb, B.; Hennecke, H. Characterization of a fixLJ-Regulated Bradyrhizobium japonicum Gene Sharing Similarity with the Escherichia coli fnr and Rhizobium meliloti fixK Genes. J. Bacteriol. 1992, 174, 2111–2120. [Google Scholar] [CrossRef]
- Cabrera, J.J.; Jiménez-Leiva, A.; Tomás-Gallardo, L.; Parejo, S.; Casado, S.; Torres, M.J.; Bedmar, E.J.; Delgado, M.J.; Mesa, S. Dissection of FixK2 Protein–DNA Interaction Unveils New Insights into Bradyrhizobium diazoefficiens Lifestyles Control. Environ. Microbiol. 2021, 23, 6194–6209. [Google Scholar] [CrossRef]
- Galinier, A.; Garnerone, A.M.; Reyrat, J.M.; Kahn, D.; Batut, J.; Boistard, P. Phosphorylation of the Rhizobium meliloti FixJ Protein Induces Its Binding to a Compound Regulatory Region at the fixK Promoter. J. Biol. Chem. 1994, 269, 23784–23789. [Google Scholar] [CrossRef]
- Lopez, O.; Morera, C.; Miranda-Rios, J.; Girard, L.; Romero, D.; Soberon, M. Regulation of Gene Expression in Response to Oxygen in Rhizobium etli: Role of FnrN in fixNOQP Expression and in Symbiotic Nitrogen Fixation. J. Bacteriol. 2001, 183, 6999–7006. [Google Scholar] [CrossRef]
- Ledbetter, R.N.; Garcia Costas, A.M.; Lubner, C.E.; Mulder, D.W.; Tokmina-Lukaszewska, M.; Artz, J.H.; Patterson, A.; Magnuson, T.S.; Jay, Z.J.; Duan, H.D.; et al. The Electron Bifurcating FixABCX Protein Complex from Azotobacter vinelandii: Generation of Low-Potential Reducing Equivalents for Nitrogenase Catalysis. Biochemistry 2017, 56, 4177–4190. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Helinski, D.R.; Ditta, G. Overlapping Transcription of the nifA Regulatory Gene in Rhizobium meliloti. Gene 1986, 50, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.; Palacios, J.M.; Imperial, J.; Ruiz-Argüeso, T. Symbiotic Autoregulation of nifA Expression in Rhizobium leguminosarum bv. viciae. J. Bacteriol. 2004, 186, 6586–6594. [Google Scholar] [CrossRef] [PubMed]
- Webb, I.U.C.; Xu, J.; Sanchez-Cañizares, C.; Karunakaran, R.; Ramachandran, V.K.; Rutten, P.J.; East, A.K.; Huang, W.E.; Watmough, N.J.; Poole, P.S. Regulation and Characterization of Mutants of fixABCX in Rhizobium leguminosarum. Mol. Plant-Microbe Interact. 2021, 34, 1167–1180. [Google Scholar] [CrossRef] [PubMed]
- Salazar, E.; Javier Díaz-Mejía, J.; Moreno-Hagelsieb, G.; Martínez-Batallar, G.; Mora, Y.; Mora, J.; Encarnación, S. Characterization of the NifA-RpoN Regulon in Rhizobium etli in Free Life and in Symbiosis with Phaseolus vulgaris. Appl. Environ. Microbiol. 2010, 76, 4510–4520. [Google Scholar] [CrossRef]
- Hauser, F.; Pessi, G.; Friberg, M.; Weber, C.; Rusca, N.; Lindemann, A.; Fischer, H.M.; Hennecke, H. Dissection of the Bradyrhizobium japonicum NifA+σ54 Regulon, and Identification of a Ferredoxin Gene (fdxN) for Symbiotic Nitrogen Fixation. Mol. Genet. Genomics 2007, 278, 255–271. [Google Scholar] [CrossRef]
- Klipp, W.; Reiländer, H.; Schlüter, A.; Krey, R.; Pühler, A. The Rhizobium meliloti fdxN Gene Encoding a Ferredoxin-like Protein Is Necessary for Nitrogen Fixation and Is Cotranscribed with nifA and nifB. Mol. Gen. Genet. 1989, 216, 293–302. [Google Scholar] [CrossRef]
- del Cerro, P.; Rolla-Santos, A.A.P.; Gomes, D.F.; Marks, B.B.; Pérez-Montaño, F.; Rodríguez-Carvajal, M.Á.; Nakatani, A.S.; Gil-Serrano, A.; Megías, M.; Ollero, F.J.; et al. Regulatory nodD1 and nodD2 Genes of Rhizobium tropici Strain CIAT 899 and Their Roles in the Early Stages of Molecular Signaling and Host-Legume Nodulation. BMC Genom. 2015, 16, 251. [Google Scholar] [CrossRef]
- Machado, D.; Pueppke, S.G.; Vinardel, J.M.; Ruiz-Sainz, J.E.; Krishnan, H.B. Expression of nodD1 and nodD2 in Sinorhizobium fredii, a Nitrogen-Fixing Symbiont of Soybean and Other Legumes. Mol. Plant-Microbe Interact. 1998, 11, 375–382. [Google Scholar] [CrossRef]
- Chen, X.C.; Feng, J.; Hou, B.H.; Li, F.Q.; Li, Q.; Hong, G.F. Modulating DNA Bending Affects NodD-Mediated Transcriptional Control in Rhizobium leguminosarum. Nucleic Acids Res. 2005, 33, 2540–2548. [Google Scholar] [CrossRef]
- Peck, M.C.; Fisher, R.F.; Long, S.R. Diverse Flavonoids Stimulate NodD1 Binding to nod Gene Promoters in Sinorhizobium meliloti. J. Bacteriol. 2006, 188, 5417–5427. [Google Scholar] [CrossRef] [PubMed]
- Rossen, L.; Shearman, C.A.; Johnston, A.W.B.; Downie, J.A. The nodD Gene of Rhizobium leguminosarum Is Autoregulatory and in the Presence of Plant Exudate Induces the nodA,B,C Genes. EMBO J. 1985, 4, 3369–3373. [Google Scholar] [CrossRef] [PubMed]
- McIver, J.; Djordjevic, M.A.; Weinman, J.J.; Bender, G.L.; Rolfe, B.G. Extension of Host Range of Rhizobium leguminosarum bv. trifolii Caused by Point Mutations in nodD That Result in Alterations in Regulatory Function and Recognition of Inducer Molecules. Mol. Plant. Microbe. Interact. 1989, 2, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.; Major, A.S.; Sullivan, J.T.; Bourke, S.D.; Kelly, S.J.; Perry, B.J.; Ronson, C.W. Rhizobium leguminosarum bv. trifolii NodD2 Enhances Competitive Nodule Colonization in the Clover-Rhizobium Symbiosis. Appl. Environ. Microbiol. 2020, 86, e01268-20. [Google Scholar] [CrossRef]
- Honma, M.A.; Asomaning, M.; Ausubel, F.M. Rhizobium meliloti nodD Genes Mediate Host-Specific Activation of nodABC. J. Bacteriol. 1990, 172, 901–911. [Google Scholar] [CrossRef]
- Ayala-García, P.; Jiménez-Guerrero, I.; Jacott, C.N.; López-Baena, F.J.; Ollero, F.J.; del Cerro, P.; Pérez-Montaño, F. The Rhizobium tropici CIAT 899 NodD2 Protein Promotes Symbiosis and Extends Rhizobial Nodulation Range by Constitutive Nodulation Factor Synthesis. J. Exp. Bot. 2022, 73, 6931–6941. [Google Scholar] [CrossRef]
- Rodpothong, P.; Sullivan, J.T.; Songsrirote, K.; Sumpton, D.; Cheung, K.W.J.T.; Thomas-Oates, J.; Radutoiu, S.; Stougaard, J.; Ronson, C.W. Nodulation Gene Mutants of Mesorhizobium loti R7A-nodZ and nolL Mutants Have Host-Specific Phenotypes on Lotus spp. Mol. Plant-Microbe Interact. 2009, 22, 1546–1554. [Google Scholar] [CrossRef]
- Kelly, S.; Sullivan, J.T.; Kawaharada, Y.; Radutoiu, S.; Ronson, C.W.; Stougaard, J. Regulation of Nod Factor Biosynthesis by Alternative NodD Proteins at Distinct Stages of Symbiosis Provides Additional Compatibility Scrutiny. Environ. Microbiol. 2018, 20, 97–110. [Google Scholar] [CrossRef]
- Fellay, R.; Hanin, M.; Montorzi, G.; Frey, J.; Freiberg, C.; Golinowski, W.; Staehelin, C.; Broughton, W.J.; Jabbouri, S. nodD2 of Rhizobium sp. NGR234 Is Involved in the Repression of the nodABC Operon. Mol. Microbiol. 1998, 27, 1039–1050. [Google Scholar] [CrossRef]
- Loh, J.; Stacey, G. Nodulation Gene Regulation in Bradyrhizobium japonicum: A Unique Integration of Global Regulatory Circuits. Appl. Environ. Microbiol. 2003, 69, 10–17. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Rodríguez-Navarro, D.-N.; Kawaharada, Y.; Rodríguez-Carvajal, M.A.; Gil-Serrano, A.; Soria-Díaz, M.E.; Pérez-Montaño, F.; Fernández-Perea, J.; Yanbo, N.; Alias-Villegas, C.; et al. Sinorhizobium fredii HH103 nolR and nodD2 Mutants Gain Capacity for Infection Thread Invasion of Lotus japonicus Gifu and Lotus Burttii. Environ. Microbiol. 2019, 21, 1718–1739. [Google Scholar] [CrossRef] [PubMed]
- Fujishige, N.A.; Lum, M.R.; De Hoff, P.L.; Whitelegge, J.P.; Faull, K.F.; Hirsch, A.M. Rhizobium Common nod Genes Are Required for Biofilm Formation. Mol. Microbiol. 2008, 67, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montaño, F.; Jiménez-Guerrero, I.; Del Cerro, P.; Baena-Ropero, I.; López-Baena, F.J.; Ollero, F.J.; Bellogín, R.; Lloret, J.; Espuny, R. The Symbiotic Biofilm of Sinorhizobium fredii SMH12, Necessary for Successful Colonization and Symbiosis of Glycine max cv Osumi, Is Regulated by Quorum Sensing Systems and Inducing Flavonoids via NodD1. PLoS ONE 2014, 9, e0105901. [Google Scholar] [CrossRef] [PubMed]
- Rumbaugh, K.P.; Sauer, K. Biofilm Dispersion. Nat. Rev. Microbiol. 2020, 18, 571–586. [Google Scholar] [CrossRef]
- Crespo-rivas, J.C.; Cuesta-berrio, L.; Ruiz-sainz, J.E. Exopolysaccharide Production by Sinorhizobium fredii HH103 Is Repressed by Genistein in a NodD1-Dependent Manner. PLoS ONE 2016, 11, e0160499. [Google Scholar] [CrossRef]
- Barnett, M.J.; Long, S.R. The Sinorhizobium meliloti SyrM Regulon: Effects on Global Gene Expression Are Mediated by syrA and nodD3. J. Bacteriol. 2015, 197, 1792–1806. [Google Scholar] [CrossRef]
- Cheng, H.P.; Walker, G.C. Succinoglycan Is Required for Initiation and Elongation of Infection Threads during Nodulation of Alfalfa by Rhizobium meliloti. J. Bacteriol. 1998, 180, 5183–5191. [Google Scholar] [CrossRef]
- Rodríguez-Navarro, D.N.; Rodríguez-Carvajal, M.A.; Acosta-Jurado, S.; Soto, M.J.; Margaret, I.; Crespo-Rivas, J.C.; Sanjuan, J.; Temprano, F.; Gil-Serrano, A.; Ruiz-Sainz, J.E.; et al. Structure and Biological Roles of Sinorhizobium fredii HH103 Exopolysaccharide. PLoS ONE 2014, 9, e115391. [Google Scholar] [CrossRef]
- Theunis, M.; Kobayashi, H.; Broughton, W.J.; Prinsen, E. Flavonoids, NodD1, NodD2, and Nod-Box NB15 Modulate Expression of the Y4wEFG Locus That Is Required for Indole-3-Acetic Acid Synthesis in Rhizobium sp. Strain NGR234. Mol. Plant-Microbe Interact. 2004, 17, 1153–1161. [Google Scholar] [CrossRef]
- Deakin, W.J.; Broughton, W.J. Symbiotic Use of Pathogenic Strategies: Rhizobial Protein Secretion Systems. Nat. Rev. Microbiol. 2009, 7, 312–320. [Google Scholar] [CrossRef]
- Barnett, M.J.; Toman, C.J.; Fisher, R.F.; Long, S.R. A Dual-Genome Symbiosis Chip for Coordinate Study of Signal Exchange and Development in a Prokaryote-Host Interaction. Proc. Natl. Acad. Sci. USA 2004, 101, 16636–16641. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Jurado, S.; Alias-Villegas, C.; Navarro-Gómez, P.; Almozara, A.; Rodríguez-Carvajal, M.A.; Medina, C.; Vinardell, J.M. Sinorhizobium fredii HH103 syrM Inactivation Affects the Expression of a Large Number of Genes, Impairs Nodulation with Soybean and Extends the Host-Range to Lotus japonicus. Environ. Microbiol. 2020, 22, 1104–1124. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, J.T.; Long, S.R. A Family of Activator Genes Regulates Expression of Rhizobium meliloti Nodulation Genes. Genetics 1989, 122, 7–18. [Google Scholar] [CrossRef]
- Kiss, E.; Mergaert, P.; Olah, B.; Kereszt, A.; Staehelin, C.; Davies, A.E.; Downie, J.A.; Kondorosi, A.; Kondorosi, E. Conservation of nolR in the Sinorhizobium and Rhizobium Genera of the Rhizobiaceae Family. Mol. Plant-Microbe Interact. 1998, 11, 1186–1195. [Google Scholar] [CrossRef]
- Lee, S.G.; Krishnan, H.B.; Jez, J.M. Structural Basis for Regulation of Rhizobial Nodulation and Symbiosis Gene Expression by the Regulatory Protein NolR. Proc. Natl. Acad. Sci. USA 2014, 111, 6509–6514. [Google Scholar] [CrossRef]
- Cren, M.; Kondorosi, A.; Kondorosi, E. NolR Controls Expression of the Rhizobium meliloti Nodulation Genes Involved in the Core Nod Factor Synthesis. Mol. Microbiol. 1995, 15, 733–747. [Google Scholar] [CrossRef]
- del Cerro, P.; Rolla-Santos, A.A.P.; Valderrama-Fernández, R.; Gil-Serrano, A.; Bellogín, R.A.; Gomes, D.F.; Pérez-Montaño, F.; Megías, M.; Hungría, M.; Ollero, F.J. NrcR, a New Transcriptional Regulator of Rhizobium tropici CIAT 899 Involved in the Legume Root-Nodule Symbiosis. PLoS ONE 2016, 11, e0154029. [Google Scholar] [CrossRef]
- Vinardell, J.M.; Ollero, F.J.; Hidalgo, A.; Lopez-Baena, F.J.; Medina, C.; Ivanov-Vangelov, K.; Parada, M.; Madinabeitia, N.; Espuny Mdel, R.; Bellogin, R.A.; et al. NoIR Regulates Diverse Symbiotic Signals of Sinorhizobium fredii HH103. Mol. Plant-Microbe Interact. 2004, 17, 676–685. [Google Scholar] [CrossRef]
- Göttfert, M.; Grob, P.; Hennecke, H. Proposed Regulatory Pathway Encoded by the nodV and nodW Genes, Determinants of Host Specificity in Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA 1990, 87, 2680–2684. [Google Scholar] [CrossRef]
- Loh, J.; Lohar, D.P.; Andersen, B.; Stacey, G. A Two-Component Regulator Mediates Population-Density-Dependent Expression of the Bradyrhizobium japonicum Nodulation Genes. J. Bacteriol. 2002, 184, 1759–1766. [Google Scholar] [CrossRef]
- Garcia, M.; Dunlap, J.; Loh, J.; Stacey, G. Phenotypic Characterization and Regulation of the nolA Gene of Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 1996, 9, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Kondorosi, E.; Pierre, M.; Cren, M.; Haumann, U.; Buiré, M.; Hoffmann, B.; Schell, J.; Kondorosi, A. Identification of NoIR, a Negative Transacting Factor Controlling the nod Regulon in Rhizobium meliloti. J. Mol. Biol. 1991, 222, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Kondorosi, E.; Gyuris, J.; Schmidt, J.; John, M.; Duda, E.; Hoffmann, B.; Schell, J.; Kondorosi, A. Positive and Negative Control of nod Gene Expression in Rhizobium meliloti Is Required for Optimal Nodulation. EMBO J. 1989, 8, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.; Gonzalez, J.E. Complex Regulation of Symbiotic Functions Is Coordinated by MucR and Quorum Sensing in Sinorhizobium meliloti. J. Bacteriol. 2011, 193, 485–496. [Google Scholar] [CrossRef]
- Bittinger, M.A.; Milner, J.L.; Saville, B.J.; Handelsman, J. rosR, a Determinant of Nodulation Competitiveness in Rhizobium etli. Mol. Plant-Microbe Interact. 1997, 10, 180–186. [Google Scholar] [CrossRef]
- Janczarek, M.; Kutkowska, J.; Piersiak, T.; Skorupska, A. Rhizobium leguminosarum bv. trifolii RosR Is Required for Interaction with Clover, Biofilm Formation and Adaptation to the Environment. BMC Microbiol. 2010, 10, 284. [Google Scholar] [CrossRef]
- Jiao, J.; Wu, L.J.; Zhang, B.; Hu, Y.; Li, Y.; Zhang, X.X.; Guo, H.J.; Liu, L.X.; Chen, W.X.; Zhang, Z.; et al. MucR Is Required for Transcriptional Activation of Conserved Ion Transporters to Support Nitrogen Fixation of Sinorhizobium fredii in Soybean Nodules. Mol. Plant-Microbe Interact. 2016, 29, 352–361. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Alias-Villegas, C.; Navarro-Gomez, P.; Zehner, S.; Del Socorro Murdoch, P.; Rodríguez-Carvajal, M.A.; Soto, M.J.; Ollero, F.J.; Ruiz-Sainz, J.E.; Göttfert, M.; et al. The Sinorhizobium fredii HH103 MucR1 Global Regulator Is Connected with the nod Regulon and Is Required for Efficient Symbiosis with Lotus burttii and Glycine max cv. Williams. Mol. Plant-Microbe Interact. 2016, 29, 700–712. [Google Scholar] [CrossRef]
- Janczarek, M. The Ros/MucR Zinc-Finger Protein Family in Bacteria: Structure and Functions. Int. J. Mol. Sci. 2022, 23, 15536. [Google Scholar] [CrossRef]
- Rachwał, K.; Matczyńska, E.; Janczarek, M. Transcriptome Profiling of a Rhizobium leguminosarum bv. trifolii rosR Mutant Reveals the Role of the Transcriptional Regulator RosR in Motility, Synthesis of Cell-Surface Components, and Other Cellular Processes. BMC Genom. 2015, 16, 1111. [Google Scholar] [CrossRef]
- Barnett, M.J.; Long, S.R. Identification and Characterization of a Gene on Rhizobium meliloti pSymA, syrB, That Negatively Affects syrM Expression. Mol. Plant Microbe Interact. 1997, 10, 550–559. [Google Scholar] [CrossRef]
- Li, M.-L.; Jiao, J.; Zhang, B.; Shi, W.-T.; Yu, W.-H.; Tian, C.-F. Global Transcriptional Repression of Diguanylate Cyclases by MucR1 Is Essential for Sinorhizobium-Soybean Symbiosis. mBio 2021, 12, e01192-21. [Google Scholar] [CrossRef] [PubMed]
- Sauviac, L.; Philippe, H.; Phok, K.; Bruand, C. An Extracytoplasmic Function Sigma Factor Acts as a General Stress Response Regulator in Sinorhizobium meliloti. J. Bacteriol. 2007, 189, 4204–4216. [Google Scholar] [CrossRef] [PubMed]
- Bertram-Drogatz, P.A.; Quester, I.; Becker, A.; Puhler, A. The Sinorhizobium meliloti MucR Protein, Which Is Essential for the Production of High-Molecular-Weight Succinoglycan Exopolysaccharide, Binds to Short DNA Regions Upstream of exoH and exoY. Mol. Gen. Genet. 1998, 257, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Schäper, S.; Wendt, H.; Bamberger, J.; Sieber, V.; Schmid, J.; Becker, A. A Bifunctional UDP-Sugar 4-Epimerase Supports Biosynthesis of Multiple Cell Surface Polysaccharides in Sinorhizobium meliloti. J. Bacteriol. 2019, 201, e00801-18. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Jiao, J.; Tian, C.-F. Adaptive Evolution of Rhizobial Symbiosis beyond Horizontal Gene Transfer: From Genome Innovation to Regulation Reconstruction. Genes 2023, 14, 274. https://doi.org/10.3390/genes14020274
Liu S, Jiao J, Tian C-F. Adaptive Evolution of Rhizobial Symbiosis beyond Horizontal Gene Transfer: From Genome Innovation to Regulation Reconstruction. Genes. 2023; 14(2):274. https://doi.org/10.3390/genes14020274
Chicago/Turabian StyleLiu, Sheng, Jian Jiao, and Chang-Fu Tian. 2023. "Adaptive Evolution of Rhizobial Symbiosis beyond Horizontal Gene Transfer: From Genome Innovation to Regulation Reconstruction" Genes 14, no. 2: 274. https://doi.org/10.3390/genes14020274
APA StyleLiu, S., Jiao, J., & Tian, C.-F. (2023). Adaptive Evolution of Rhizobial Symbiosis beyond Horizontal Gene Transfer: From Genome Innovation to Regulation Reconstruction. Genes, 14(2), 274. https://doi.org/10.3390/genes14020274





