Abstract
Ophiocordyceps lanpingensis (O. lanpingensis) belongs to the genus Ophiocordyceps, which is often found in Yunnan Province, China. This species is pharmacologically important for the treatment of renal disorders induced by oxidative stress and an inadequate immune response. In the present study, the mitogenome of O. lanpingensis was determined to be a circular molecule 117,560 bp in length, and to have 31% G + C content and 69% A + T content. This mitogenome comprised 82% of the whole genome that codes for significant genes. The protein-coding regions of the O. lanpingensis mitogenome, containing 24 protein-coding genes, were associated with respiratory chain complexes, such as 3 ATP-synthase complex F0 subunits (atp6, atp8, and atp9), 2 complex IV subunits/cytochrome c oxidases (cox2 and cox3), 1 complex III subunit (cob), 4 electron transport complex I subunits/NADH dehydrogenase complex subunits (nad1, nad4, nad5, and nad6), 2 ribosomal RNAs (rns, rnl), and 11 hypothetical/predicted proteins, i.e., orf609, orf495, orf815, orf47, orf150, orf147, orf292, orf127, orf349, orf452, and orf100. It was noted that all genes were positioned on the same strand. Further, 13 mitochondrial genes with respiratory chain complexes, which presented maximum similarity with other fungal species of Ophiocordyceps, were investigated. O. lanpingensis was compared with previously sequenced species within Ophiocordycepitaceae. Comparative analysis indicated that O. lanpingensis was more closely related to O. sinensis, which is one of the most remarkable and expensive herbs due to its limited availability and the fact that it is difficult to culture. Therefore, O. lanpingensis is an important medicinal resource that can be effectively used for medicinal purposes. More extensive metabolomics research is recommended for O. lanpingensis.
1. Introduction
Mitochondria establish a fully developed eukaryotic cellular system and perform the most important functions in cellular pathways, such as respiration and energy production [1]. A feature that differentiates mitochondria (mt) from the nucleus is the presence of numerous mitochondrial genomes (mitogenomes) that encrypt messages for the coding of a variety of genes to perform different functions [2]. Due to its high copy number and rapidly conserved inherited genetic functions, as well as its fast evolution, mitochondrial DNA (mt-DNA) has been extensively acknowledged as a molecular marker present in the evolutionary trees of a large number of fungi and mushrooms [3]. Through the study of ascomycetes, researchers have discovered that mt-DNAs are usually circular and significantly encoded by the same DNA strand [4].
Fungi are a large group of eukaryotic entities that are classified as microorganisms (for example, yeasts, molds, and mushrooms) and are ubiquitously scattered across the globe [5]. In this study, the modern taxonomy of Cordyceps senu lato was tabulated, including three families of Cordyceps s. l. and their genera, which consist of a number of species. All the species in this study belong to the Ophiocordycipitaceae family, which consists of 477 species [6]. The study of medicinal mushrooms and fungal species is highly established in China. Some fungal species are considered traditional Chinese herbs. As the knowledge of Cordyceps has increased and its important secondary metabolites have been screened [7], it has begun to be investigated for its medicinal potential in relation to diabetes, thrombosis, cancer, pathogenic infections, blood pressure management, asthma, and lung and kidney diseases, as well as for its ability to enhance immunity and metabolic activities [8].
O. sinensis, previously known as C. sinensis, is a well-known Chinese caterpillar fungus [9]. Considered an important medicinal fungus, it is widely accepted as being a traditional Chinese herbal medicine and is considered a significant remedy for renal diseases, as well as bronchitis, pneumonia, cough, and asthma [10]. The laboratory investigation of O. sinensis has proved its medicinal status, as it may be beneficial for the treatment of several kidney diseases, although its actual mechanism of action is not clearly understood [11]. O. sinensis may work as a modulator of the immune response, and may manage oxidative stress, as well as other renal functions [12].
Due to its high significance and extensive medicinal use, the wild resources of O. sinensis are frequently diminishing. It requires large-scale production and is artificially cultured, making it difficult to fulfill the medicinal demand for it [13]. O. lanpingensis has been identified as an important species of the genus Ophiocordyceps; as it is closely related to O. sinensis, they might share similar properties [14]. O. lanpingensis is an outdated therapeutic option for the handling of renal and urinary abnormalities. However, it has been noted that the chemical compositions of O. lanpingensis and O. sinensis are both unique, but still seem quite similar to one another [12]. Moreover, O. lanpingensis’ easy artificial culture procedure means it may be an alternative to O. sinensis that could potentially fulfill current medicinal needs [14]. O. lanpingensis can help heal pathological kidney injuries and diminish unusual intensities of the kidney index, creatine kinase, serum creatinine, blood urea nitrogen, and malondialdehyde, as well as increase the cell-mediated immune response and the intensities of immunoglobulin G, glutathione, and superoxide dismutase. O. lanpingensis at a high dose has presented more significant results in model animals with acute renal failure than those treated with a low dose of O. lanpingensis. O. lanpingensis may improve the abnormalities of glycerol-prompted acute kidney injury/acute renal failure in model animals by suppressing oxidative stress and improving the cell-mediated immune response [12].
First, this study was systematically designed. Then, the identification and isolation of O. lanpingensis from Yunnan Province, China, were performed, followed by the genome sequencing, assembly, and annotation of its mitogenome. Its gene contents were calculated, its tRNA structures were identified, and its genome organization was studied. Moreover, base pair compositions, calculations of the mitogenome size, the identification of protein-coding genes (PCGs), gene arrangements, and information on amino acids associated with the tRNA gene of O. lanpingensis and other Ophiocordyceps species were also analyzed. Phylogenetic relationships between Ophiocordyceps species were also established based on mt PCGs. Our study of the mitogenome of O. lanpingensis may provide a pathway for further investigations into its taxonomic classification, targeted gene/protein pathways, and conservation genetics, which in turn may help to enhance the study of other closely related species within Ophiocordyceps.
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
O. lanpingensis was isolated from the Biluo Snow Mountains, Yingpan Town, Lanping County, Yunnan Province, China. The O. lanpingensis specimen is shown in Figure 1a. Morphological estimations and other molecular assumptions regarding important characteristics of Ophiocordyceps have been documented in previous studies [15]. The specimen YHH-OLA0054 of O. lanpingensis was deposited in the Yunnan Herbal Herbarium (YHH) of Yunnan University. The strain YFCC 1606 isolated from YHH-OLA0054 was deposited in the Yunnan Fungal Culture Collection (YFCC) at Yunnan University. Mitogenome DNA extraction was performed using a fungal DNA Kit (#D3390-00, Omega Bio-Tek, Norcross, GA, USA), following the manufacturer’s instructions. The integrity of the genomic DNA was calculated with one percent agarose gel electrophoresis application, while the procedural quantitates were calculated using NanoDrop (Wilmington, DE, USA).
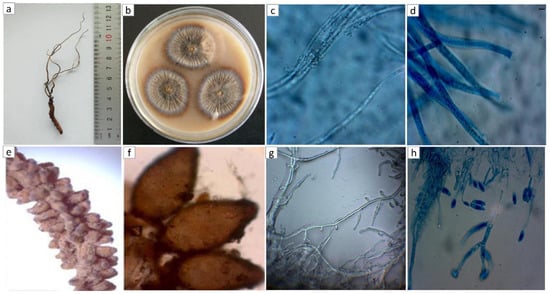
Figure 1.
Morphological characteristics of O. lanpingensis: the fresh specimen (a), colonies (b), asci- and ascospores (c,d), fertile part and perithecia (e,f), and mycelia and conidia of O. lanpingensis (g,h).
2.2. Genome Library, Sequencing, Assembly, and Annotation
An Illumina TruSeq library was generated using the IlluminaTruseq™ DNA Sample Preparation Kit (BGI, Shenzhen, China) using the manufacturer’s protocols. Then, genome sequencing was performed on the Illumina HiSeq 4000 platform for the 2 × 150 bp sequencing of paired-end reads. Clean data with no unpaired reads were retrieved, as well as high-quality reads having values < Q20. Then, retrieved clean data were used to assemble the following mitogenomes using the SPAdes 3.13.0 genome assembler [16], and gaps among the contig were filled using MITObim V1.9 [17]. The assembled strain was named YFCC 1606. The coverage was 500X.
2.3. Annotation of the Mitogenome
MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) (accessed on 5 February 2022) was used to annotate the mitogenome of O. lanpingensis. The MITOS tool [18] also assisted in mitogenome information extraction, and this was designed on the basis of the particular study of the corresponding genetic code (GC). Uncertain results were manually adjusted via sequence alignments with homologous intronless genes from closely related species. The originally annotated O. lanpingensis PCG, rRNA, or tRNA genes were also analyzed via alignment with the previously identified Ophiocordyceps mitogenome. Mitogenome sequences were scanned and ORF frames were studied using InterProScan software [19]. The tRNAscan-SE 2.0.5 program [20] was used to obtain information on the tRNA genes. Graphical mapping of the mitogenome of O. lanpingensis was carried out with the help of the OrganellarGenomeDraw (OGDRAW) tool [21]. Analysis of codon usage (CU) was performed using Sequence Manipulation Suite software [22], based on GC-4. A comparison of the arrangement of genes in O. lanpingensis and other reported Ophiocordyceps species, as well as genomic synteny estimation, was performed with Mauve v2.4.0 [23].
2.4. Mitogenomic Comparison of Six Ophiocordyceps Species
DNASTAR laserene 7.1, an important bioinformatics tool, was used to evaluate the nucleotide base composition of six mitogenomes (http://www.dnastar.com/t-dnastar-lasergene.aspx) (accessed on 1 April 2022). Comparison in terms of nucleotide length, as well as the CU encoding information of RNAs of the complete mitogenomes of O. lanpingensis, O. sinensis, Hirsutella vermicola (H. vermicola), H. thompsonii, H. rhossiliensis, and H. minnesotensis, was computed using MEGA 6.0 software [24]. Differences in the G-nucleotide base composition and C-nucleotide base composition on the strand disproportionateness were determined using GC and AT skew calculations. These important replication phenomena, wherein prokaryotes present a relative excess of nucleotides, were calculated using the following equations:
AT skew = [A − T]/[A + T]
GC skew = [G − C]/[G + C]
The occurrence of CU was noted using Sequence Manipulation Suite sequence estimating software. The overall mean genetic distances between 15 core protein-coding genes (PCGs) of 6 species were estimated with respect to the Kimura-2-parameter (K2P) substitution model in MEGA 6.0 and using GC [24]. Finally, the synteny of six mitogenomes was performed using the multiple genome alignment application Mauve v2.4.0 [23].
2.5. Phylogenetic Analysis
To determine the evolutionary relationship of Ophiocordyceps, phylogenetic analysis was conducted using the 14-mitochondrial-PCG dataset, derived from 53 taxa within Ascomycota. Sequence alignment of 14 PCGs was performed using MEGA 6.0. Bayesian interference (BI) was employed to generate the phylogenetic tree based on the 14 concatenated PCGs. The BI analysis was performed with MrBayes v3.1.2 for three million generations using the model GTR + G + I [25]. Trees were sampled every hundredth generation. The initial twenty-five percent of trees were discarded as burn-in, and the remaining trees were used to create a consensus tree using the sumt command.
3. Results
3.1. Morphological Characteristics of O. lanpingensis
It is important to reveal the morphological features of O. lanpingensis that contribute to the ecology of fungal diversity; therefore, the microscopic features of O. lanpingensis were observed in detail in our previous study [26]. The graphical representation of O. lanpingensis used in that study is repeated here (Figure 1). O. lanpingensis contains several dark-pigmented stromata that are simple and usually fibrous. Its productive (fertile) parts are cylindrical in shape and have a tapered, sterile terminal portion. Its perithecia are oval and apparent, and attach to its stroma at a right angle. Its asci are cylindrical and have a hemispherical ascus cap. Its ascospores are cylindrical and multi-septate, and have indistinct septation (Figure 1).
3.2. Mitogenomic Characteristics of O. lanpingensis
As it is important to understand the genetic characteristics of O. lanpingensis, in this research, the complete mitogenome of O. lanpingensis was studied (Table 1). The mitogenome of O. lanpingensis was found to be an obvious typical circular molecule 117,560 bp in length (Figure 2). This mitogenome showed similar characteristics to the mitogenome of O. sinensis: the complete mitogenome size of O. sinensis was found to be 157,510 bp, which was 39,950 bp larger than that of O. lanpingensis. Further, the estimated G + C contents of the O. lanpingensis mitogenome accounted for as low as 31% of its total contents, which was comparable to O. sinensis (30.2%); the A + T contents of O. lanpingensis accounted for 69%. The number of PCGs in O. lanpingensis was 24 and in O. sinensis was 15. Moreover, the G + C contents of the standard PCGs and RNA genes were 23.9% and 35.66%, respectively, in the mitogenome sequence of O. lanpingensis. Further, it was estimated that the mitogenome of O. lanpingensis was highly complex/compact and that it presented high genome capacity, with 82% of its genome coding for functional gene regions. The number of rRNA/tRNAs (%), which is important for the protein synthesis of O. lanpingensis, was found to be 2/23; for O. sinensis, this was 2/27.

Table 1.
General features in the mitogenome of O. lanpingensis.
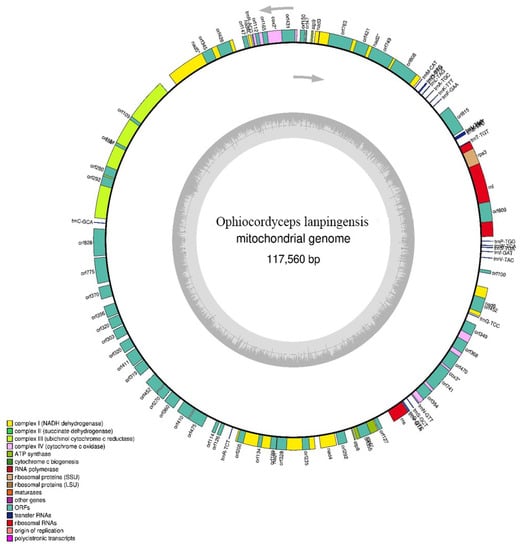
Figure 2.
Circular map of the complete mitogenome of O. lanpingensis constructed using Organellar Genome DRAW. All functional genes were in the same strand. The mitogenome coordinates had 24 standard protein-coding genes (PCGs), 23 transfer RNA (tRNA) genes, and 2 ribosomal RNA (rRNA) genes.
The functional genes found in the mitogenome of O. lanpingensis were associated with respiratory chain (RC) complexes (Table 2), for example, three ATP-synthase complex F0 subunits (atp6, atp8, and atp9); two complex IV subunits/cytochrome c oxidases, i.e., cox2 and cox3; one complex III subunit, i.e., cob; four electron transport complex I subunits/NADH dehydrogenase complex subunits, i.e., nad1, nad4, nad5, and nad6; and eleven ORFs (hypothetical proteins), i.e., orf609, orf495, orf815, orf47, orf150, orf147, orf292, orf127, orf349, orf452, and orf100. These functionally important genes were all found on the same positively oriented strand (Figure 2). It is remarkable that thirteen respective mt genes associated with the RC complexes were conserved and presented the same similarity likelihood as those of other filamentous fungal species [27].

Table 2.
Summary of the gene content of the O. lanpingensis mitogenome.
The mitogenome of O. lanpingensis was found to contain two rRNA genes, rns and rnl, while 25 tRNA genes were detected (as shown in Table 2), coding for a total of 20 amino acids. This enforces the importance of the tRNA ingress phenomenon in relation to the cytoplasm. The presence of tRNA-W was found to be conserved among O. langpingensis and O. sinensis, which code according to GC-4 (Fox, 1987). Information regarding the genetic components of the mitogenome of O. langpingensis is given in Table 3, which presents the genes’ start and stop positions, along with their lengths.

Table 3.
Summary of genetic components of O. lanpingensis mitogenome.
3.3. Codon Usage in the Mitogenome of O. lanpingensis
The CU patterns of the functional mt genes were found to be highly significant with respect to evolutionary and mechanistic analysis [28]. The CU patterns of the important PCGs, such as atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad5, and nad6, were determined. Further, the intronic regions in the intergenic regions in these 14 PCGs, such as atp6 (1 intron), atp8, atp9, cob (5 introns), cox1 (12 introns), cox2 (2 introns), cox3 (4 introns), nad1 (3 introns), nad2 (5 introns), nad3, nad4, nad4L, and nad5 (3 introns), and in the 11 ORFs (orf609, orf495, orf815, orf47, orf150, orf147, orf292, orf127, orf349, orf452, and orf100), were also determined. It was observed that in the case of the 11 ORFs, the start codon AUG most frequently appeared. Further, only orf47 started with “UCU”, only orf150 started with “UAU”, and only orf452 started with “UUU” (Table 4).

Table 4.
Codon usage of PCGs in the mitogenome of O. lanpingensis.
It was observed that the most frequently used codon among the PCGs was leucine, followed by isoleucine and lysine (Table 4). The codons for serine, arginine, tyrosine, asparagine, and phenylalanine, as well as the stop codon, were also represented frequently, whereas histidine and methionine were the least used codons (Table 4 and Figure 3). It was determined that the most often used codons were UAA for leucine, AUU for isoleucine, AAA for lysine, AGU for serine, AGA for arginine, UAU for tyrosine, AAU for asparagine, UUU for phenylalanine, and CAU for histidine. UAA and UAG were the only stop codons represented in the mitogenome (Figure 3).
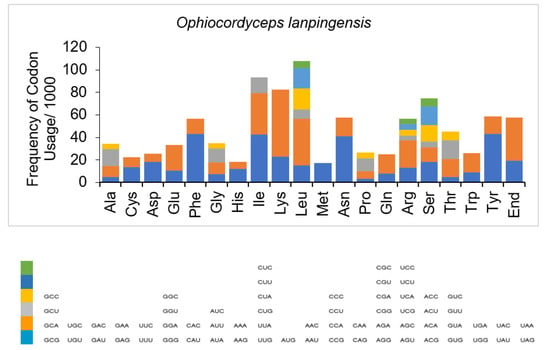
Figure 3.
Codon usage analysis of the mitogenome of O. lanpingensis. The frequency of codon usage is plotted on the y axis.
3.4. Transfer RNAs and Ribosomal RNAs
tRNA genes are useful as linkers between mRNA and proteins and are involved in the translation process [29]. tRNAscan–SE, a widely accepted tool for predicting tRNA genes in mitogenomes, was used in this study to predict the tRNA genes in the mitogenome of O. lanpingensis. A cloverleaf representation of each tRNA was obtained, and 20 tRNAs were found, which, through rough clustering estimation, were divided into two groups of 14 amino acids within the range of 70 to 84 bp in length. The output showed that, among the predicted tRNAs, few were found in the form of multiple copies; the trnM-CAT gene for methionine presented three copies, while the rest of the determined tRNA structures showed only one copy, as shown in Table 5 and Figure 4.

Table 5.
tRNAs in the mitogenome of O. lanpingensis.
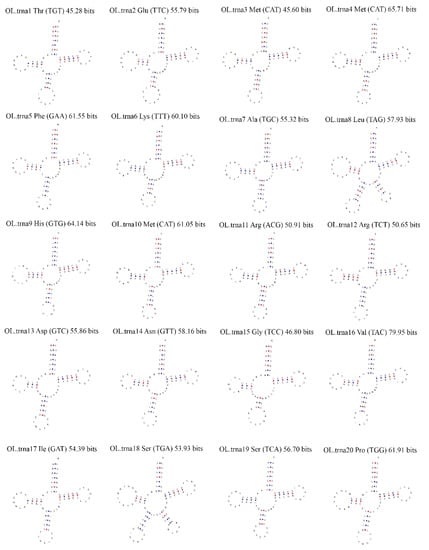
Figure 4.
Cloverleaf representation of predicted secondary structures of tRNA genes from the O. lanpingensis mitogenome. Each tRNA is labeled with the abbreviation of its corresponding amino acid. The tRNA structural plots were generated using MITOS software (http://mitos.bioinf.uni-leipzig.de/index.py).
3.5. Mitogenomic Comparison of Six Ophiocordyceps Species
The complete mitogenomes of six Ophiocordyceps species were typical circular molecules with total lengths ranging from 52,245 to 157,509 bp (Figure 5). The mitogenome sizes varied widely, with the 157,509 bp mitogenome of O. sinensis being the largest, while that of H. minnesotensis, at 52,245 bp, was the smallest. The gene-coding regions of these mitogenomes contained genes for rnL, ATP6, ATP8, ATP9, NAD2, NAD3, NAD4, NAD5, and COX2, which are involved in energy metabolism. These were evolutionarily conserved among all the mitogenomes, revealing positive evolutionary selection for these genes. Genes for COX1 were absent only in H. minessotensis. Similarly, rns, COX3, and NAD6 were commonly conserved among all the species except O. sinensis. NAD1 was found to be unique to O. lanpingensis. In addition, several ORFs were identified in the six mitogenomes, with their number ranging from 4 to 66 across the genomes. The smallest number of ORFs was found in H. minnesotensis, whereas the highest number was found in O. sinensis. All the genes were located on the positive or forward strand, except for ORF 12, ORF 352, and ORF 481 in O. sinensis, which were located on the reverse strand.
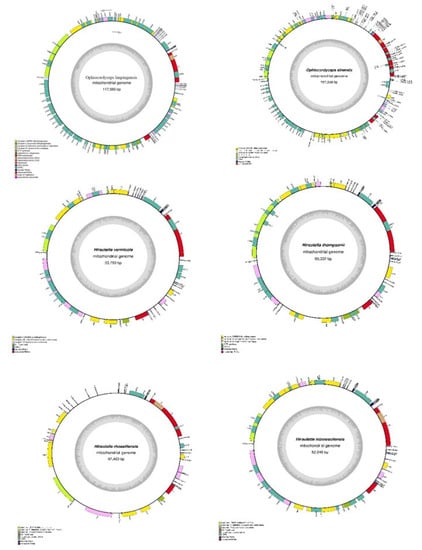
Figure 5.
Circle maps of mitogenomes of O. lanpingensis and five related species. Genes are represented by differently colored blocks, as indicated in the legend below the maps.
3.6. Codon Usage Analysis of the Mitogenomes from Six Ophiocordyceps Species
Next, the CU analyses of the six mitogenomes, which are shown in Figure 6, were compared. Leucine was the most frequently coded region, followed by isoleucine and lysine. However, O. lanpingensis and O. sinensis clearly exhibited similar patterns of CU frequency compared to the other four mitogenomes.
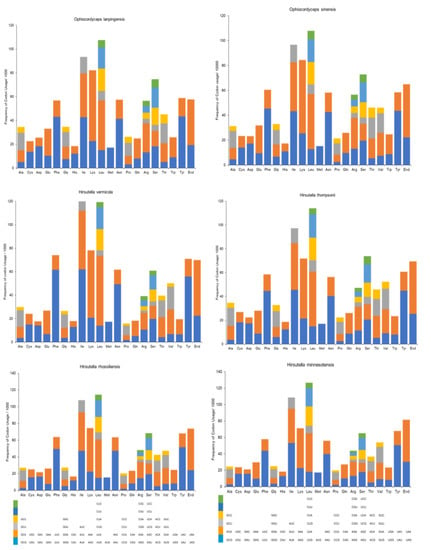
Figure 6.
Codon usage analysis of the mitogenomes from six Ophiocordyceps species. Frequency of codon usage is plotted on the y axis.
3.7. Gene Arrangement Analysis of Six Ophiocordyceps Species
Gene order and rearrangement in mitogenomes have great significance for biological evolution. In this study, each of the four Ophiocordyceps mitogenomes contained two rRNA genes, the small subunit ribosomal RNA gene (rns) and the large subunit ribosomal RNA gene (rnl). There were no significant differences in the number of rRNA genes among the six species, suggesting that these genes are experiencing positive evolutionary selection pressures. Five different groups of gene arrangement were detected in the six Ophiocordyceps mitogenomes (Figure 7). The results indicate that the Ophiocordyceps species had undergone large-scale gene rearrangements in their mitogenomes during evolution. Large-scale gene rearrangements were observed in the mitogenome of H. thompsonni, another Ophiocordycipitaceae species. The mt gene order of O. lanpingensis was very much like that of O. sinensis. Hence, the gene order and sequence homology of these two species, O. lanpingensis and O. sinensis, revealed that they are experiencing positive natural selection pressures and are closely related. Their sequences and structural homology suggest that they are similar in function.
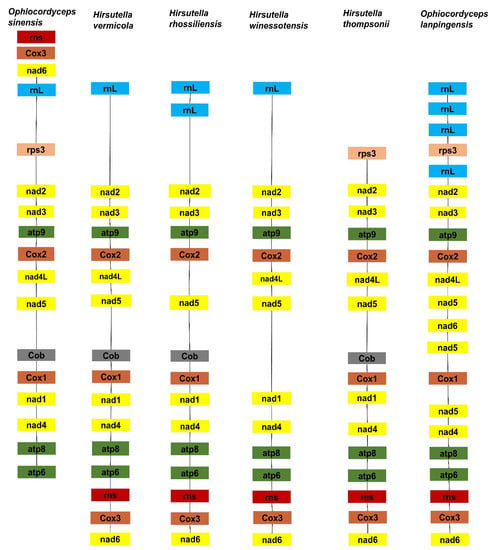
Figure 7.
The gene arrangement of mitogenomes of O. lanpingensis and other related species.
3.8. Synteny Analysis among Six Ophiocordyceps Species
Synteny analysis indicated that the six Ophiocordyceps mitogenomes were divisible into different homologous regions, where the sizes and relative positions of these regions were highly variable (Figure 8). The locations and sizes of these homologous regions varied among the six mitogenomes. In Figure 8, the homologous regions are connected via colored lines, which in turn connect the six mitogenomes, whereas black bar boxes show the genes with similar orders and sequence conservation among the six mitogenomes. Gene order and synteny analysis revealed that 13 genes were conserved across 6 mitogenomes. Meanwhile, the gene orders of O. lanpingensis and O. sinensis were highly conserved compared to the other four species.
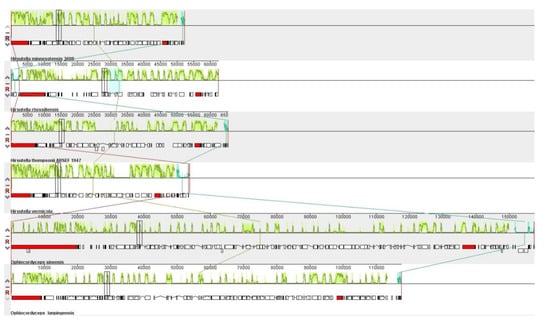
Figure 8.
Mitogenome collinearity /synteny progress of six Ophiocordyceps species. Mauve alignment determined the homologous regions of the six mitogenomes, which are connected by colored lines to explain the sequence of gene arrangement. The sizes and relative positions of the homologous fragments varied across the mitogenomes.
3.9. Phylogenetic Analysis of Six Ophiocordyceps Species
The O. lanpingensis mitogenome data were compared with previously published data on other fungal mitogenomes to comprehensively investigate the evolutionary relationships of the fungal species studied. The mitogenome of O. lanpingensis, along with five other mitogenomes, namely those of O. sinensis, H. minnesotensis, H. vermicola, H. rhossiliensis, and H. thomsonii, is presented in the Ophiocordycipitaceae family of the BI phylogenetic tree (section highlighted in gray). This tree was constructed to scale, with branch lengths in the same units as those of the evolutionary distances being used to infer the tree. The evolutionary distances were calculated using the maximum composite likelihood technique and are expressed in terms of the number of base substitutions per site. This analysis involved six nucleotide sequences. The positions of codons were estimated using the 1st + 2nd + 3rd + Noncoding formula. It was confirmed that all uncertain positions were discarded against each sequence pair. There was a total of 157,539 positions in the final dataset. Phylogenetic evolutionary investigations were performed using Clustal X2.0 and MEGA 6.0 software [24].
In this study, the sequence alignment and phylogenetic analysis were followed by the analysis of the parameters explained by Wang et al., 2020 [15]. The phylogenetic tree constructed using BI showed that O. lanpingensis shares its lineage with O. sinensis. As shown in Figure 9, regarding the evolutionary aspects of the fungal mitogenomes, the analysis indicated that O. lanpingensis and O. sinensis were more closely related than other species. O. lanpingensis and O. sinensis have previously been placed within the family Ophiocordycipitaceae, which includes species such as H. rhossiliensis, H. vermicola, H. thompsonii, and H. minnesotensis (Figure 9). Meanwhile, species in the Clavicipitaceae and Cordycipitaceae families have evolved at a higher evolutionary rate with regard to their mitochondrial genes than Ophiocordycipitaceae.
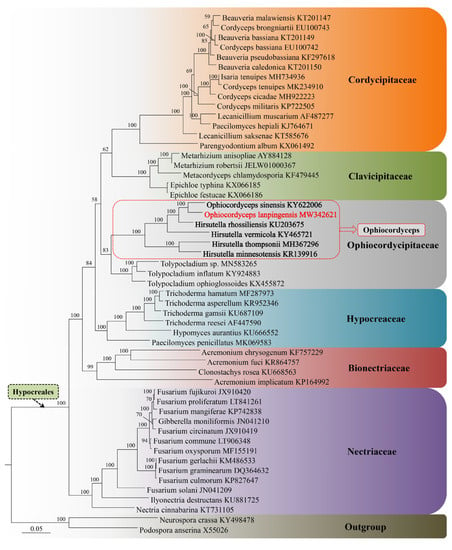
Figure 9.
Phylogenetic placement of O. lanpingensis in the order Hypocreales, inferred from mitogenomes based on Bayesian inference.
4. Discussion
Mitogenomic analysis has been reported to be a successful technique in several studies, and has demonstrated its benefits for the phylogenetic classification and estimation of evolutionary relationships in rapidly growing populations [30]. Through the increase in genomic knowledge and the advanced technology of next-generation sequencing, the sequencing procedures for the mitogenomes of fungal species have been enhanced, although the investigation of fungal mitogenomes remains more difficult than that of plants and animals [31]. As of July 2019, only 612 mitogenomes of fungi had been reported in the NCBI repository, and 488 of the 612 mitogenomes were from Ascomycetes species [32]. Therefore, it is highly recommended more mitogenomes of fungal species are explored in order to gain a deeper understanding of the fungal kingdom and its evolutionary significance.
In the present study, the first mitogenome of the important species O. lanpingensis was sequenced and analyzed in order to reveal its particular characteristics. During the comparative analysis of six Ophiocordyceps mitogenomes, it was observed that the size of the O. sinensis mitogenome was the largest, at 157,539 bp [33], while the O. lanpingensis mitogenome at 117,560 bp was the next most complex and largest Ophiocordyceps mitogenome. Further, the nucleotide lengths and CU of the complete mitogenomes of O. lanpingensis, O. sinensis, H. vermicola, H. thompsonii, H. rhossiliensis, and H. minnesotensis were computed, and the results show that leucine was the most frequently coded region among the mitogenomes, followed by isoleucine and lysine. However, O. lanpingensis and O. sinensis clearly exhibited similar patterns of CU frequency compared to the other four mitogenomes. A total of 25 tRNA genes were retrieved via the O. lanpingensis mitogenome analysis, and these were found to play significant roles in decoding mRNAs into proteins, following the translation mechanism. For the very first time, in this study, the complete mitogenome of O. lanpingensis was described. It was found to be a circular molecule 117,560 bp in length; its overall G + C content was 31%, while its A + T content was 69%. In addition, the G + C content of O. sinensis was found to be 30.8% and its A + T content was about 71%. These values are therefore highly similar among these two species. The mitogenome of O. lanpingensis was found to be a generally compacted and very bulky genome, with 82% of the genome-coding region being for functional genes. It was observed that the protein-coding regions of the whole mitogenome encompassed the genes associated with RC complexes, and these genes were all located on the same strand with a clockwise orientation. Thirteen representative mt genes were involved in the RC complexes, showing maximum similarity with species in the filamentous fungus group. Further, O. lanpingensis was compared with previously sequenced species within Ophiocordycepitaceae. Comparative analysis was used to compare the gene order, synteny, and circular genome arrangements of these species. This analysis indicated that O. lanpingensis shares more similar features with O. sinensis than the other species.
Four complex 1 (NADH) genes were found to be expressed in O. langpingensis, while three genes, namely nad1, nad6, and nad4L, were expressed in O. sinensis. One complex III (ubiquinol cytochrome c reductase) gene (cob) was common in both species; three complex IV (cytochrome oxidase) genes, i.e., cox1, cox2, and cox3, were common in both species; three ATP synthase genes, i.e., atp6, atp8, and atp9, were also common in both species. Two ribosomal RNAs (rRNAs) were also common in both species. The tRNA genes of the fungal mitogenomes studied have been proposed to play an important part in the transcription of novel repeats and in the recombination proceedings [34]. Therefore, in this study, 25 tRNAs in O. lanpingensis were found and analyzed, while 27 tRNAs were found to be expressed in O. sinensis in a previous study [35]. Based on a combined gene dataset (14 PCGs), the phylogenetic information of Ophiocordyceps was constructed using Bayesian inference. O. lanpingensis has been assigned as a sister species to O. sinensis. In this study, it was observed that O. lanpingensis and O. sinensis share a lineage with H. rhossiliensis and H. vermicola, while H. thompsonii and H. minnesotensis form a separate clade (Figure 9).
5. Conclusions
O. lanpingensis, a species of the genus Ophiocordyceps, is commonly found in Yunnan Province, China. It is pharmacologically important in the treatment of renal disorders induced by oxidative stress and inadequate immune response. In this study, the complete mitogenome of O. lanpingensis was described. It was found to be a circular molecule 117,560 bp in length, with its overall G + C content being 31% and its A + T content being 69%. This mitogenome was found to be a generally compact and high-capacity genome, with 82% of its genome coding for functional genes. The mitogenome was found to consist of 20 amino acids, while 24 PCGs were identified within it. The protein-coding regions of the whole mitogenome were found to contain genes involved in RC complexes, such as three ATP-synthase complex F0 subunits (atp6, atp8, and atp9), two complex IV subunits/cytochrome c oxidases (cox2 and cox3), one complex III subunit (cob), four electron transport complex I subunits/NADH dehydrogenase complex subunits (nad1, nad4, nad5, and nad6), one ribosomal RNA (rns, rnl), and eleven hypothetical proteins (orf609, orf495, orf815, orf47, orf150, orf147, orf292, orf127, orf349, orf452, and orf100). These genes were all located on the same strand. Thirteen representative mt genes were found to be involved in the RC complexes, showing high degrees of similarity with those of other species of filamentous fungi. Further, O. lanpingensis was compared with previously sequenced species within Ophiocordycepitaceae. Comparative analysis was used to compare the gene order, synteny, and circular genome arrangements of these species. This analysis showed that O. lanpingensis is more closely related to O. sinensis than to the other species. O. sinensis is an expensive fungus, owing to its low availability and the fact that it is difficult to culture. Therefore, O. lanpingenesis is of great importance to the management of traditional medicinal and natural supplemental needs. In the future, a more extensive metabolomics study of O. lanpingensis is recommended so its natural metabolites and their targeted pathways, which may be useful for the management of different diseases, can be more comprehensively identified.
Author Contributions
All authors contributed to the design and development of the final draft of this manuscript. D.W. and H.Y. performed experimental analysis (sample collection and DNA extraction); S.B., Y.W. and G.M. performed annotation, other analyses of the mitochondrial genome, and the comparison of the six mitogenomes. All authors contributed to the writing, review, and editing of the final draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 31870017, No. 32060007) and International Cooperation Project of Science and Technology Department of Yunnan Province (No. 202103AM140032).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Supplementary data can be provided on demand by the corresponding author.
Acknowledgments
All authors are thankful to Yunnan University for providing the platform for these research activities.
Conflicts of Interest
There are no conflict of interest, and all the authors agreed to the publication of this paper.
References
- Hamilton, G. The hidden risks for ‘three-person’ babies. Nat. News 2015, 525, 444. [Google Scholar]
- Chen, X.J.; Butow, R.A. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 2005, 6, 815–825. [Google Scholar]
- Chen, S.; Wang, Y.; Zhu, K.; Yu, H. Mitogenomics, phylogeny and morphology reveal Ophiocordyceps pingbianensis sp. nov., an entomopathogenic fungus from China. Life 2021, 11, 686. [Google Scholar]
- Abuduaini, A.; Wang, Y.-B.; Zhou, H.-Y.; Kang, R.-P.; Ding, M.-L.; Jiang, Y.; Suo, F.-Y.; Huang, L.-D. The complete mitochondrial genome of Ophiocordyceps gracilis and its comparison with related species. IMA Fungus 2021, 12, 1–14. [Google Scholar]
- Wang, L.; Zhang, S.; Li, J.-H.; Zhang, Y.-J. Mitochondrial genome, comparative analysis and evolutionary insights into the entomopathogenic fungus Hirsutella thompsonii. Environ. Microbiol. 2018, 20, 3393–3405. [Google Scholar]
- Bibi, S.; Wang, Y.-B.; Tang, D.-X.; Kamal, M.A.; Yu, H. Prospects for Discovering the Secondary Metabolites of Cordyceps Sensu Lato by the Integrated Strategy. Med. Chem. (Los Angeles) 2021, 17, 97–120. [Google Scholar]
- Bibi, S.; Hasan, M.M.; Wang, Y.-B.; Papadakos, S.P.; Yu, H. Cordycepin as a Promising Inhibitor of SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp). Curr. Med. Chem. 2022, 29, 152–162. [Google Scholar] [CrossRef]
- Zhao, Y.-Y.; Li, H.-T.; Feng, Y.-L.; Bai, X.; Lin, R.-C. Urinary metabonomic study of the surface layer of Poria cocos as an effective treatment for chronic renal injury in rats. J. Ethnopharmacol. 2013, 148, 403–410. [Google Scholar]
- Sung, G.H.; Hywel Jones, N.L.; Sung, J.M.; Luangsaard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the Clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar]
- Zhu, J.-S.; Halpern, G.M.; Jones, K. The scientific rediscovery of a precious ancient Chinese herbal regimen: Cordyceps sinensis Part II. J. Altern. Complement. Med. 1998, 4, 429–457. [Google Scholar]
- Chiu, C.-H.; Chyau, C.-C.; Chen, C.-C.; Lin, C.-H.; Cheng, C.-H.; Mong, M.-C. Polysaccharide extract of Cordyceps sobolifera attenuates renal injury in endotoxemic rats. Food Chem. Toxicol. 2014, 69, 281–288. [Google Scholar]
- Zhang, Y.; Du, Y.; Yu, H.; Zhou, Y.; Ge, F. Protective effects of ophiocordyceps lanpingensis on glycerol-induced acute renal failure in mice. J. Immunol. Res. 2017, 2017, 2012585. [Google Scholar]
- Kaushik, V.; Singh, A.; Arya, A.; Sindhu, S.C.; Sindhu, A.; Singh, A. Enhanced production of cordycepin in Ophiocordyceps sinensis using growth supplements under submerged conditions. Biotechnol. Rep. 2020, 28, e00557. [Google Scholar]
- Chen, Z.-H.; Dai, Y.-D.; Yu, H.; Yang, K.; Yang, Z.-L.; Yuan, F.; Zeng, W.-B. Systematic analyses of Ophiocordyceps lanpingensis sp. nov., a new species of Ophiocordyceps in China. Microbiol. Res. 2013, 168, 525–532. [Google Scholar]
- Wang, Y.-B.; Wang, Y.; Fan, Q.; Duan, D.-E.; Zhang, G.-D.; Dai, R.-Q.; Dai, Y.-D.; Zeng, W.-B.; Chen, Z.-H.; Li, D.-D.; et al. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): New taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 2020, 103, 1–46. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar]
- Hahn, C.; Bachmann, L.; Chevreux, B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—A baiting and iterative mapping approach. Nucleic Acids Res. 2013, 41, e129. [Google Scholar]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA genes in genomic sequences. In Gene Prediction; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–14. [Google Scholar]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar]
- Chen, Z.; Xu, L.; Yu, H.; Zeng, W.; Dai, Y.; Wang, Y. Phylogeny and active ingredients of artificial Ophiocordyceps lanpingensis ascomata. AIP Conf. Proc. 2018, 1956, 20011. [Google Scholar]
- Yuan, X.-L.; Cao, M.; Li, P.-P.; Cheng, S.; Liu, X.-M.; Du, Y.-M.; Zhang, Z.-F.; Shen, G.-M.; Zhang, P. The mitochondrial genome of Arthrinium arundinis and its phylogenetic position within Sordariomycetes. Int. J. Biol. Macromol. 2019, 121, 956–963. [Google Scholar]
- Chen, Z.; Nie, H.; Wang, Y.; Pei, H.; Li, S.; Zhang, L.; Hua, J. Rapid evolutionary divergence of diploid and allotetraploid Gossypium mitochondrial genomes. BMC Genom. 2017, 18, 1–15. [Google Scholar]
- Lant, J.T.; Berg, M.D.; Sze, D.H.W.; Hoffman, K.S.; Akinpelu, I.C.; Turk, M.A.; Heinemann, I.U.; Duennwald, M.L.; Brandl, C.J.; O’Donoghue, P. Visualizing tRNA-dependent mistranslation in human cells. RNA Biol. 2018, 15, 567–575. [Google Scholar]
- Wang, T.; Zhang, S.; Pei, T.; Yu, Z.; Liu, J. Tick mitochondrial genomes: Structural characteristics and phylogenetic implications. Parasites Vectors 2019, 12, 1–15. [Google Scholar]
- Li, Q.; Wang, Q.; Jin, X.; Chen, Z.; Xiong, C.; Li, P.; Zhao, J.; Huang, W. The first complete mitochondrial genome from the family Hygrophoraceae (Hygrophorus russula) by next-generation sequencing and phylogenetic implications. Int. J. Biol. Macromol. 2019, 122, 1313–1320. [Google Scholar]
- Chen, C.; Li, Q.; Fu, R.; Wang, J.; Xiong, C.; Fan, Z.; Hu, R.; Zhang, H.; Lu, D. Characterization of the mitochondrial genome of the pathogenic fungus Scytalidium auriculariicola (Leotiomycetes) and insights into its phylogenetics. Sci. Rep. 2019, 9, 1–12. [Google Scholar]
- Li, Y.; Hu, X.-D.; Yang, R.-H.; Hsiang, T.; Wang, K.; Liang, D.-Q.; Liang, F.; Cao, D.-M.; Zhou, F.; Wen, G.; et al. Complete mitochondrial genome of the medicinal fungus Ophiocordyceps sinensis. Sci. Rep. 2015, 5, 1–11. [Google Scholar]
- Saccone, C.; Gissi, C.; Reyes, A.; Larizza, A.; Sbisà, E.; Pesole, G. Mitochondrial DNA in metazoa: Degree of freedom in a frozen event. Gene 2002, 286, 3–12. [Google Scholar]
- Kang, X.; Hu, L.; Shen, P.; Li, R.; Liu, D. SMRT sequencing revealed mitogenome characteristics and mitogenome-wide DNA modification pattern in Ophiocordyceps sinensis. Front. Microbiol. 2017, 8, 1422. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).