Genome-Wide Identification and Functional Analysis of NAP1 in Triticum aestivum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of the Whole Genome of the TaNAP1 Family
2.2. Physical and Chemical Properties of TaNAP1 Genes
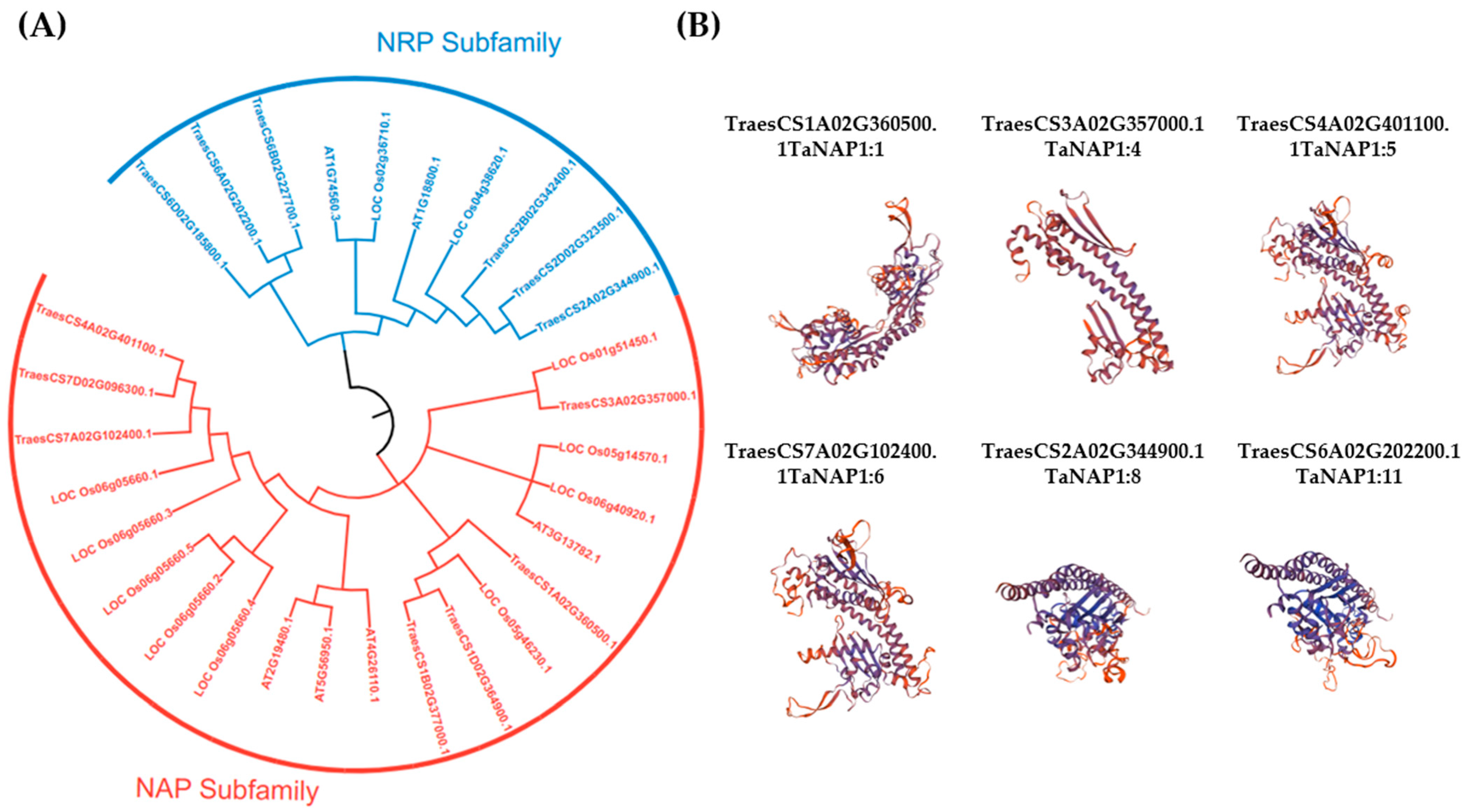
2.3. Multiple Sequence Alignments and Phylogenetic Tree Construction
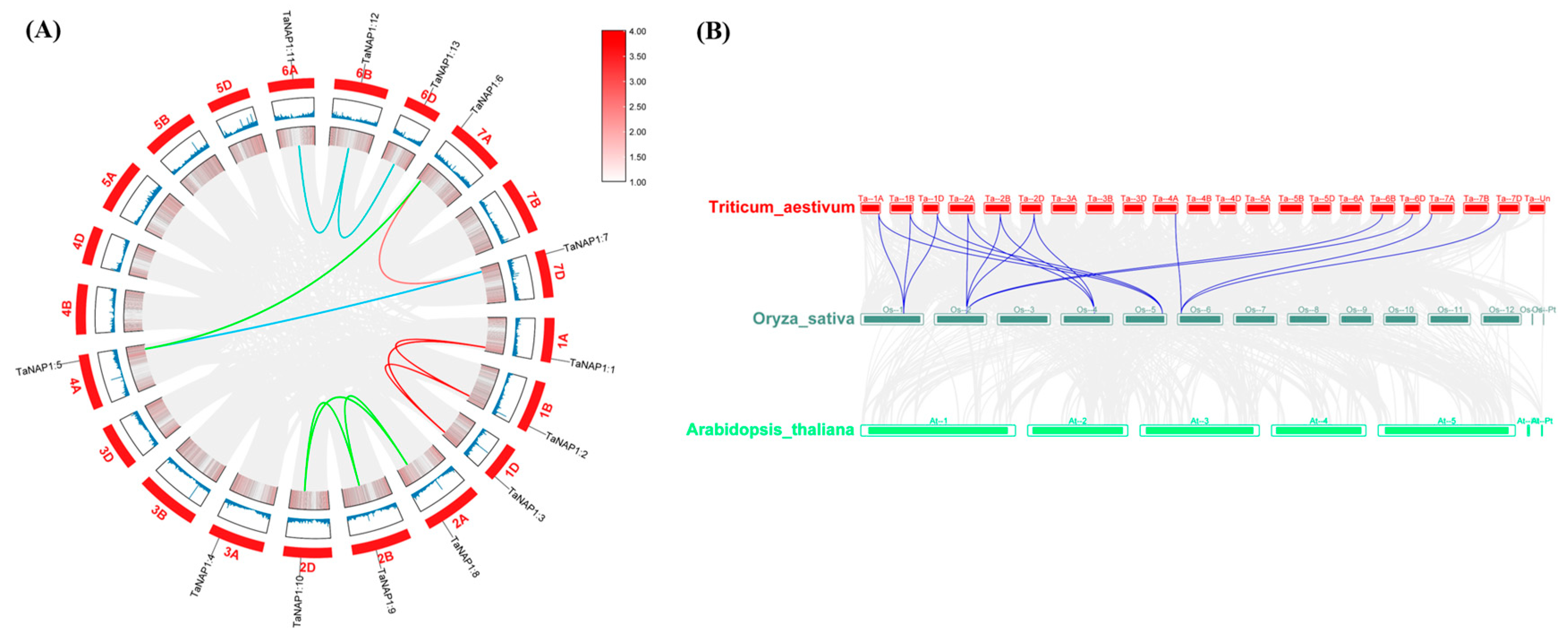
2.4. The Chromosomal Location, Synteny Analysis, and Duplication of TaNAP1
2.5. Calculation of Ka/Ks Values
2.6. Structural Analysis of the TaNAP1 Gene and Predicted Tertiary Structure of Proteins
2.7. Tissues Expression Profiles of TaNAP1
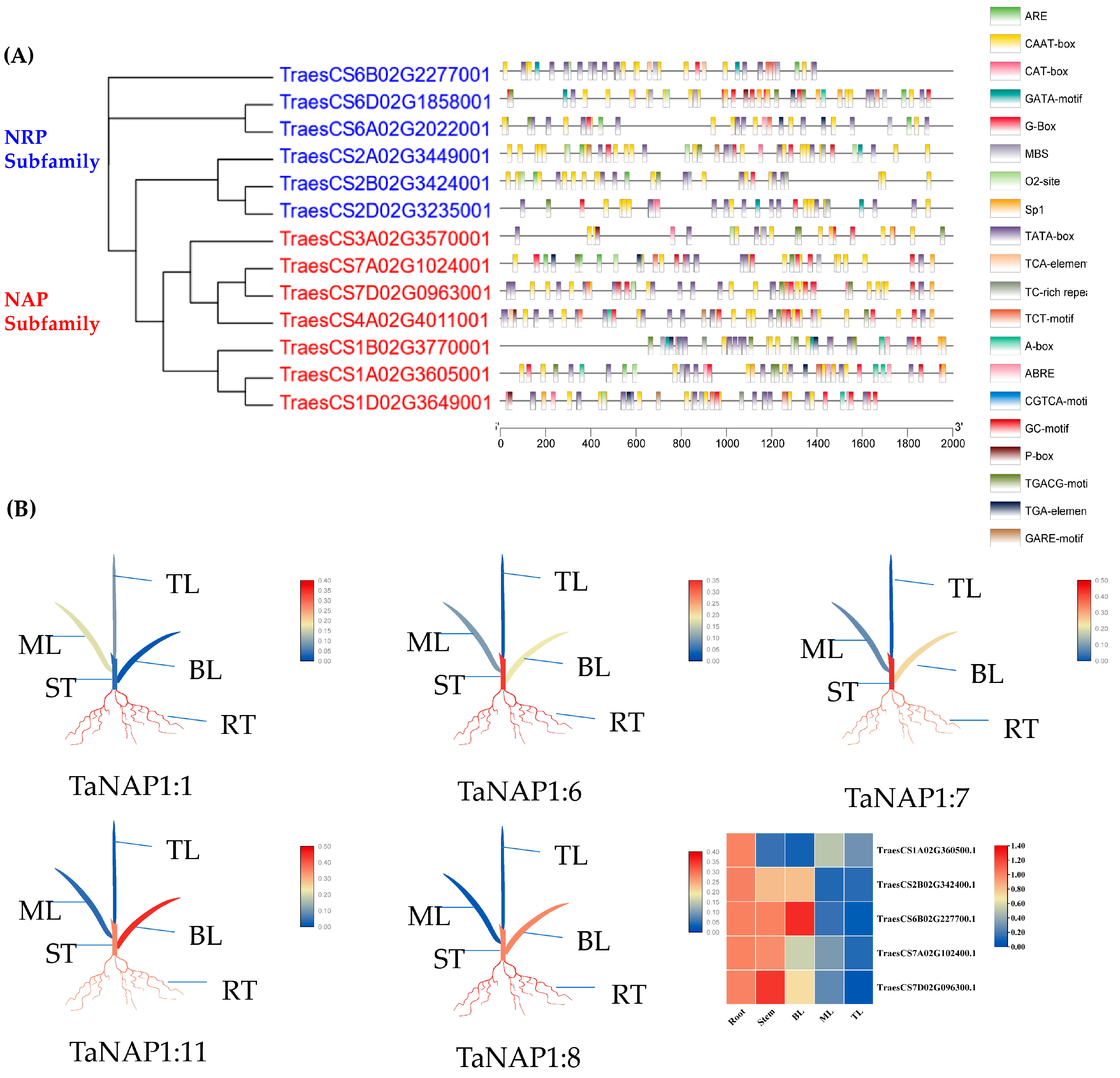
2.8. Cis-Acting Element Prediction
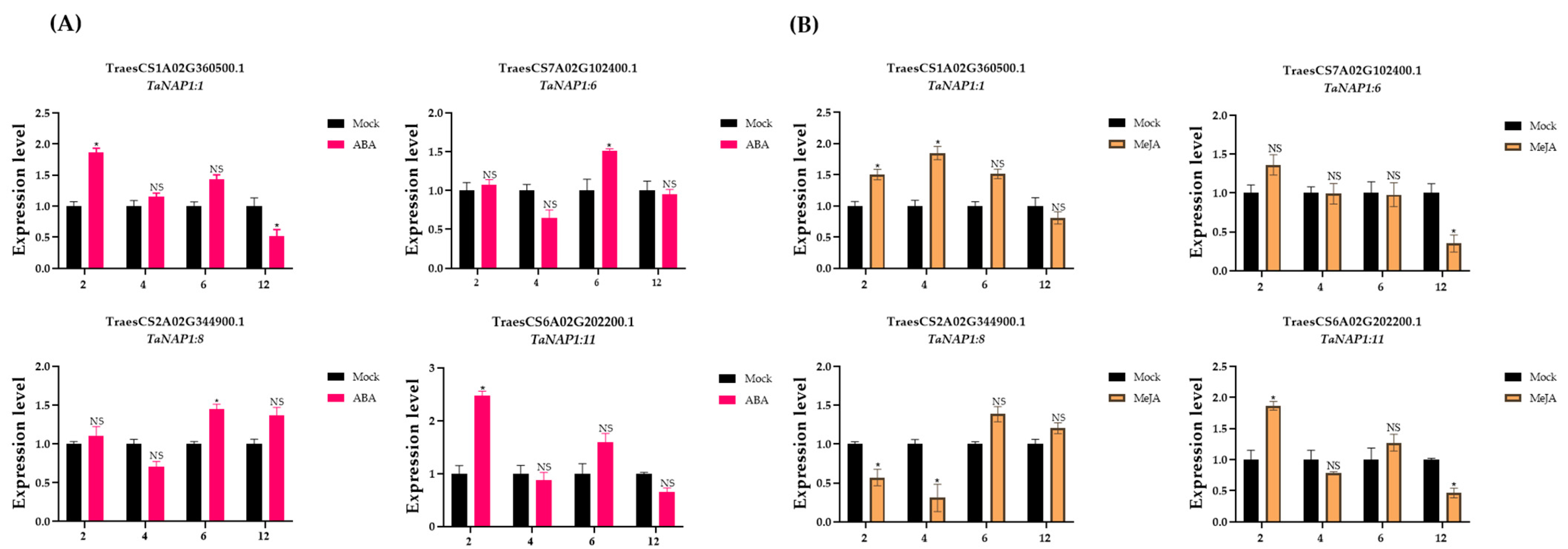
2.9. Expression of TaNAP1 under Abscisic Acid and Methyl Jasmonate Treatments
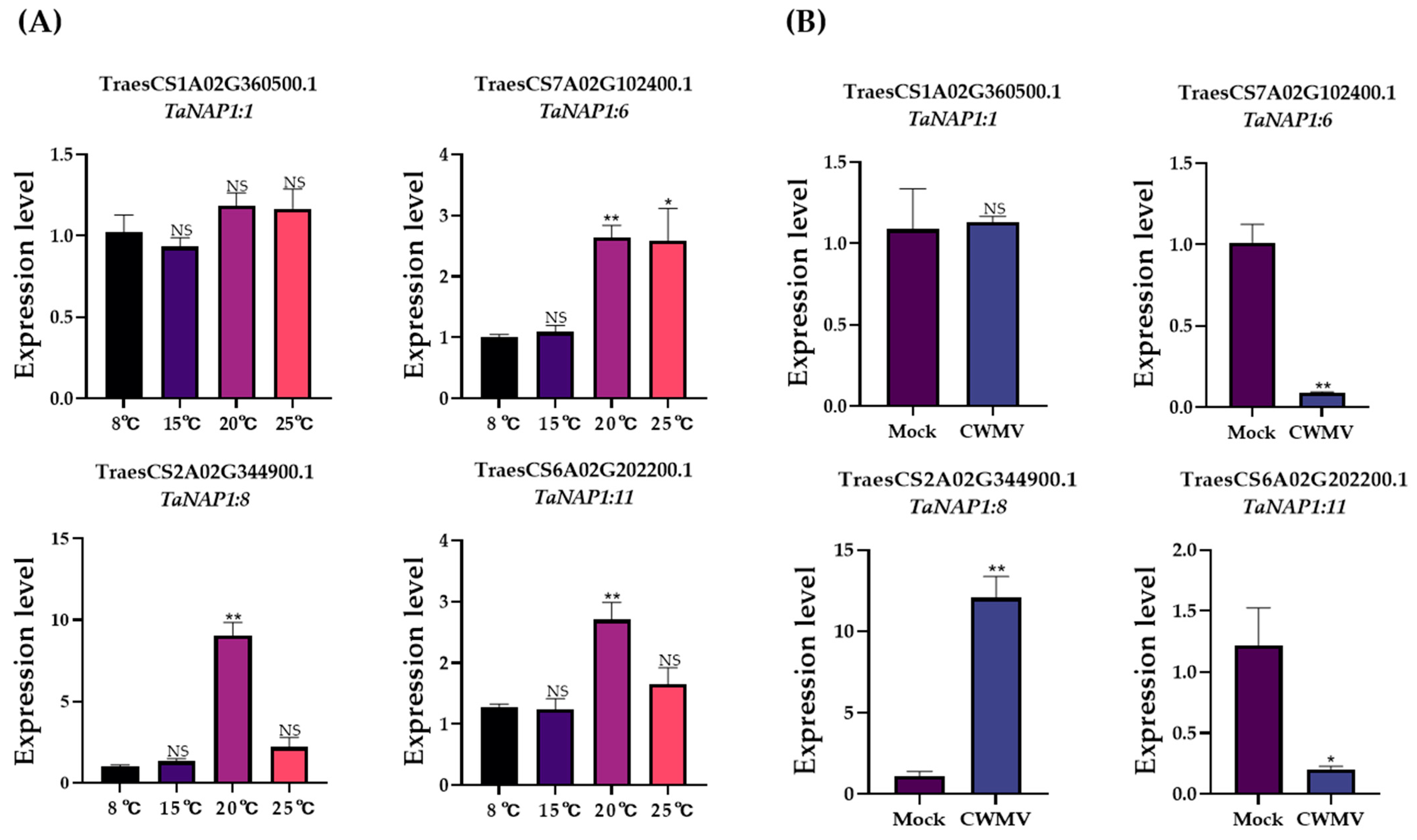
2.10. Expression of TaNAP1 after CWMV Infection
2.11. Expression Analysis of TaNAP1 by qRT-PCR
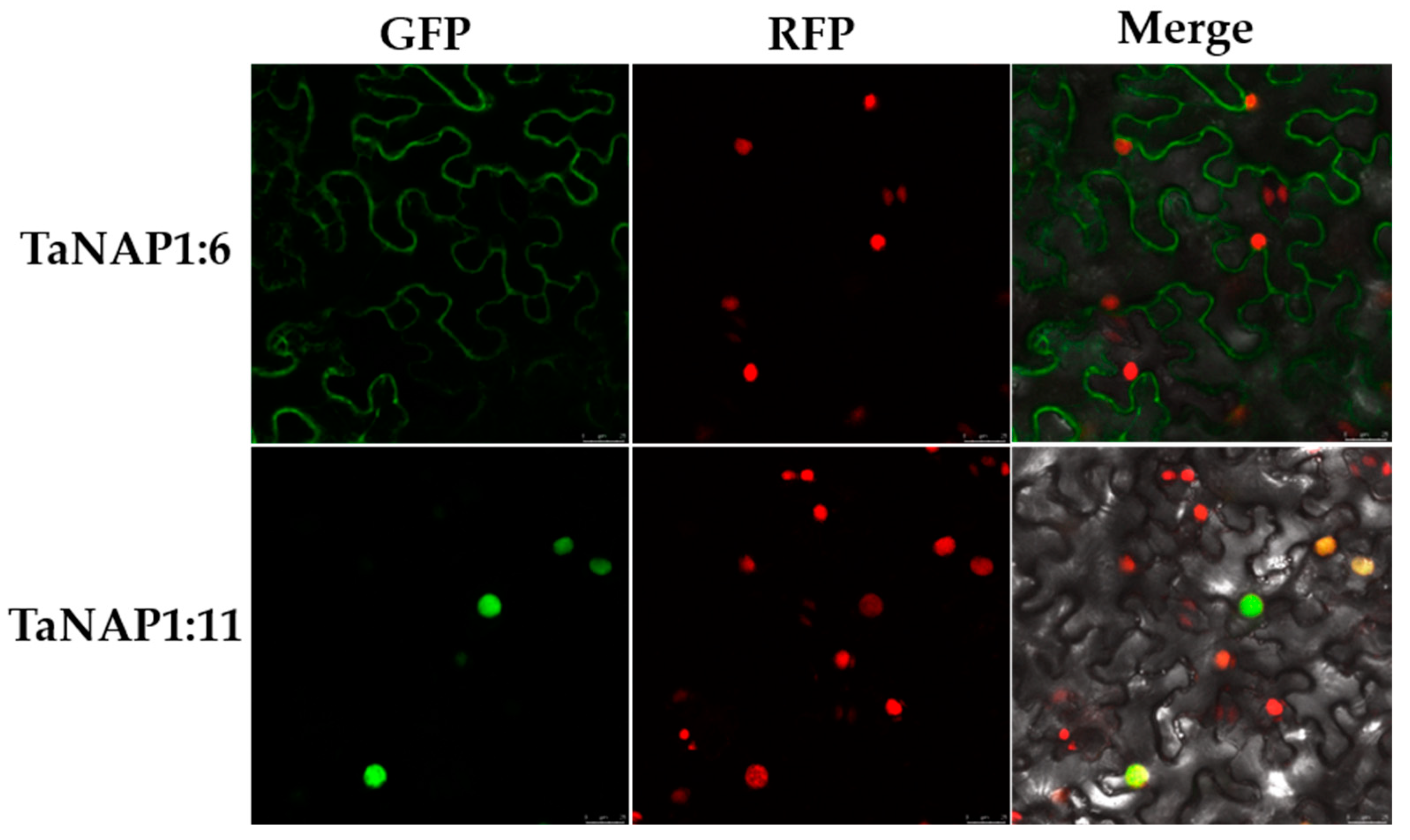
2.12. Plant Growth and TaNAP1 Subcellular Localization Assay
3. Results
3.1. Identification and Characterization of NAP1 in Triticum aestivum
3.2. Phylogenetic Analysis of TaNAP1 and the Tertiary Structure Models
3.3. Genetic Structure and Analysis of Conserved Patterns of NAP1
3.4. The Chromosomal Location, Synteny Analysis, and Duplication Events of TaNAP1
3.5. Analysis of the TaNAP1 Promoter Region
3.6. Expression Profile of TaNAP1 in Wheat at the Three-Leaf Stage
3.7. Analysis of TaNAP1 Expression under Different Stresses
3.8. Subcellular Localization Analysis of TaNAP1
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Paralogous Pairs | Ka | Ks | Ka/Ks | T (Mya) | |
|---|---|---|---|---|---|
| TraesCS1A02G360500.1 | TraesCS1B02G377000.1 | 0.010514 | 0.110014 | 0.095572 | 6.044715 |
| TraesCS1A02G360500.1 | TraesCS1D02G364900.1 | 0 | 0.076105 | 0 | 4.181585 |
| TraesCS1B02G377000.1 | TraesCS1D02G364900.1 | 0.010515 | 0.131817 | 0.079771 | 7.242715 |
| TraesCS2A02G344900.1 | TraesCS2B02G342400.1 | 0.013223 | 0.0274 | 0.482603 | 1.505511 |
| TraesCS2A02G344900.1 | TraesCS2D02G323500.1 | 0.006588 | 0.020386 | 0.323159 | 1.120127 |
| TraesCS2B02G342400.1 | TraesCS2D02G323500.1 | 0.00659 | 0.034253 | 0.19239 | 1.882007 |
| TraesCS4A02G401100.1 | TraesCS7A02G102400.1 | 0.020113 | 0.08873 | 0.226677 | 4.875255 |
| TraesCS4A02G401100.1 | TraesCS7D02G096300.1 | 0.006641 | 0.061125 | 0.108645 | 3.358501 |
| TraesCS6A02G202200.1 | TraesCS6B02G227700.1 | 0.001668 | 0.013202 | 0.126352 | 0.725366 |
| TraesCS6B02G227700.1 | TraesCS6D02G185800.1 | 0.001668 | 0.02664 | 0.062615 | 1.463729 |
| TraesCS7A02G102400.1 | TraesCS7D02G096300.1 | 0.014472 | 0.084035 | 0.172211 | 4.617296 |
References
- Park, Y.J.; Luger, K. The structure of nucleosome assembly protein 1. Proc. Natl. Acad. Sci. USA 2006, 103, 1248–1253. [Google Scholar] [CrossRef] [PubMed]
- Laskey, R.A.; Honda, B.M.; Mills, A.D.; Finch, J.T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 1978, 275, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Das, C.; Tyler, J.K.; Churchill, M.E. The histone shuffle: Histone chaperones in an energetic dance. Trends Biochem. Sci. 2010, 35, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, D.R.; Murray, A.W. NAP1 acts with Clb1 to perform mitotic functions and to suppress polar bud growth in budding yeast. J. Cell Biol. 1995, 130, 675–685. [Google Scholar] [CrossRef]
- Shimizu, Y.; Akashi, T.; Okuda, A.; Kikuchi, A.; Fukui, K. NBP1 (Nap1 binding protein 1), an essential gene for G2/M transition of Saccharomyces cerevisiae, encodes a protein of distinct sub-nuclear localization. Gene 2000, 246, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Shikama, N.; Chan, H.M.; Krstic-Demonacos, M.; Smith, L.; Lee, C.W.; Cairns, W.; La Thangue, N.B. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell Biol. 2000, 20, 8933–8943. [Google Scholar] [CrossRef]
- Mizuguchi, G.; Shen, X.; Landry, J.; Wu, W.H.; Sen, S.; Wu, C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 2004, 303, 343–348. [Google Scholar] [CrossRef]
- Yoon, H.W.; Kim, M.C.; Lee, S.Y.; Hwang, I.; Bahk, J.D.; Hong, J.C.; Ishimi, Y.; Cho, M.J. Molecular cloning and functional characterization of a cDNA encoding nucleosome assembly protein 1 (NAP-1) from soybean. Mol. Gen. Genet. MGG 1995, 249, 465–473. [Google Scholar] [CrossRef]
- Dong, A.; Zhu, Y.; Yu, Y.; Cao, K.; Sun, C.; Shen, W.H. Regulation of biosynthesis and intracellular localization of rice and tobacco homologues of nucleosome assembly protein 1. Planta 2003, 216, 561–570. [Google Scholar] [CrossRef]
- Liu, Z.; Zhu, Y.; Gao, J.; Yu, F.; Dong, A.; Shen, W.H. Molecular and reverse genetic characterization of NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) genes unravels their function in transcription and nucleotide excision repair in Arabidopsis thaliana. Plant J. 2009, 59, 27–38. [Google Scholar] [CrossRef]
- Zhu, Y.; Dong, A.; Meyer, D.; Pichon, O.; Renou, J.P.; Cao, K.; Shen, W.H. Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 2006, 18, 2879–2892. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar Singh, A.; Chandrakant Bobde, R.; Vasudevan, D. Structural Characterization of Arabidopsis thaliana NAP1-Related Protein 2 (AtNRP2) and Comparison with its Homolog AtNRP1. Molecules 2019, 24, 2258. [Google Scholar] [CrossRef]
- Barna, B.; Gemes, K.; Domoki, M.; Bernula, D.; Ferenc, G.; Balint, B.; Nagy, I.; Feher, A. Arabidopsis NAP-related proteins (NRPs) contribute to the coordination of plant growth, developmental rate, and age-related pathogen resistance under short days. Plant Sci. 2018, 267, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, Y.; Liu, W.; Cao, M.; Massart, S.; Wang, X. Identification, Characterization and Full-Length Sequence Analysis of a Novel Polerovirus Associated with Wheat Leaf Yellowing Disease. Front. Microbiol. 2017, 8, 1689. [Google Scholar] [CrossRef]
- Sanfacon, H. Grand Challenge in Plant Virology: Understanding the Impact of Plant Viruses in Model Plants, in Agricultural Crops, and in Complex Ecosystems. Front. Microbiol. 2017, 8, 860. [Google Scholar] [CrossRef]
- Diao, A.; Chen, J.; Ye, R.; Zheng, T.; Yu, S.; Antoniw, J.F.; Adams, M.J. Complete sequence and genome properties of Chinese wheat mosaic virus, a new furovirus from China. J. Gen. Virol. 1999, 80, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Antoniw, J.F.; Kreuze, J. Virgaviridae: A new family of rod-shaped plant viruses. Arch. Virol. 2009, 154, 1967–1972. [Google Scholar] [CrossRef]
- Andika, I.B.; Zheng, S.; Tan, Z.; Sun, L.; Kondo, H.; Zhou, X.; Chen, J. Endoplasmic reticulum export and vesicle formation of the movement protein of Chinese wheat mosaic virus are regulated by two transmembrane domains and depend on the secretory pathway. Virology 2013, 435, 493–503. [Google Scholar] [CrossRef]
- Sun, L.; Andika, I.B.; Kondo, H.; Chen, J. Identification of the amino acid residues and domains in the cysteine-rich protein of Chinese wheat mosaic virus that are important for RNA silencing suppression and subcellular localization. Mol. Plant Pathol. 2013, 14, 265–278. [Google Scholar] [CrossRef]
- Sun, L.; Andika, I.B.; Shen, J.; Yang, D.; Ratti, C.; Chen, J. The CUG-initiated larger form coat protein of Chinese wheat mosaic virus binds to the cysteine-rich RNA silencing suppressor. Virus Res. 2013, 177, 66–74. [Google Scholar] [CrossRef]
- Tripathi, A.K.; Singh, K.; Pareek, A.; Singla-Pareek, S.L. Histone chaperones in Arabidopsis and rice: Genome-wide identification, phylogeny, architecture and transcriptional regulation. BMC Plant Biol. 2015, 15, 42. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. TIG 2002, 18, 486. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Muthusamy, S.K.; Dalal, M.; Chinnusamy, V.; Bansal, K.C. Genome-wide identification and analysis of biotic and abiotic stress regulation of small heat shock protein (HSP20) family genes in bread wheat. J. Plant Physiol. 2017, 211, 100–113. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Liu, T.-t.; Xu, M.-z.; Gao, S.-q.; Zhang, Y.; Hu, Y.; Jin, P.; Cai, L.-n.; Cheng, Y.; Chen, J.-p.; Yang, J.; et al. Genome-wide identification and analysis of the regulation wheat DnaJ family genes following wheat yellow mosaic virus infection. J. Integr. Agric. 2022, 21, 153–169. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, T.; Li, J.; Wu, N.; Wu, G.; Yang, J.; Chen, X.; He, L.; Chen, J. Chinese wheat mosaic virus-derived vsiRNA-20 can regulate virus infection in wheat through inhibition of vacuolar- (H+)-PPase induced cell death. New Phytol. 2020, 226, 205–220. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Chen, X.; Yang, J.; Zhang, T.; Li, J.; Zhang, S.; Zhong, K.; Zhang, H.; Chen, J.; Yang, J. Rice black-streaked dwarf virus-encoded P5-1 regulates the ubiquitination activity of SCF E3 ligases and inhibits jasmonate signaling to benefit its infection in rice. New Phytol. 2020, 225, 896–912. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, P.; Zhong, K.; Zhang, F.; Xu, M.; He, L.; Jin, P.; Chen, J.; Yang, J. Wheat Yellow Mosaic Virus NIb Interacting with Host Light Induced Protein (LIP) Facilitates Its Infection through Perturbing the Abscisic Acid Pathway in Wheat. Biology 2019, 8, 80. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, J.; Feng, H.; Liu, S.; Liu, P.; Chen, X.; Yang, J.; He, L.; Yang, J.; Chen, J. Phosphorylated viral protein evades plant immunity through interfering the function of RNA-binding protein. PLoS Pathog. 2022, 18, e10104122022. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhu, Y.; Dong, A.; Shen, W.H. Histone H2A/H2B chaperones: From molecules to chromatin-based functions in plant growth and development. Plant J. 2015, 83, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Yang, D.; Jin, Y.; Liu, X.; Zhang, Z.; Gu, L.; Zhang, H. Genome-Wide Identification of NAP1 and Function Analysis in Moso Bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2022, 23, 6491. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Yoshida, Y. Evolutionary Origin of JAZ Proteins and Jasmonate Signaling. Mol. Plant 2019, 12, 153–155. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Asayama, M. Regulatory system for light-responsive gene expression in photosynthesizing bacteria: Cis-elements and trans-acting factors in transcription and post-transcription. Biosci. Biotechnol. Biochem. 2006, 70, 565–573. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Himmelbach, A.; Liu, L.; Zierold, U.; Altschmied, L.; Maucher, H.; Beier, F.; Müller, D.; Hensel, G.; Heise, A.; Schützendübel, A.; et al. Promoters of the barley germin-like GER4 gene cluster enable strong transgene expression in response to pathogen attack. Plant Cell 2010, 22, 937–952. [Google Scholar] [CrossRef]
- Wang, D.R.; Yang, K.; Wang, X.; You, C.X. A C2H2-type zinc finger transcription factor, MdZAT17, acts as a positive regulator in response to salt stress. J. Plant Physiol. 2022, 275, 153737. [Google Scholar] [CrossRef]
- Rahim, A.A.; Uzair, M.; Rehman, N.; Rehman, O.U.; Zahra, N.; Khan, M.R. Genome-Wide Identification and Characterization of Receptor-Like Protein Kinase 1 (RPK1) Gene Family in Triticum aestivum Under Drought Stress. Front. Genet. 2022, 13, 912251. [Google Scholar] [CrossRef]
- Islam, W.; Naveed, H.; Zaynab, M.; Huang, Z.; Chen, H.Y.H. Plant defense against virus diseases; growth hormones in highlights. Plant Signal. Behav. 2019, 14, 1596719. [Google Scholar] [CrossRef] [PubMed]
- Fraser, R.S.S.; Whenham, R.J. Abscisic acid metabolism in tomato plants infected with tobacco mosaic virus: Relationships with growth, symptoms and the Tm-1 gene for TMV resistance. Physiol. Mol. Plant Pathol. 1989, 34, 215–226. [Google Scholar] [CrossRef]
- Liu, C.; Tian, S.; Lv, X.; Pu, Y.; Peng, H.; Fan, G.; Ma, X.; Ma, L.; Sun, X. Nicotiana benthamiana asparagine synthetase associates with IP-L and confers resistance against tobacco mosaic virus via the asparagine-induced salicylic acid signalling pathway. Mol. Plant Pathol. 2022, 23, 60–77. [Google Scholar] [CrossRef]
- Ryu, C.M.; Murphy, J.F.; Mysore, K.S.; Kloepper, J.W. Plant growth-promoting rhizobacteria systemically protect Arabidopsis thaliana against Cucumber mosaic virus by a salicylic acid and NPR1-independent and jasmonic acid-dependent signaling pathway. Plant J. 2004, 39, 381–392. [Google Scholar] [CrossRef]
- Tan, X.; Zhang, H.; Yang, Z.; Wei, Z.; Li, Y.; Chen, J.; Sun, Z. NF-YA transcription factors suppress jasmonic acid-mediated antiviral defense and facilitate viral infection in rice. PLoS Pathog. 2022, 18, e10105482022. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, K.; Xu, K.; Zhang, K.; Jin, X.; Yang, M.; Zhang, Y.; Wang, X.; Han, C.; Yu, J.; et al. Barley stripe mosaic virus infection requires PKA-mediated phosphorylation of γb for suppression of both RNA silencing and the host cell death response. New Phytol. 2018, 218, 1570–1585. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Shi, C.; Hu, H.; Zhang, Z.; Wang, Z.; Chen, Z.; Feng, H.; Liu, P.; Guo, J.; Lu, Q.; et al. N6-methyladenosine RNA modification promotes viral genomic RNA stability and infection. Nat. Commun. 2022, 13, 6576. [Google Scholar] [CrossRef] [PubMed]

| Name | Gene ID | Exons | Gene Location | CDS Length (bp) | Size (aa) | MW (kDa) | PI | Protein Location |
|---|---|---|---|---|---|---|---|---|
| TaNAP1:1 | TraesCS1A02G360500.1 | 10 | 1A-541632666-541636078 | 1194 | 397 | 44.6 | 4.35 | Nucleus |
| TaNAP1:2 | TraesCS1B02G377000.1 | 11 | 1B-608533160-608536582 | 1080 | 359 | 41.1 | 4.33 | Nucleus |
| TaNAP1:3 | TraesCS1D02G364900.1 | 11 | 1D-445542135-445545423 | 1101 | 366 | 41.7 | 4.36 | Nucleus |
| TaNAP1:4 | TraesCS3A02G357000.1 | 6 | 3A-604998850-605001934 | 744 | 247 | 28.3 | 7.05 | Nucleus |
| TaNAP1:5 | TraesCS4A02G401100.1 | 12 | 4A-675131549-675135314 | 1149 | 382 | 42.8 | 4.23 | Nucleus |
| TaNAP1:6 | TraesCS7A02G102400.1 | 12 | 7A-62879024-62882679 | 1152 | 383 | 43.1 | 4.25 | Nucleus |
| TaNAP1:7 | TraesCS7D02G096300.1 | 11 | 7D-58500798-58504536 | 1149 | 382 | 42.9 | 4.24 | Nucleus |
| TaNAP1:8 | TraesCS2A02G344900.1 | 10 | 2A-582637903-582642714 | 762 | 253 | 29.4 | 4.13 | Nucleus |
| TaNAP1:9 | TraesCS2B02G342400.1 | 10 | 2B-488198352-488204713 | 762 | 253 | 29.47 | 4.17 | Nucleus |
| TaNAP1:10 | TraesCS2D02G323500.1 | 10 | 2D-416401053-416406189 | 762 | 253 | 29.46 | 4.13 | Nucleus |
| TaNAP1:11 | TraesCS6A02G202200.1 | 10 | 6A-337723056-337727746 | 756 | 251 | 28.8 | 4.26 | Nucleus |
| TaNAP1:12 | TraesCS6B02G227700.1 | 10 | 6B-353554146-353558991 | 756 | 251 | 28.8 | 4.26 | Nucleus |
| TaNAP1:13 | TraesCS6D02G185800.1 | 10 | 6D-242216366-242222032 | 756 | 251 | 28.8 | 4.26 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, H.; Wu, M.; Wang, Z.; Wang, X.; Chen, J.; Yang, J.; Liu, P. Genome-Wide Identification and Functional Analysis of NAP1 in Triticum aestivum. Genes 2023, 14, 1041. https://doi.org/10.3390/genes14051041
Feng H, Wu M, Wang Z, Wang X, Chen J, Yang J, Liu P. Genome-Wide Identification and Functional Analysis of NAP1 in Triticum aestivum. Genes. 2023; 14(5):1041. https://doi.org/10.3390/genes14051041
Chicago/Turabian StyleFeng, Huimin, Mila Wu, Ziqiong Wang, Xia Wang, Jianping Chen, Jian Yang, and Peng Liu. 2023. "Genome-Wide Identification and Functional Analysis of NAP1 in Triticum aestivum" Genes 14, no. 5: 1041. https://doi.org/10.3390/genes14051041
APA StyleFeng, H., Wu, M., Wang, Z., Wang, X., Chen, J., Yang, J., & Liu, P. (2023). Genome-Wide Identification and Functional Analysis of NAP1 in Triticum aestivum. Genes, 14(5), 1041. https://doi.org/10.3390/genes14051041






