Chromosome-Level Assembly of Flowering Cherry (Prunus campanulata) Provides Insight into Anthocyanin Accumulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Sample
2.2. Genome Size and Heterozygosity Analysis
2.3. Genome Sequencing
2.4. Genome Assembly and Assessments
2.5. Genome Annotation
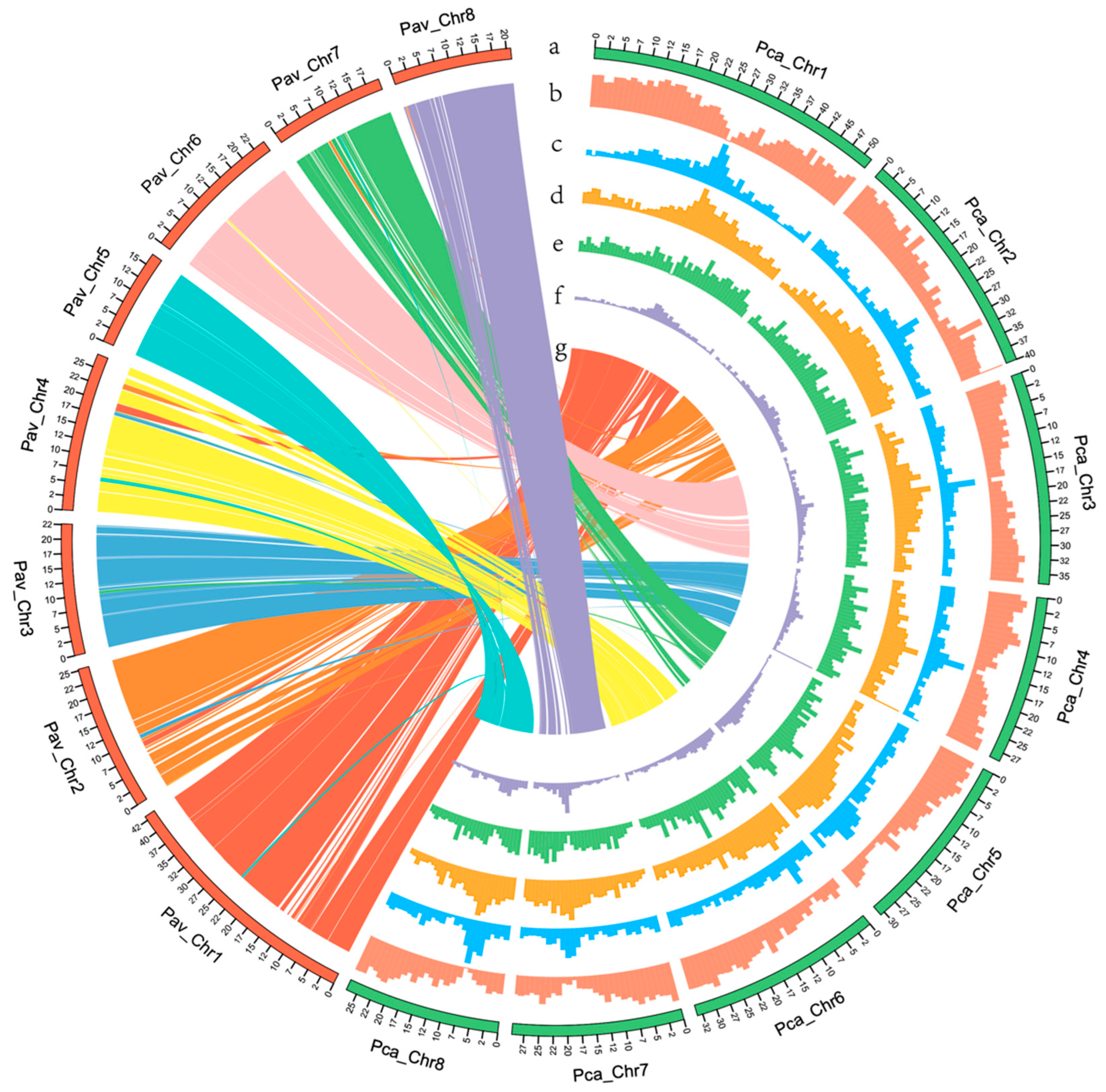
2.6. Synteny Analysis
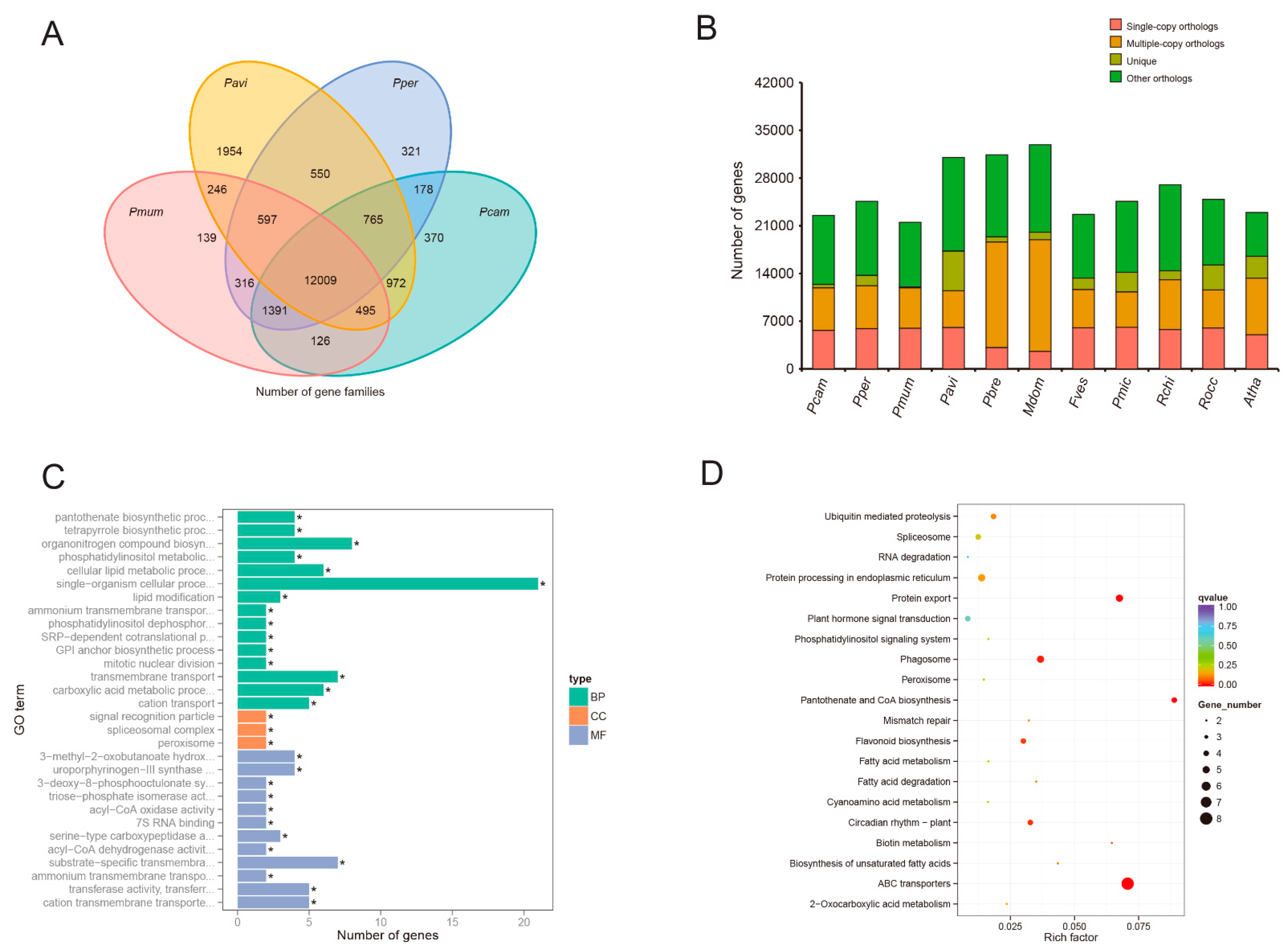
2.7. Gene Family Analyses and Evolution
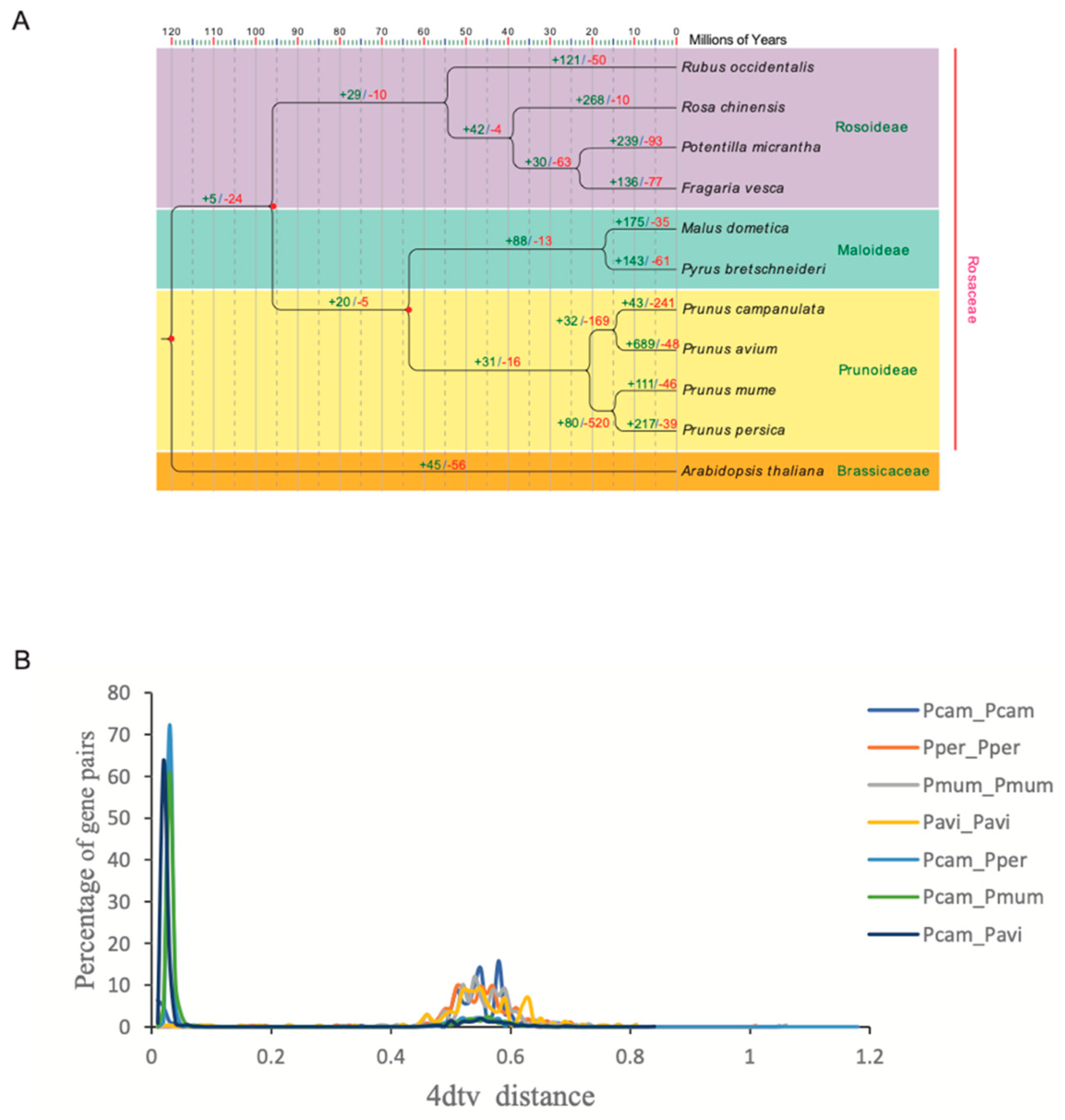
2.8. Phylogenetic Analyses
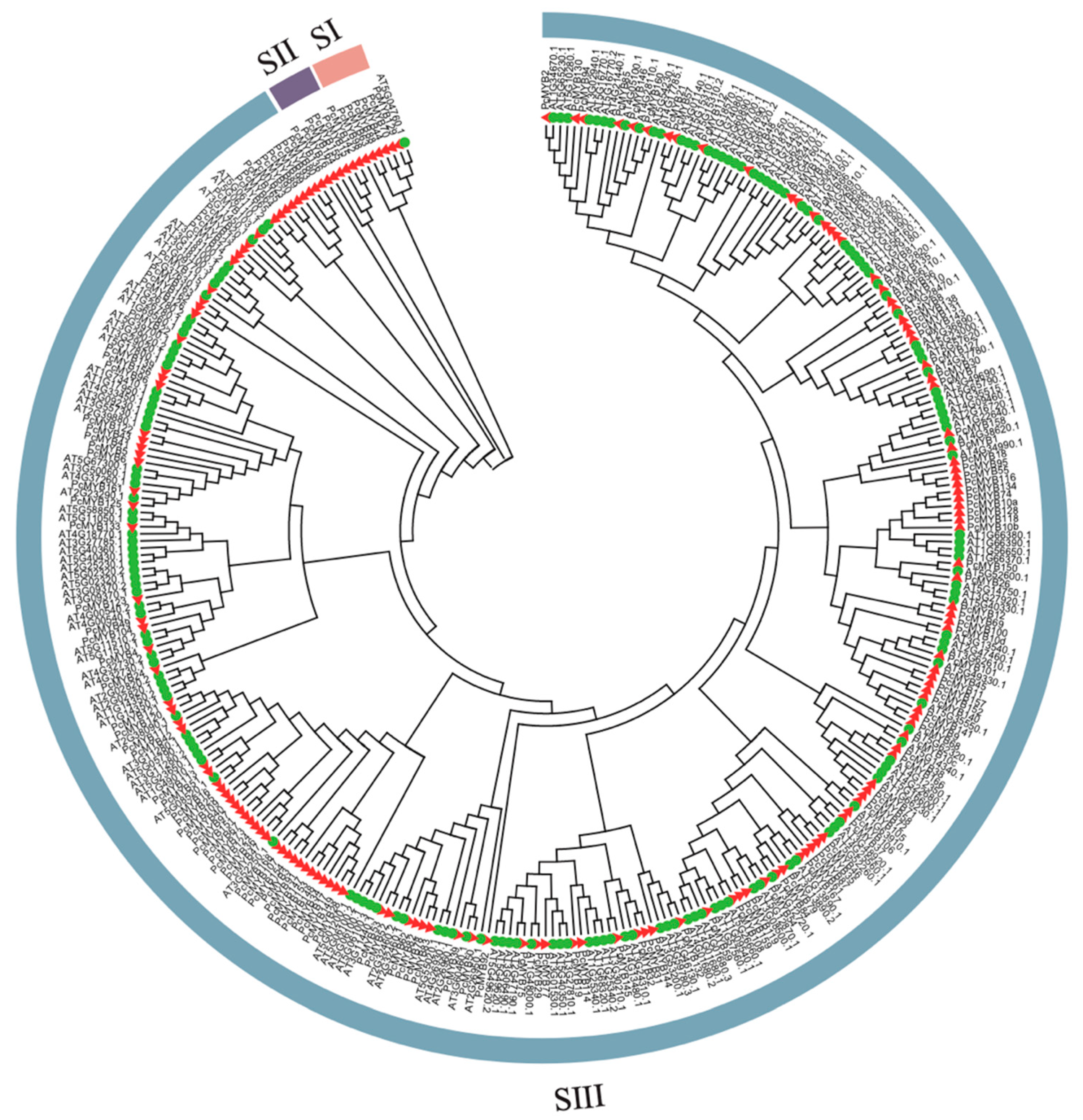
2.9. Identification of MYB Family Genes
2.10. Chromosome Distribution and Phylogenetic Analysis
2.11. Transcriptomic Analysis
3. Results
3.1. Genome Sequencing and Assembly
3.2. Assessment of Genome Quality
3.3. Genome Annotation
3.4. Gene Family Analyses and Evolution
3.5. Identification and Phylogenetic Analysis of MYB Family Genes
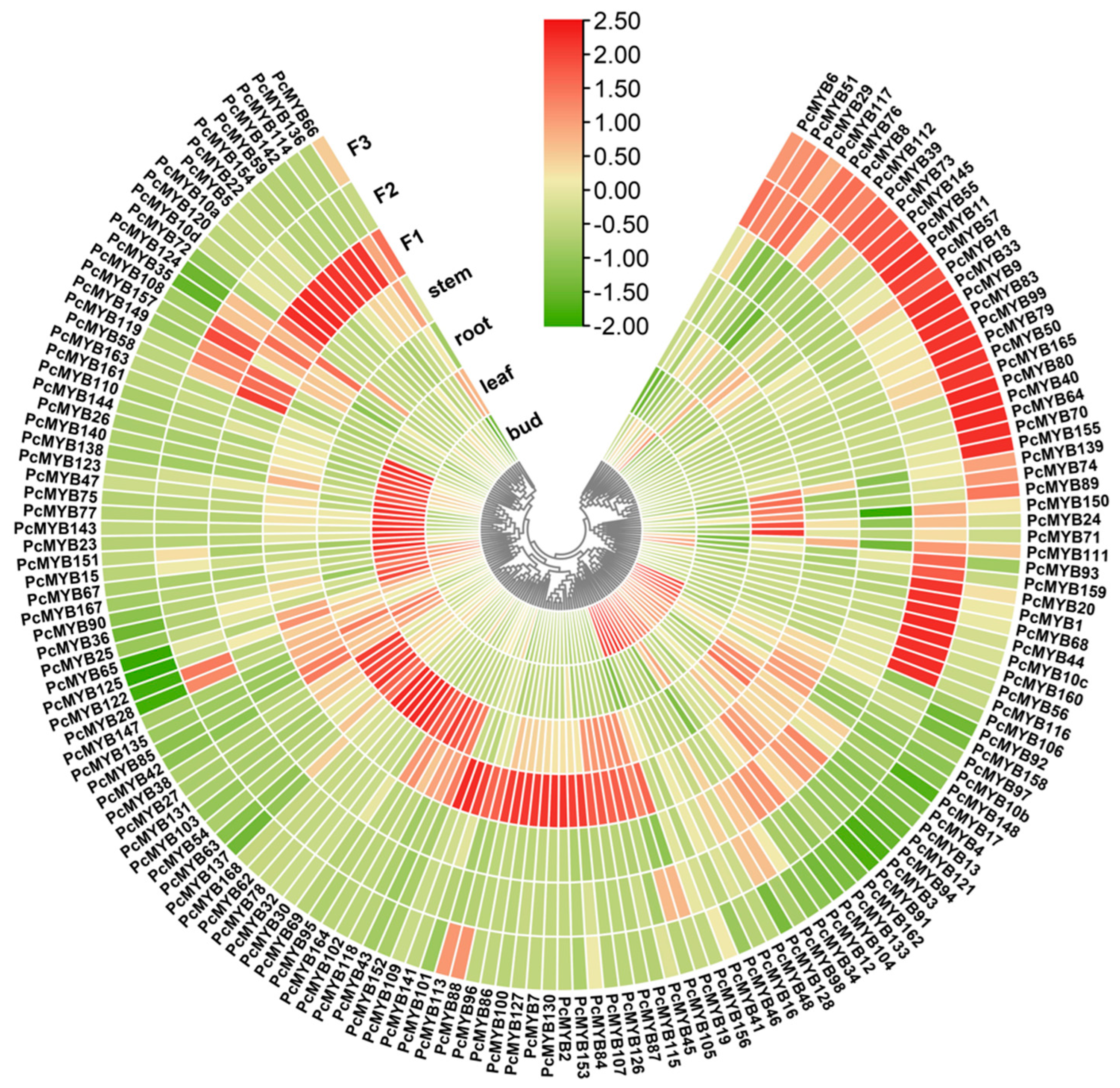
3.6. MYB Gene Expression Profiles in Diverse Tissues
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bortiri, E.; Oh, S.H.; Jiang, J.; Baggett, S.; Granger, A.; Weeks, C.; Buckingham, M.; Potter, D.; Parfitt, D.E. Phylogeny and Systematics of Prunus (Rosaceae) as Determined by Sequence Analysis of ITS and the Chloroplast trnL-trnF Spacer DNA. Syst. Bot. 2001, 26, 797–807. [Google Scholar] [CrossRef]
- Shi, S.; Li, J.; Sun, J.; Yu, J.; Zhou, S. Phylogeny and Classification of Prunus sensu lato (Rosaceae). J. Integr. Plant Biol. 2013, 55, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Matsumoto, A.; Yoshimura, K.; Katsuki, T.; Iwamoto, K.; Kawahara, T.; Mukai, Y.; Tsuda, Y.; Ishio, S.; Nakamura, K.; et al. Origins of Japanese Flowering Cherry (Prunus subgenus Cerasus) Cultivars Revealed Using Nuclear SSR Markers. Tree Genet. Genomes 2014, 10, 477–487. [Google Scholar] [CrossRef]
- Yu, D.J.; Li, C.L. China Flora; Science Press: Beijing, China, 1986; ISBN 13031.3176. [Google Scholar]
- Chien, C.T.; Chen, S.Y.; Yang, J.C. Effect of Stratification and Drying on the Germination and Storage of Prunus campanulata Seeds. Taiwan J. For. Sci. 2002, 17, 413–420. [Google Scholar] [CrossRef]
- Kato, S.; Matsumoto, A.; Yoshimura, K.; Katsuki, T.; Iwamoto, K.; Tsuda, Y.; Ishio, S.; Nakamura, K.; Moriwaki, K.; Shiroishi, T.; et al. Clone Identification in Japanese Flowering Cherry (Prunus Subgenus Cerasus) Cultivars Using Nuclear SSR Markers. Breed. Sci. 2012, 62, 248–255. [Google Scholar] [CrossRef]
- Kanazawa, Y.; Kameyama, Y.; JingXiu, L.; Hamano, C.; Suzuki, K. Genetic Relationship Between Early-Flowering Cherry Cultivars and Regional Populations of Prunus Campanulata. Hortic. Res. Jpn. 2016, 15, 129–138. [Google Scholar] [CrossRef]
- Guo, Y.; Kramer, M.; Pooler, M. Screening Ornamental Cherry (Prunus) Taxa for Resistance to Infection by Blumeriella Jaapii. HortSci. 2018, 53, 200–203. [Google Scholar] [CrossRef]
- Charlesworth, D.; Wright, S.I. Breeding Systems and Genome Evolution. Curr. Opin. Genet. Dev. 2001, 11, 685–690. [Google Scholar] [CrossRef]
- Tao, R.; Iezzoni, A.F. The S-RNase-Based Gametophytic Self-Incompatibility System in Prunus Exhibits Distinct Genetic and Molecular Features. Sci. Hortic. 2010, 124, 423–433. [Google Scholar] [CrossRef]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The Genome Sequence of Sweet Cherry (Prunus avium) for Use in Genomics-Assisted Breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef] [Green Version]
- Baek, S.; Choi, K.; Kim, G.B.; Yu, H.J.; Cho, A.; Jang, H.; Kim, C.; Kim, H.J.; Chang, K.S.; Kim, J.H.; et al. Draft Genome Sequence of Wild Prunus yedoensis Reveals Massive Inter-Specific Hybridization between Sympatric Flowering Cherries. Genome Biol. 2018, 19, 1–17. [Google Scholar] [CrossRef]
- Yi, X.G.; Yu, X.Q.; Chen, J.; Zhang, M.; Liu, S.W.; Zhu, H.; Li, M.; Duan, Y.F.; Chen, L.; Wu, L.; et al. The Genome of Chinese Flowering Cherry (Cerasus serrulata) Provides New Insights into Cerasus Species. Hortic. Res. 2020, 7, 165. [Google Scholar] [CrossRef] [PubMed]
- Wöhner, T.W.; Emeriewen, O.F.; Wittenberg, A.H.J.; Schneiders, H.; Vrijenhoek, I.; Halász, J.; Hrotkó, K.; Hoff, K.J.; Gabriel, L.; Lempe, J.; et al. The Draft Chromosome-Level Genome Assembly of Tetraploid Ground Cherry (Prunus fruticosa Pall.) from Long Reads. Genomics 2021, 113, 4173–4183. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Kingsford, C. A Fast, Lock-Free Approach for Efficient Parallel Counting of Occurrences of k-Mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef]
- Liu, B.H.; Shi, Y.J.; Yuan, J.Y.; Hu, X.S.; Zhang, H.; Li, N.; Li, Z.Y.; Chen, Y.X.; Mu, D.S.; Fan, W. Estimation of Genomic Characteristics by Analyzing K-Mer Frequency in De Novo Genome Projects. arXiv 2020, arXiv:1308.2012. [Google Scholar]
- Chin, C.S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased Diploid Genome Assembly with Single-Molecule Real-Time Sequencing. Nat Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, Finished Microbial Genome Assemblies from Long-Read SMRT Sequencing Data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Adey, A.; Kitzman, J.O.; Burton, J.N.; Daza, R.; Kumar, A.; Christiansen, L.; Ronaghi, M.; Amini, S.; Gunderson, K.L.; Steemers, F.J.; et al. In Vitro, Long-Range Sequence Information for de Novo Genome Assembly via Transposase Contiguity. Genome Res. 2014, 24, 2041–2049. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burton, J.N.; Adey, A.; Patwardhan, R.P.; Qiu, R.; Kitzman, J.O.; Shendure, J. Chromosome-Scale Scaffolding of de Novo Genome Assemblies Based on Chromatin Interactions. Nat. Biotechnol. 2013, 31, 1119–1125. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Parra, G.; Bradnam, K.; Korf, I. CEGMA: A Pipeline to Accurately Annotate Core Genes in Eukaryotic Genomes. Bioinformatics 2007, 23, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Hao, W. LTR_FINDER: An Efficient Tool for the Prediction of Full-Length LTR Retrotransposons. Nucl. Acids Res. 2007, 35, W265–W268. [Google Scholar] [CrossRef]
- Price, A.L.; Jones, N.C.; Pevzner, P.A. De Novo Identification of Repeat Families in Large Genomes. Bioinformatics 2005, 21, i351–i358. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R. RepeatModeler-1.0.11. Inst. Sys-Tems Biol. Available online: http://www.repeatmasker.org/RepeatModeler (accessed on 9 April 2019).
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a Database of Eukaryotic Repetitive Elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open 4.0. Inst. Sys-Tems Biol. Available online: http://www.repeatmasker.org/RepeatMasker (accessed on 3 September 2019).
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using Repeat Masker to Identify Repetitive Elements in Genomic Sequences. Curr. Protoc. Bioinforma. 2009, 25, 4.10.1–4.10.14. [Google Scholar] [CrossRef]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. Augustus: Ab Initio Prediction of Alternative Transcripts. Nucl. Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef]
- Burge, C. Identification of Genes in Human Genomic DNA. Ph.D. Thesis, Stanford University, Stanford, CA, USA, 1997. [Google Scholar]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two Open Source Ab Initio Eukaryotic Gene-Finders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haussler, D.K.D.; Eeckman, M.G.R.F.H. A Generalized Hidden Markov Model for the Recognition of Human Genes in DNA. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology, St. Louis, MI, USA, 12-15 June 1996; pp. 134–142. [Google Scholar] [PubMed]
- Korf, I. Gene Finding in Novel Genomes. BMC Bioinform. 2004, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome Initiative Analysis of the Genome Sequence of the Flowering Plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [CrossRef] [PubMed]
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F.; et al. The High-Quality Draft Genome of Peach (Prunus persica) Identifies Unique Patterns of Genetic Diversity, Domestication and Genome Evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, W.; Sun, L.; Zhao, F.; Huang, B.; Yang, W.; Tao, Y.; Wang, J.; Yuan, Z.; Fan, G.; et al. The Genome of Prunus mume. Nat. Commun. 2012, 3, 1318. [Google Scholar] [CrossRef] [PubMed]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The Genome of the Domesticated Apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The Genome of the Pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef]
- Shulaev, V.; Sargent, D.J.; Crowhurst, R.N.; Mockler, T.C.; Folkerts, O.; Delcher, A.L.; Jaiswal, P.; Mockaitis, K.; Liston, A.; Mane, S.P.; et al. The Genome of Woodland Strawberry (Fragaria vesca). Nat. Genet. 2011, 43, 109–116. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 1–9. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate Alignment of Transcriptomes in the Presence of Insertions, Deletions and Gene Fusions. Genome Biol. 2013, 14, 1–13. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated Eukaryotic Gene Structure Annotation Using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. UniProt: A Worldwide Hub of Protein Knowledge. Nucl. Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
- Zdobnov, E.M.; Apweiler, R. InterProScan–an Integration Platform for the Signature-Recognition Methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucl. Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. TRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucl. Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-Fold Faster RNA Homology Searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucl. Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Delcher, A.L.; Salzberg, S.L.; Phillippy, A.M. Using MUMmer to Identify Similar Regions in Large Sequence Sets. Curr. Protoc. Bioinforma. 2003, 1, 10.3.1–10.3.18. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Itai, A.; Isobe, S. Genome Sequencing and Analysis of Two Early-Flowering Cherry (Cerasus × Kanzakura) Varieties, ‘Kawazu-Zakura’ and ‘Atami-Zakura. DNA Res. 2021, 28, dsab026. [Google Scholar] [CrossRef]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Buti, M.; Moretto, M.; Barghini, E.; Mascagni, F.; Natali, L.; Brilli, M.; Lomsadze, A.; Sonego, P.; Giongo, L.; Alonge, M. The Genome Sequence and Transcriptome of Potentilla micrantha and Their Comparison to Fragaria vesca (the Woodland Strawberry). GigaScience 2018, 7, giy010. [Google Scholar] [CrossRef] [PubMed]
- Hibrand Saint-Oyant, L.; Ruttink, T.; Hamama, L.; Kirov, I.; Lakhwani, D.; Zhou, N.-N.; Bourke, P.M.; Daccord, N.; Leus, L.; Schulz, D. A High-Quality Genome Sequence of Rosa chinensis to Elucidate Ornamental Traits. Nat. Plants 2018, 4, 473–484. [Google Scholar] [CrossRef]
- VanBuren, R.; Wai, C.M.; Colle, M.; Wang, J.; Sullivan, S.; Bushakra, J.M.; Liachko, I.; Vining, K.J.; Dossett, M.; Finn, C.E. A Near Complete, Chromosome-Scale Assembly of the Black Raspberry (Rubus occidentalis) Genome. Gigascience 2018, 7, giy094. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucl. Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [Green Version]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A Computational Tool for the Study of Gene Family Evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Rambaut, A. Se-Al Sequence Alignment Editor, Version 2.0 A11. Available online: http://tree.bio.ed.ac.uk/software/seal (accessed on 2 February 2021).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC Results into a Web-Based, Interactive, and Extensible FastQ Quality Control Tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Nat. Preced. 2010, 11, 1. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Tuan, P.A.; Bai, S.; Yaegaki, H.; Tamura, T.; Hihara, S.; Moriguchi, T.; Oda, K. The Crucial Role of PpMYB10.1 in Anthocyanin Accumulation in Peach and Relationships between Its Allelic Type and Skin Color Phenotype. BMC Plant Biol. 2015, 15, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Hao, R.; Xu, Z.; Yang, W.; Wang, J.; Cheng, T.; Pan, H.; Zhang, Q. Isolation and Functional Characterization of a R2R3-MYB Regulator of Prunus mume Anthocyanin Biosynthetic Pathway. Plant Cell Tissue Organ Cult. PCTOC 2017, 131, 417–429. [Google Scholar] [CrossRef]
- Lee, S.; Wen, J. A Phylogenetic Analysis of Prunus and the Amygdaloideae (Rosaceae) Using ITS Sequences of Nuclear Ribosomal DNA. Am. J. Bot. 2001, 88, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Chin, S.W.; Shaw, J.; Haberle, R.; Wen, J.; Potter, D. Diversification of Almonds, Peaches, Plums and Cherries—Molecular Systematics and Biogeographic History of Prunus (Rosaceae). Mol. Phylogenet. Evol. 2014, 76, 34–48. [Google Scholar] [CrossRef]
- Chin, S.; Lutz, S.; Wen, J.; Potter, D.A. The Bitter and the Sweet: Inference of Homology and Evolution of Leaf Glands in Prunus (Rosaceae) through Anatomy, Micromorphology, and Ancestral–Character State Reconstruction. Int. J. Plant Sci. 2013, 174, 27–46. [Google Scholar] [CrossRef]
- Bradshaw, A.D. Evolutionary Significance of Phenotypic Plasticity in Plants. Adv. Genet. 1965, 13, 115–155. [Google Scholar] [CrossRef]
- Sun, Y.; Shang, L.; Zhu, Q.H.; Fan, L.; Guo, L. Twenty Years of Plant Genome Sequencing: Achievements and Challenges. Trends Plant Sci. 2022, 27, 391–401. [Google Scholar] [CrossRef]
- Pryszcz, L.P.; Gabaldón, T. Redundans: An Assembly Pipeline for Highly Heterozygous Genomes. Nucl. Acids Res. 2016, 44, e113. [Google Scholar] [CrossRef]
- Michael, T.P.; VanBuren, R. Building Near-Complete Plant Genomes. Curr. Opin. Plant Biol. 2020, 54, 26–33. [Google Scholar] [CrossRef]
- Wang, J.; Liu, W.; Zhu, D.; Hong, P.; Zhang, S.; Xiao, S.; Tan, Y.; Chen, X.; Xu, L.; Zong, X.; et al. Chromosome-Scale Genome Assembly of Sweet Cherry (Prunus avium L.) cv. Tieton Obtained Using Long-Read and Hi-C Sequencing. Hortic. Res. 2020, 7, 1–11. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Wang, H. The Contributions of Transposable Elements to the Structure, Function, and Evolution of Plant Genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef]
- Lee, S.-I.; Kim, N.-S. Transposable Elements and Genome Size Variations in Plants. Genom. Inform. 2014, 12, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Tenaillon, M.I.; Hufford, M.B.; Gaut, B.S.; Ross-Ibarra, J. Genome Size and Transposable Element Content as Determined by High-Throughput Sequencing in Maize and Zea Luxurians. Genome Biol. Evol. 2011, 3, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.F.; Wen, C.H.; Wang, C.T.; Chu, F.H. Transcriptome and Flower Genes Analysis of Prunus campanulata Maxim. J. Hortic. Sci. Biotechnol. 2020, 95, 44–52. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB Gene Family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, N.A.; Glover, B.J. MYB–BHLH–WD40 Protein Complex and the Evolution of Cellular Diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis Transcription Factor MYB77 Modulates Auxin Signal Transduction. Plant Cell 2007, 19, 2440–2453. [Google Scholar] [CrossRef]
- Xiao, R.; Zhang, C.; Guo, X.; Li, H.; Lu, H. MYB Transcription Factors and Its Regulation in Secondary Cell Wall Formation and Lignin Biosynthesis during Xylem Development. Int. J. Mol. Sci. 2021, 22, 3560. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB Transcription Factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB Transcription Factors in Transcriptional Regulation of Anthocyanin Biosynthesis in Horticultural Plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef]
- Cooke, J.E.; Eriksson, M.E.; Junttila, O. The Dynamic Nature of Bud Dormancy in Trees: Environmental Control and Molecular Mechanisms. Plant Cell Environ. 2012, 35, 1707–1728. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The R2R3 MYB Transcription Factor PavMYB10.1 Involves in Anthocyanin Biosynthesis and Determines Fruit Skin Colour in Sweet Cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef] [PubMed]
- Starkevič, P.; Paukštytė, J.; Kazanavičiūtė, V.; Denkovskienė, E.; Stanys, V.; Bendokas, V.; Siksnianas, T.; Razanskiene, A.; Razanskas, R. Expression and Anthocyanin Biosynthesis-Modulating Potential of Sweet Cherry (Prunus avium L.) MYB10 and BHLH Genes. PLoS ONE 2015, 10, e0126991. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Zhang, F.; Tao, Y.; Song, S.; Fang, J. Reference Gene Selection for Quantitative Real-Time PCR Normalization in Different Cherry Genotypes, Developmental Stages and Organs. Sci. Hortic. 2015, 181, 182–188. [Google Scholar] [CrossRef]
- Zhou, H.; Peng, Q.; Zhao, J.; Owiti, A.; Ren, F.; Liao, L.; Wang, L.; Deng, X.; Jiang, Q.; Han, Y. Multiple R2R3-MYB Transcription Factors Involved in the Regulation of Anthocyanin Accumulation in Peach Flower. Front. Plant Sci. 2016, 7, 1557. [Google Scholar] [CrossRef] [PubMed]
- Rahim, M.; Busatto, N.; Trainotti, L. Regulation of Anthocyanin Biosynthesis in Peach Fruits. Planta 2014, 240, 913–929. [Google Scholar] [CrossRef]
- Xi, W.; Feng, J.; Liu, Y.; Zhang, S.; Zhao, G. The R2R3-MYB Transcription Factor PaMYB10 is Involved in Anthocyanin Biosynthesis in Apricots and Determines Red Blushed Skin. BMC Plant Biol. 2019, 19, 1–14. [Google Scholar] [CrossRef]
- Fiol, A.; García-Gómez, B.E.; Jurado-Ruiz, F.; Alexiou, K.; Howad, W.; Aranzana, M.J. Characterization of Japanese Plum (Prunus salicina) PsMYB10 Alleles Reveals Structural Variation and Polymorphisms Correlating with Fruit Skin Color. Front. Plant Sci. 2021, 12, 655267. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, H.; Lin-Wang, K.; Vimolmangkang, S.; Espley, R.V.; Wang, L.; Allan, A.C.; Han, Y. Transcriptome analysis and transient transformation suggest an ancient duplicated MYB transcription factor as a candidate gene for leaf red coloration in peach. BMC Plant Biol. 2014, 14, 1–13. [Google Scholar] [CrossRef] [Green Version]



| Parameter | P. campanulata | P. avium “Satonishiki” | P. yedoensis | C. serrulata | P. fruticosa |
|---|---|---|---|---|---|
| Sequencing platform | PacBio, Illumina, 10× Genomics, Hi-C | Illumina | PacBio, Illumina | Nanopore, Illumina, Hi-C | Nanopore, Hi-C |
| Heterozygosity percentage | 0.54% | — | — | 1.67% | — |
| N50 length (contigs) (Mb) | 2.02 | — | 0.133 | 1.56 | 0.533 |
| N50 length (scaffolds) (Mb) | 33.44 | 0.219 | 0.198 | 31.12 | 43.82 |
| No. of protein-coding genes | 28,319 | 43,349 | 41,294 | 29,094 | 58,880 |
| Chromosome level/Total size of assigned scaffolds (Mb) | Yes/283.62 | Yes/191.70 | No | Yes/252.25 | Yes/— |
| No. of scaffolds | 370 | 10, 148 | 3, 185 | 67 | — |
| Total length of assembled scaffolds (Mb) | 300.48 | 272.36 | 323.78 | 265.4 | — |
| No. of contigs | 687 | 51,877 | 4292 | 182 | 1275 |
| Total size of assembled contigs (Mb) | 299.15 | — | 318.74 | 263.16 | 366.5 |
| Busco assessment | C:96.6% [S:92.8%,D:3.8%], F:0.8%,M:2.6%,n:1440 | C:96% [S:78.3%,D:17.7%], F:1.8%,M:2.2%,n:956 | N/A | C:94.7% [S:83.8%,D:10.9%], F:1.86%,M:3.47%,n:1614 | C:96.4% [S:94.1%,D:2.3%], F:1.3%,M:2.3%,n:1614 |
| Type | De novo + Repbase 1 | TE Proteins 2 | Combined TEs 3 | |||
|---|---|---|---|---|---|---|
| Length(bp) | % in Genome | Length(bp) | % in Genome | Length(bp) | % in Genome | |
| DNA | 46,577,999 | 15.52 | 8,655,257 | 2.88 | 48,545,281 | 16.18 |
| LINE 4 | 5,778,324 | 1.93 | 2,431,202 | 0.81 | 7,025,267 | 2.34 |
| SINE 5 | 14,544 | 0 | 0 | 0 | 14,544 | 0 |
| LTR 6 | 92,381,320 | 30.79 | 22,553,446 | 7.52 | 93,460,219 | 31.15 |
| Unknown 7 | 16,239,753 | 5.41 | 0 | 0 | 16,239,753 | 5.41 |
| Total | 157,524,477 | 52.49 | 33,566,604 | 11.19 | 160,006,931 | 53.32 |
| Method | Gene Set | Number | Average Transcript Length (bp) | Average CDS Length (bp) | Average Exons Per Gene | Average Exon Length (bp) | Average Intron Length (bp) |
|---|---|---|---|---|---|---|---|
| De novo | Augustus | 21,837 | 2714.22 | 1201.26 | 4.98 | 241.3 | 380.31 |
| GlimmerHMM | 35,772 | 6382.00 | 796.8 | 3.41 | 234.01 | 2322.32 | |
| SNAP | 22,256 | 3128.31 | 721.53 | 4.17 | 172.89 | 758.43 | |
| Geneid | 31,597 | 3987.08 | 914.41 | 4.49 | 203.78 | 881.1 | |
| Genscan | 20,493 | 8917.35 | 1370.01 | 6.42 | 213.25 | 1391.37 | |
| Homolog | Arabidopsis_thaliana | 26,286 | 2299.76 | 1045.17 | 3.79 | 276.13 | 450.47 |
| Fragaria_vesca | 21,218 | 3082.18 | 1347.82 | 4.46 | 302.29 | 501.45 | |
| Malus_domestica | 25,135 | 2556.85 | 1263.62 | 4.2 | 300.84 | 404.10 | |
| Prunus_avium | 27,359 | 2019.39 | 891.10 | 3.68 | 242.34 | 421.46 | |
| Prunus_mume | 24,960 | 2764.26 | 1273 | 4.5 | 283.05 | 426.38 | |
| Prunus_persica | 29,626 | 2176.78 | 1082.37 | 4.01 | 269.93 | 363.61 | |
| Pyrus_bretschneideri | 21,837 | 2893.41 | 1292.81 | 4.42 | 292.21 | 467.42 | |
| RNAseq | PASA | 70,332 | 2899.39 | 1019.99 | 4.79 | 212.94 | 495.89 |
| Cufflinks | 47,625 | 5826.71 | 2195.12 | 6.17 | 355.93 | 702.80 | |
| EVM | 30,644 | 3031.10 | 1161.66 | 4.53 | 256.69 | 530.25 | |
| Pasa-update | 30,431 | 3022.99 | 1181.12 | 4.56 | 258.99 | 517.31 | |
| Final set | 28,291 | 3135.45 | 1226.50 | 4.75 | 258.41 | 509.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, D.; Li, X.; Li, Y.; Zhou, S.; Zhou, Q.; Liu, X.; Shen, X. Chromosome-Level Assembly of Flowering Cherry (Prunus campanulata) Provides Insight into Anthocyanin Accumulation. Genes 2023, 14, 389. https://doi.org/10.3390/genes14020389
Jiang D, Li X, Li Y, Zhou S, Zhou Q, Liu X, Shen X. Chromosome-Level Assembly of Flowering Cherry (Prunus campanulata) Provides Insight into Anthocyanin Accumulation. Genes. 2023; 14(2):389. https://doi.org/10.3390/genes14020389
Chicago/Turabian StyleJiang, Dongyue, Xiangkong Li, Yingang Li, Shiliang Zhou, Qi Zhou, Xinhong Liu, and Xin Shen. 2023. "Chromosome-Level Assembly of Flowering Cherry (Prunus campanulata) Provides Insight into Anthocyanin Accumulation" Genes 14, no. 2: 389. https://doi.org/10.3390/genes14020389
APA StyleJiang, D., Li, X., Li, Y., Zhou, S., Zhou, Q., Liu, X., & Shen, X. (2023). Chromosome-Level Assembly of Flowering Cherry (Prunus campanulata) Provides Insight into Anthocyanin Accumulation. Genes, 14(2), 389. https://doi.org/10.3390/genes14020389





