Identification and Functional Characterization of WRKY, PHD and MYB Three Salt Stress Responsive Gene Families in Mungbean (Vigna radiata L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Three Gene Family Members in Mungbean
2.2. Evolutionary Tree, Conserved Motifs, Gene Structure, and Promoter Analysis
2.3. Chromosomal Position, Gene Duplication, and Collinearity Analysis of Genes in the Same Gene Family
2.4. Plant Material Treatment and Gene EXPRESSION Pattern Analysis
2.5. qRT-PCR Analysis of Candidate Genes of Three Gene Families under Salt Stress
2.6. Protein Interaction Network Analysis of Differentially Expressed Genes
3. Result
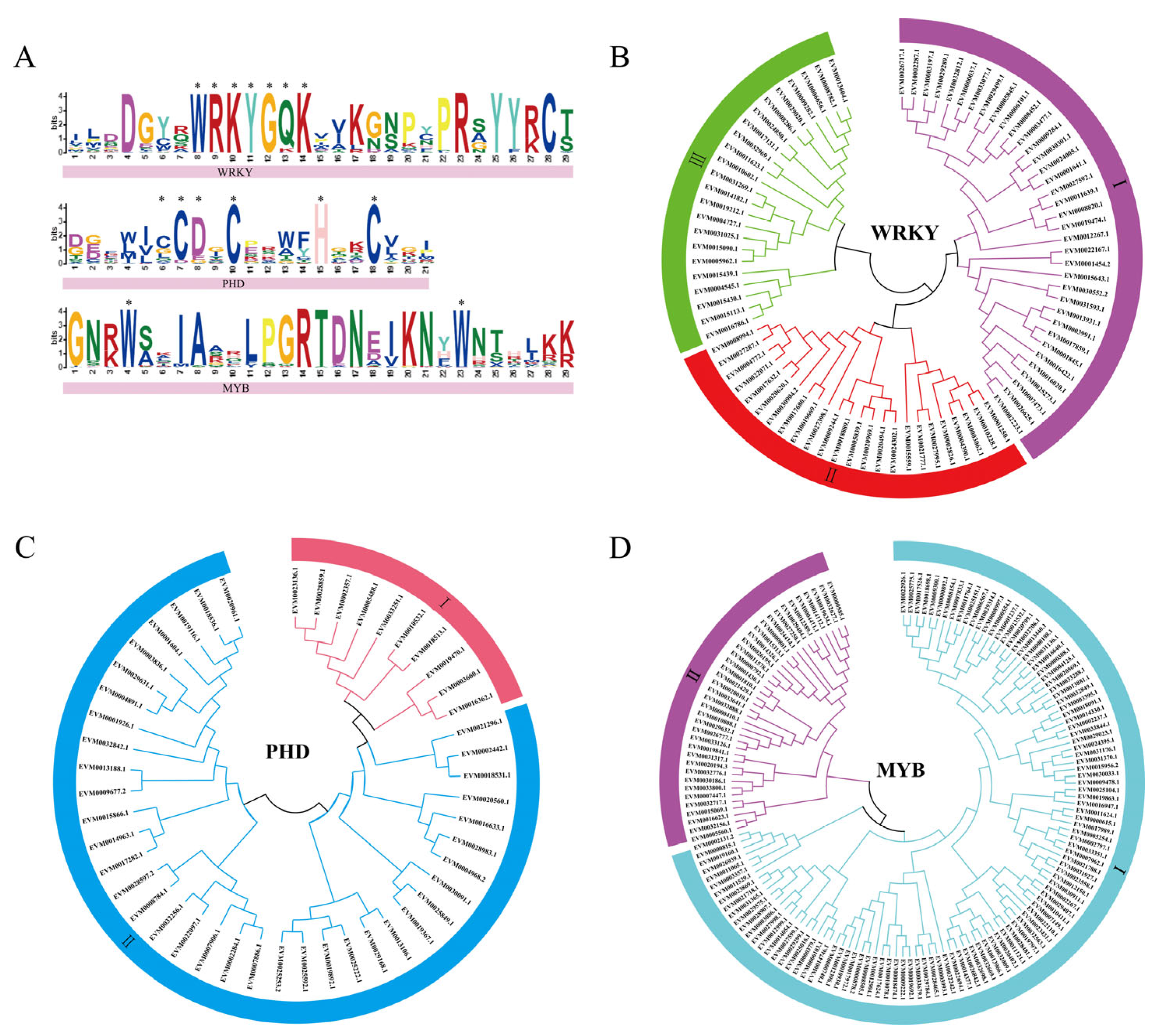
3.1. Mungbean WRKY, PHD, and MYB Three Gene Family Members
3.2. Phylogenetic Tree and Sequence Structure Analysis of Three Gene Families in Mungbean
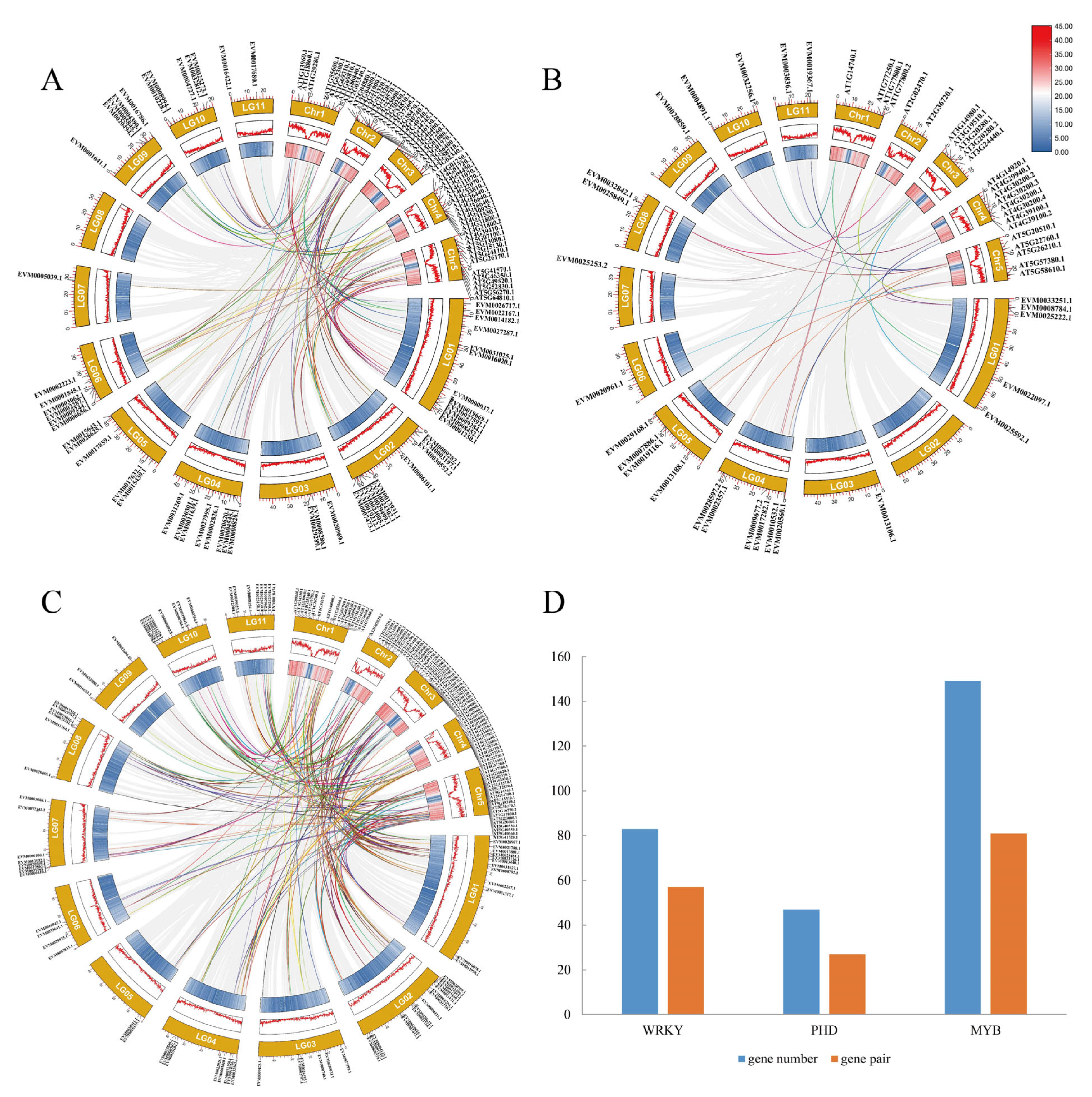
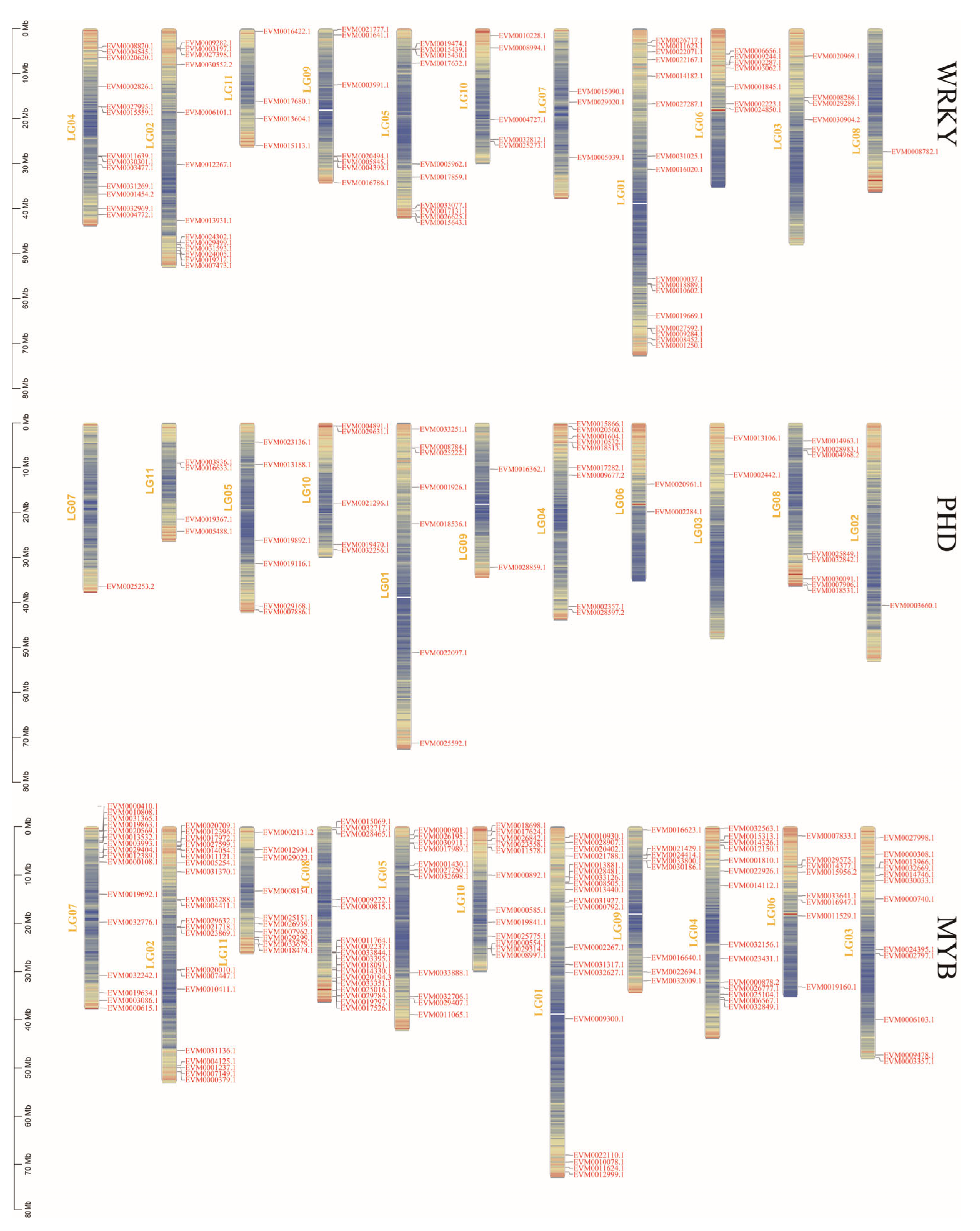
3.3. Collinearity Analysis and Chromosome Mapping of the Three Gene Families in Mungbeans
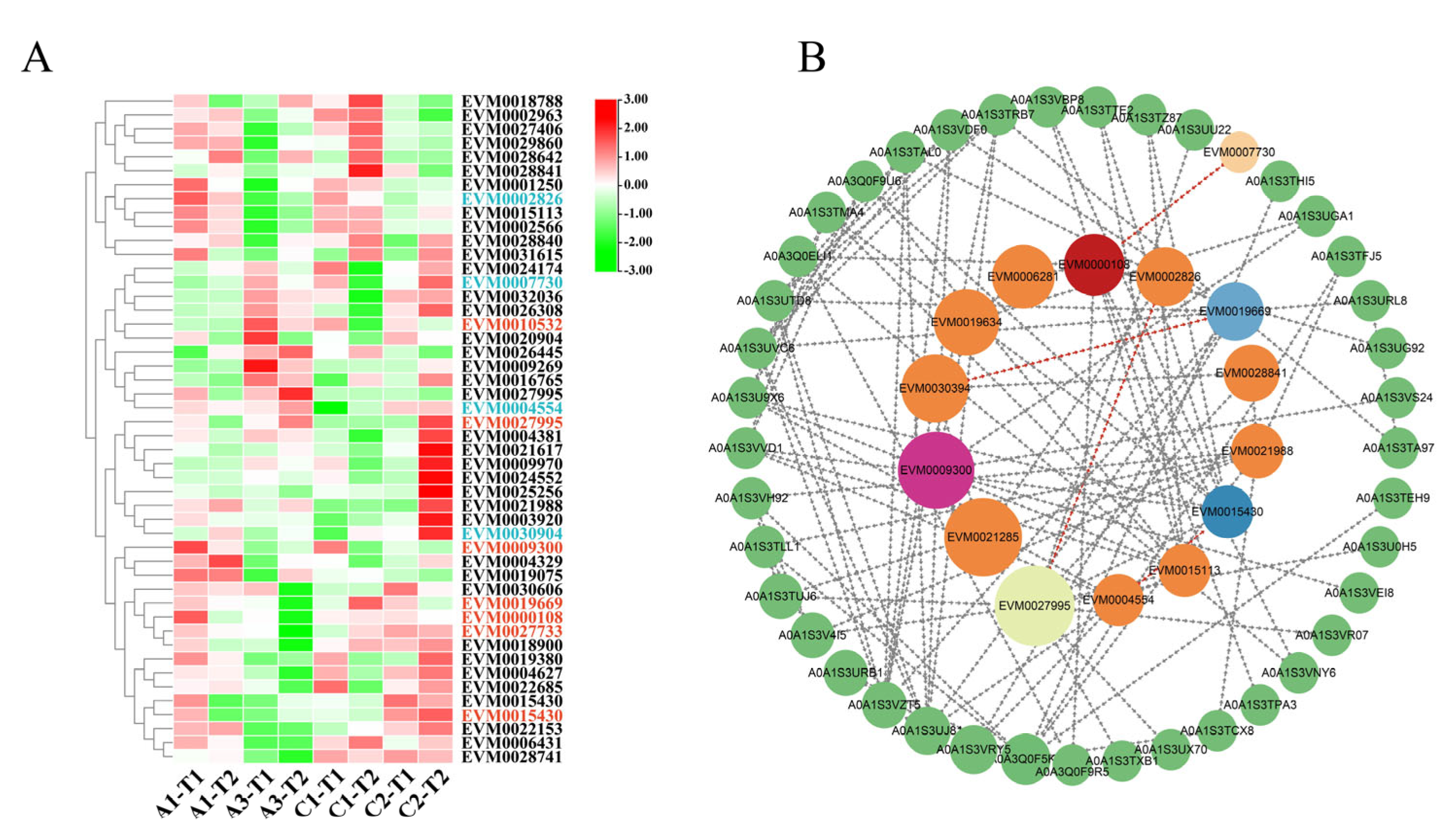
3.4. Gene Expression and Protein Interaction Analysis of VrWRKY, VrPHD, and VrMYB Differentially Expressed Genes
4. Discussion
4.1. Identification of New Members of VrWRKYs, VrPHDs, and VrMYBs in Mungbean
4.2. Evolutionary Relationships of WRKY, PHD, MYB Genes in Mungbean
4.3. Differential Expression Genes and Protein Interactions of WRKY, PHD, and MYB in Mungbeans under Salt Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kang, Y.J.; Kim, S.K.; Kim, M.Y.; Lestari, P.; Kim, K.H.; Ha, B.K.; Jun, T.H.; Hwang, W.J.; Lee, T.; Lee, J.; et al. Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 2014, 5, 5443. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.L.; Vastolo, V.; Ciccarelli, M.; Albano, L.; Macchia, P.E.; Ungaro, P. Dietary polyphenols and chromatin remodeling. Crit. Rev. Food Sci. Nutr. 2017, 57, 2589–2599. [Google Scholar] [CrossRef]
- Boamah, D.; Lin, T.; Poppinga, F.A.; Basu, S.; Rahman, S.; Essel, F.; Chakravarty, S. Characteristics of a PHD Finger Subtype. Biochemistry 2018, 57, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.Y.; Wang, Y.X.; You, C.J. Structural and functional characteristics of plant PHD domain-containing proteins. Yi Chuan 2021, 43, 323–339. [Google Scholar] [CrossRef]
- Tameling, W.I.; Elzinga, S.D.; Darmin, P.S.; Vossen, J.H.; Takken, F.L.; Haring, M.A.; Cornelissen, B.J. The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 2002, 14, 2929–2939. [Google Scholar] [CrossRef]
- Guo, Y.H.; Yu, Y.P.; Wang, D.; Wu, C.A.; Yang, G.D.; Huang, J.G.; Zheng, C.C. GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 2009, 183, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.K.; Pandit, E.; Nayak, D.K.; Behera, L.; Mohapatra, T. Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol. 2019, 19, 352. [Google Scholar] [CrossRef]
- Reboledo, G.; Agorio, A.; Ponce De Leon, I. Moss transcription factors regulating development and defense responses to stress. J. Exp. Bot. 2022, 73, 4546–4561. [Google Scholar] [CrossRef]
- Bo, C.; Cai, R.; Fang, X.; Wu, H.; Ma, Z.; Yuan, H.; Cheng, B.; Fan, J.; Ma, Q. Transcription factor ZmWRKY20 interacts with ZmWRKY115 to repress expression of ZmbZIP111 for salt tolerance in maize. Plant J. 2022, 111, 1660–13675. [Google Scholar] [CrossRef]
- Wu, Y.; Wen, J.; Xia, Y.; Zhang, L.; Du, H. Evolution and functional diversification of R2R3-MYB transcription factors in plants. Hortic. Res. 2022, 9, uhac058. [Google Scholar] [CrossRef]
- Aasland, R.; Gibson, T.J.; Stewart, A.F. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995, 20, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Kachirskaia, I.; Walter, K.L.; Kuo, J.H.; Lake, A.; Davrazou, F.; Chan, S.M.; Martin, D.G.; Fingerman, I.M.; Briggs, S.D.; et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 2007, 282, 2450–2455. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, M.; Oelmuller, R. WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY Transcription Factor Family in Model Plants and Crops. Crit. Rev. Plant Sci. 2018, 36, 311–335. [Google Scholar] [CrossRef]
- Klempnauer, K.H.; Gonda, T.J.; Bishop, J.M. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: The architecture of a transduced oncogene. Cell 1982, 31, 453–463. [Google Scholar] [CrossRef]
- Lipsick, J.S. One billion years of Myb. Oncogene 1996, 13, 223–235. [Google Scholar]
- Rosinski, J.A.; Atchley, W.R. Molecular evolution of the Myb family of transcription factors: Evidence for polyphyletic origin. J. Mol. Evol. 1998, 46, 74–83. [Google Scholar] [CrossRef]
- Kranz, H.; Scholz, K.; Weisshaar, B. c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J. 2000, 21, 231–235. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.K.; Rao, G.Y. Insights into the Diversification and Evolution of R2R3-MYB Transcription Factors in Plants. Plant Physiol. 2020, 183, 637–655. [Google Scholar] [CrossRef]
- Prouse, M.B.; Campbell, M.M. The interaction between MYB proteins and their target DNA binding sites. Biochim. Biophys. Acta 2012, 1819, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999, 41, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Gomez, J.; Talle, B.; Wilson, Z.A. Increased expression of the MALE STERILITY1 transcription factor gene results in temperature-sensitive male sterility in barley. J. Exp. Bot. 2020, 71, 6328–6339. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, H.; Wang, Y.; Li, F.; Xiang, Y. Genome-wide identification of PHD-finger genes and expression pattern analysis under various treatments in moso bamboo (Phyllostachys edulis). Plant Physiol. Biochem. 2018, 123, 378–391. [Google Scholar] [CrossRef]
- Tanaka, N.; Uraguchi, S.; Kajikawa, M.; Saito, A.; Ohmori, Y.; Fujiwara, T. A rice PHD-finger protein OsTITANIA, is a growth regulator that functions through elevating expression of transporter genes for multiple metals. Plant J. 2018, 96, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Kim, J.; Bordiya, Y.; Qiao, H.; Sung, S. Abscisic acid negatively regulates the Polycomb-mediated H3K27me3 through the PHD-finger protein, VIL1. New Phytol. 2022, 235, 1057–1069. [Google Scholar] [CrossRef]
- Han, Z.; Wang, J.; Wang, X.; Zhang, X.; Cheng, Y.; Cai, Z.; Nian, H.; Ma, Q. GmWRKY21, a Soybean WRKY Transcription Factor Gene, Enhances the Tolerance to Aluminum Stress in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 833326. [Google Scholar] [CrossRef]
- Hung, F.Y.; Shih, Y.H.; Lin, P.Y.; Feng, Y.R.; Li, C.; Wu, K. WRKY63 transcriptional activation of COOLAIR and COLDAIR regulates vernalization-induced flowering. Plant Physiol. 2022, 190, 532–547. [Google Scholar] [CrossRef]
- Wang, Z.; Gao, M.; Li, Y.; Zhang, J.; Su, H.; Cao, M.; Liu, Z.; Zhang, X.; Zhao, B.; Guo, Y.D.; et al. SlWRKY37 positively regulates jasmonic acid- and dark-induced leaf senescence in tomato. J. Exp. Bot. 2022, 73, 6207–6225. [Google Scholar] [CrossRef]
- Jiang, H.; Zhou, L.J.; Gao, H.N.; Wang, X.F.; Li, Z.W.; Li, Y.Y. The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple. Plant Physiol. 2022, 189, 2044–2060. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, B.; Chen, Z.; Wu, M.; Chao, D.; Wei, Q.; Xin, Y.; Li, L.; Ming, Z.; Xia, J. Three OsMYB36 members redundantly regulate Casparian strip formation at the root endodermis. Plant Cell 2022, 34, 2948–2968. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Shi, Y.; Wang, R.; Feng, Y.; Wang, L.; Zhang, H.; Shi, X.; Jing, G.; Deng, P.; Song, T.; et al. The transcription factor OsMYBc and an E3 ligase regulate expression of a K+ transporter during salt stress. Plant Physiol. 2020, 190, 843–859. [Google Scholar] [CrossRef]
- Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Seki, M.; Miura, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol. Biol. 2000, 42, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sola, M.A.; Arbona, V.; Gomez-Cadenas, A.; Rodriguez-Concepcion, M.; Rodriguez-Villalon, A. A root specific induction of carotenoid biosynthesis contributes to ABA production upon salt stress in arabidopsis. PLoS ONE 2014, 9, e90765. [Google Scholar] [CrossRef] [PubMed]
- Barrero, J.M.; Rodriguez, P.L.; Quesada, V.; Piqueras, P.; Ponce, M.R.; Micol, J.L. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 2006, 29, 2000–2008. [Google Scholar] [CrossRef]
- Endo, A.; Sawada, Y.; Takahashi, H.; Okamoto, M.; Ikegami, K.; Koiwai, H.; Seo, M.; Toyomasu, T.; Mitsuhashi, W.; Shinozaki, K.; et al. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 2008, 147, 1984–1993. [Google Scholar] [CrossRef]
- Venema, K.; Quintero, F.J.; Pardo, J.M.; Donaire, J.P. The arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J. Biol. Chem. 2002, 277, 2413–2418. [Google Scholar] [CrossRef]
- Leidi, E.O.; Barragan, V.; Rubio, L.; El-Hamdaoui, A.; Ruiz, M.T.; Cubero, B.; Fernandez, J.A.; Bressan, R.A.; Hasegawa, P.M.; Quintero, F.J.; et al. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 2010, 61, 495–506. [Google Scholar] [CrossRef]
- Galvez, F.J.; Baghour, M.; Hao, G.; Cagnac, O.; Rodriguez-Rosales, M.P.; Venema, K. Expression of LeNHX isoforms in response to salt stress in salt sensitive and salt tolerant tomato species. Plant Physiol. Biochem. 2012, 51, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Bassil, E.; Ohto, M.A.; Esumi, T.; Tajima, H.; Zhu, Z.; Cagnac, O.; Belmonte, M.; Peleg, Z.; Yamaguchi, T.; Blumwald, E. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 2011, 23, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Li, Z.; Wang, Y.; Wang, J.; Xiao, M.; Liu, H.; Quan, R.; Zhang, H.; Huang, R.; Zhu, L.; et al. Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance. Plant Biotechnol. J. 2022, 20, 468–484. [Google Scholar] [CrossRef] [PubMed]
- Hoang, X.L.T.; Nhi, D.N.H.; Thu, N.B.A.; Thao, N.P.; Tran, L.P. Transcription Factors and Their Roles in Signal Transduction in Plants under Abiotic Stresses. Curr. Genom. 2017, 18, 483–497. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, J.; Li, Z.; Qiao, J.; Quan, R.; Wang, J.; Huang, R.; Qin, H. SALT AND ABA RESPONSE ERF1 improves seed germination and salt tolerance by repressing ABA signaling in rice. Plant Physiol. 2022, 189, 1110–1127. [Google Scholar] [CrossRef]
- Duan, L.; Dietrich, D.; Ng, C.H.; Chan, P.M.; Bhalerao, R.; Bennett, M.J.; Dinneny, J.R. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 2013, 25, 324–341. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Hoefsloot, H.C.; Mol, S.; Feron, R.; de Boer, G.J.; Haring, M.A.; Testerink, C. Capturing Arabidopsis root architecture dynamics with ROOT-FIT reveals diversity in responses to salinity. Plant Physiol. 2014, 166, 1387–1402. [Google Scholar] [CrossRef]
- Kawa, D.; Julkowska, M.M.; Sommerfeld, H.M.; Ter Horst, A.; Haring, M.A.; Testerink, C. Phosphate-Dependent Root System Architecture Responses to Salt Stress. Plant Physiol. 2016, 172, 690–706. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Koevoets, I.T.; Mol, S.; Hoefsloot, H.; Feron, R.; Tester, M.A.; Keurentjes, J.J.B.; Korte, A.; Haring, M.A.; de Boer, G.J.; et al. Genetic Components of Root Architecture Remodeling in Response to Salt Stress. Plant Cell 2017, 29, 3198–3213. [Google Scholar] [CrossRef]
- Yan, Q.; Wang, Q.; Xuzhen, C.; Wang, L.; Somta, P.; Xue, C.; Chen, J.; Wu, R.; Lin, Y.; Yuan, X.; et al. High-quality genome assembly, annotation and evolutionary analysis of the mungbean (Vigna radiata) genome. Authorea 2020. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic. Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic. Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, K.; Sheng, S.; Wang, M.; Hua, P.; Wang, Y.; Chen, P.; Wang, K.; Zhao, M.; Wang, Y.; et al. Transcriptome analysis of MYB transcription factors family and PgMYB genes involved in salt stress resistance in Panax ginseng. BMC Plant Biol. 2022, 22, 479. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic. Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Li, Y.; Qin, P.; Sun, A.; Xiao, W.; Chen, F.; He, Y.; Yu, K.; Li, Y.; Zhang, M.; Guo, X. Genome-wide identification, new classification, expression analysis and screening of drought & heat resistance related candidates in the RING zinc finger gene family of bread wheat (Triticum aestivum L.). BMC Genom. 2022, 23, 696. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Liu, J.; Xue, C.; Li, Y.; Yan, Q.; Chen, J.; Wu, R.; Zhang, X.; Chen, X.; Yuan, X. Genetic analysis and identification of VrFRO8, a salt tolerance-related gene in mungbean. Gene 2022, 836, 146658. [Google Scholar] [CrossRef]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef]
- Liu, J.; Lin, Y.; Chen, J.; Xue, C.; Wu, R.; Yan, Q.; Chen, X.; Yuan, X. Identification and Clarification of VrCYCA1: A Key Genic Male Sterility-Related Gene in Mungbean by Multi-Omics Analysis. Agriculture 2022, 12, 686. [Google Scholar] [CrossRef]
- Lin, Y.; Laosatit, K.; Chen, J.; Yuan, X.; Wu, R.; Amkul, K.; Chen, X.; Somta, P. Mapping and Functional Characterization of Stigma Exposed 1, a DUF1005 Gene Controlling Petal and Stigma Cells in Mungbean (Vigna radiata). Front. Plant Sci. 2020, 11, 575922. [Google Scholar] [CrossRef]
- Liu, J.; Lin, Y.; Chen, J.; Xue, C.; Wu, R.; Yan, Q.; Chen, X.; Yuan, X. Genome-wide association studies provide genetic insights into natural variation of seed-size-related traits in mungbean. Front. Plant Sci. 2022, 13, 997988. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Zhang, Y.Q.; Tao, J.J.; Chen, H.W.; Li, Q.T.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, J.S.; Chen, S.Y. The Alfin-like homeodomain finger protein AL5 suppresses multiple negative factors to confer abiotic stress tolerance in Arabidopsis. Plant J. 2015, 81, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Wang, J.; Gao, C.; Jin, C.; Li, D.; Peng, D.; Du, G.; Li, Y.; Chen, M. Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 2018, 268, 47–53. [Google Scholar] [CrossRef]
- Loreti, E.; Betti, F.; Ladera-Carmona, M.J.; Fontana, F.; Novi, G.; Valeri, M.C.; Perata, P. Argonaute1 and argonaute4 Regulate Gene Expression and Hypoxia Tolerance. Plant Physiol. 2020, 182, 287–300. [Google Scholar] [CrossRef]
- Jiang, S.C.; Mei, C.; Liang, S.; Yu, Y.T.; Lu, K.; Wu, Z.; Wang, X.F.; Zhang, D.P. Crucial roles of the pentatricopeptide repeat protein SOAR1 in Arabidopsis response to drought, salt and cold stresses. Plant Mol. Biol. 2015, 88, 369–385. [Google Scholar] [CrossRef]
- Huang, Z.; Song, L.; Xiao, Y.; Zhong, X.; Wang, J.; Xu, W.; Jiang, C.Z. Overexpression of Myrothamnus flabellifolia MfWRKY41 confers drought and salinity tolerance by enhancing root system and antioxidation ability in Arabidopsis. Front Plant Sci. 2022, 13, 967352. [Google Scholar] [CrossRef]
- Wei, Y.L.; Jin, J.P.; Liang, D.; Gao, J.; Li, J.; Xie, Q.; Lu, C.Q.; Yang, F.X.; Zhu, G.F. Genome-wide identification of Cymbidium sinense WRKY gene family and the importance of its Group III members in response to abiotic stress. Front. Plant Sci. 2022, 13, 969010. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zheng, L.; Qanmber, G.; Guo, M.; Wang, Z.; Yang, Z. Response of phytohormone mediated plant homeodomain (PHD) family to abiotic stress in upland cotton (Gossypium hirsutum spp.). BMC Plant Biol. 2021, 21, 13. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Qiu, G. Cloning and activity analysis of the promoter of nucleotide exchange factor gene ZjFes1 from the seagrasses Zostera japonica. Sci. Rep. 2020, 10, 17291. [Google Scholar] [CrossRef]
- Liu, H.; Cui, P.; Zhang, B.; Zhu, J.; Liu, C.; Li, Q. Binding of the transcription factor MYC2-like to the ABRE of the OsCYP2 promoter enhances salt tolerance in Oryza sativa. PLoS ONE 2022, 17, e0276075. [Google Scholar] [CrossRef]
- Ai, G.; Zhang, D.; Huang, R.; Zhang, S.; Li, W.; Ahiakpa, J.K.; Zhang, J. Genome-Wide Identification and Molecular Characterization of the Growth-Regulating Factors-Interacting Factor Gene Family in Tomato. Genes 2020, 11, 1435. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Zhang, X.; Li, Z.; Zhao, Y.; Lohaus, R.; Chang, X.; Dong, W.; Ho, S.Y.W.; Liu, X.; et al. The water lily genome and the early evolution of flowering plants. Nature 2020, 577, 79–84. [Google Scholar] [CrossRef]
- Wang, H.; Yin, X.; Du, D.; Liang, Z.; Han, Z.; Nian, H.; Ma, Q. GsMYB7 encoding a R2R3-type MYB transcription factor enhances the tolerance to aluminum stress in soybean (Glycine max L.). BMC Genom. 2022, 23, 529. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, Y.; Chi, Y.; Fan, B.; Chen, Z. Characterization of Soybean WRKY Gene Family and Identification of Soybean WRKY Genes that Promote Resistance to Soybean Cyst Nematode. Sci. Rep. 2017, 7, 17804. [Google Scholar] [CrossRef]
- Hou, J.; Fan, W.; Ma, R.; Li, B.; Yuan, Z.; Huang, W.; Wu, Y.; Hu, Q.; Lin, C.; Zhao, X.; et al. MALE STERILITY 3 encodes a plant homeodomain-finger protein for male fertility in soybean. J. Integr. Plant Biol. 2022, 64, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Higashi, K.; Ishiga, Y.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana. Mol. Genet Genom. 2008, 279, 303–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene ID | Gene ID | Ka | Ks | Ka/Ks | Gene ID | Gene ID | Ka | Ks | Ka/Ks |
|---|---|---|---|---|---|---|---|---|---|
| EVM0031994.1 | EVM0021777.1 | 0.377257 | 1.355966 | 0.27822 | EVM0019212.1 | EVM0004727.1 | 0.494769 | 1.555822 | 0.318012 |
| EVM0001250.1 | EVM0010228.1 | 0.318741 | 0.71844 | 0.443657 | EVM0016020.1 | EVM0025273.1 | 0.210695 | 0.922716 | 0.228343 |
| EVM0027287.1 | EVM0008994.1 | 0.127341 | 1.112803 | 0.114433 | EVM0007473.1 | EVM0025273.1 | 0.323461 | 1.935039 | 0.16716 |
| EVM0008820.1 | EVM0019474.1 | 0.272679 | 0.805485 | 0.338528 | EVM0027592.1 | EVM0011639.1 | 0.288415 | 1.085786 | 0.265628 |
| EVM0021285.1 | EVM0020969.1 | 0.499739 | 2.310074 | 0.21633 | EVM0024302.1 | EVM0020494.1 | 0.2786 | 1.016181 | 0.274163 |
| EVM0020620.1 | EVM0017632.1 | 0.162551 | 0.814435 | 0.199587 | EVM0014182.1 | EVM0031025.1 | 0.495803 | 2.475382 | 0.200294 |
| EVM0026717.1 | EVM0002287.1 | 0.24299 | 0.971014 | 0.250244 | EVM0009284.1 | EVM0030301.1 | 0.13935 | 0.797171 | 0.174806 |
| EVM0002826.1 | EVM0003062.1 | 0.465142 | 5.29056 | 0.087919 | EVM0029499.1 | EVM0005845.1 | 0.174814 | 0.905242 | 0.193113 |
| EVM0031593.1 | EVM0026645.1 | 0.657167 | 1.357884 | 0.483964 | EVM0027995.1 | EVM0000159.1 | 0.515184 | 0.848937 | 0.606858 |
| EVM0017859.1 | EVM0001845.1 | 0.158668 | 0.775746 | 0.204536 | EVM0020969.1 | EVM0005039.1 | 0.337596 | 1.225598 | 0.275454 |
| EVM0011623.1 | EVM0008286.1 | 0.457181 | 2.158146 | 0.21184 | EVM0003062.1 | EVM0004390.1 | 0.276607 | 0.929997 | 0.297428 |
| EVM0009282.1 | EVM0008286.1 | 0.326785 | 0.903277 | 0.361778 | EVM0001250.1 | EVM0004390.1 | 0.504291 | 2.157071 | 0.233785 |
| EVM0003197.1 | EVM0029289.1 | 0.181362 | 0.521552 | 0.347734 | EVM0008452.1 | EVM0003477.1 | 0.297527 | 1.291118 | 0.230441 |
| EVM0019669.1 | EVM0017680.1 | 0.424596 | 1.498066 | 0.28343 | EVM0004545.1 | EVM0016786.1 | 0.362343 | 1.866473 | 0.194132 |
| EVM0026625.1 | EVM0002223.1 | 0.141808 | 0.598734 | 0.236846 | EVM0014182.1 | EVM0031269.1 | 0.23133 | 0.976648 | 0.236861 |
| EVM0017131.1 | EVM0024850.1 | 0.235397 | 0.771602 | 0.305076 | EVM0031025.1 | EVM0031269.1 | 0.499192 | 1.702519 | 0.293208 |
| EVM0006656.1 | EVM0013604.1 | 0.343103 | 2.769751 | 0.123875 | EVM0011623.1 | EVM0032969.1 | 0.257186 | 0.927936 | 0.277159 |
| EVM0014182.1 | EVM0004727.1 | 0.460905 | 4.070824 | 0.113222 | EVM0022071.1 | EVM0004772.1 | 0.176798 | 0.992875 | 0.178067 |
| EVM0031025.1 | EVM0004727.1 | 0.297638 | 0.753707 | 0.394898 | EVM0021285.1 | EVM0024302.1 | 0.524252 | 1.414758 | 0.37056 |
| EVM0031269.1 | EVM0004727.1 | 0.432249 | 1.959744 | 0.220564 | EVM0014182.1 | EVM0019212.1 | 0.442443 | 2.983577 | 0.148293 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Liu, J.; Xue, C.; Lin, Y.; Yan, Q.; Chen, J.; Wu, R.; Chen, X.; Yuan, X. Identification and Functional Characterization of WRKY, PHD and MYB Three Salt Stress Responsive Gene Families in Mungbean (Vigna radiata L.). Genes 2023, 14, 463. https://doi.org/10.3390/genes14020463
Li S, Liu J, Xue C, Lin Y, Yan Q, Chen J, Wu R, Chen X, Yuan X. Identification and Functional Characterization of WRKY, PHD and MYB Three Salt Stress Responsive Gene Families in Mungbean (Vigna radiata L.). Genes. 2023; 14(2):463. https://doi.org/10.3390/genes14020463
Chicago/Turabian StyleLi, Shicong, Jinyang Liu, Chenchen Xue, Yun Lin, Qiang Yan, Jingbin Chen, Ranran Wu, Xin Chen, and Xingxing Yuan. 2023. "Identification and Functional Characterization of WRKY, PHD and MYB Three Salt Stress Responsive Gene Families in Mungbean (Vigna radiata L.)" Genes 14, no. 2: 463. https://doi.org/10.3390/genes14020463





