Alternative Splicing of NAC Transcription Factor Gene CmNST1 Is Associated with Naked Seed Mutation in Pumpkin, Cucurbita moschata
Abstract
1. Introduction
2. Materials and Methods
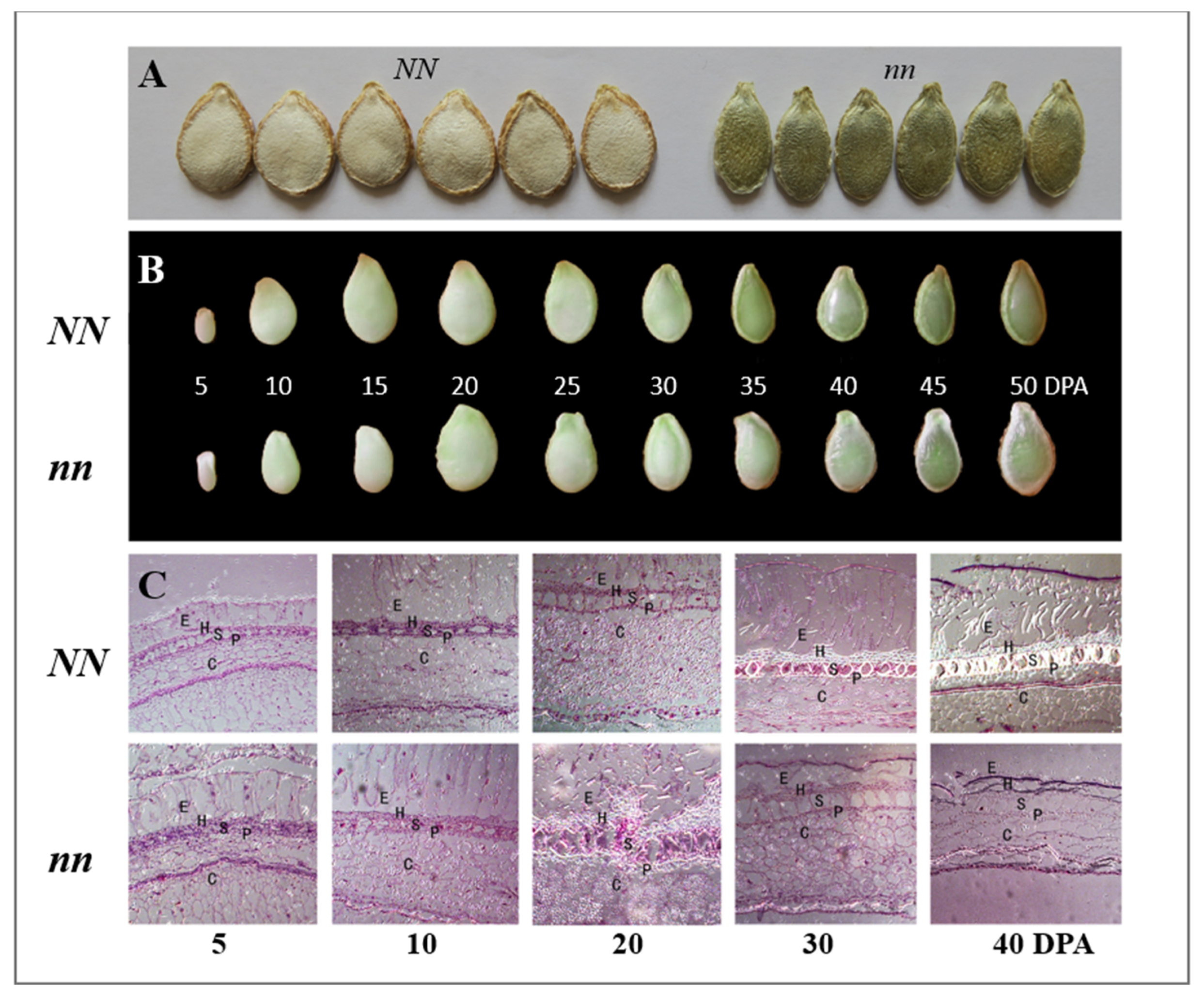
2.1. Plant Materials and Phenotyping of Seed Coat
2.2. Bulked Segregant Analysis (BSA) and Resequencing (BSA-Seq)
2.3. Linkage Mapping and Identification of Candidate Gene for the CmN Locus
2.4. Sequence Analysis of CmN Candidate Gene
2.5. Quantitative Real-Time PCR (qPCR)
2.6. Bulked Segregant RNA-Seq (BSR-Seq)
3. Results
3.1. Phenotypic Characterization and Inheritance of Naked Seed Mutation
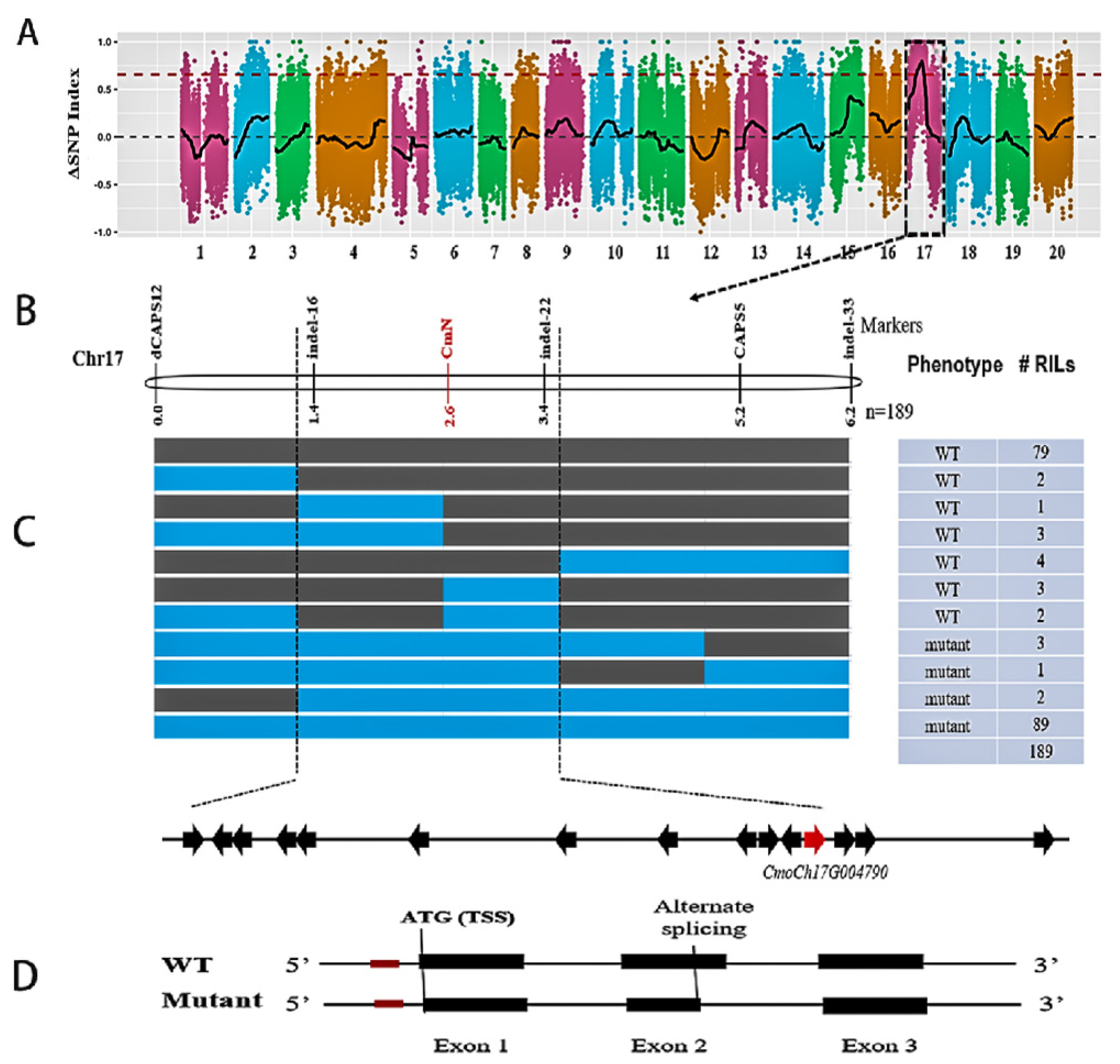
3.2. BSA-Seq Analysis
3.3. Linkage Mapping and Candidate Gene Identification for the N Locus
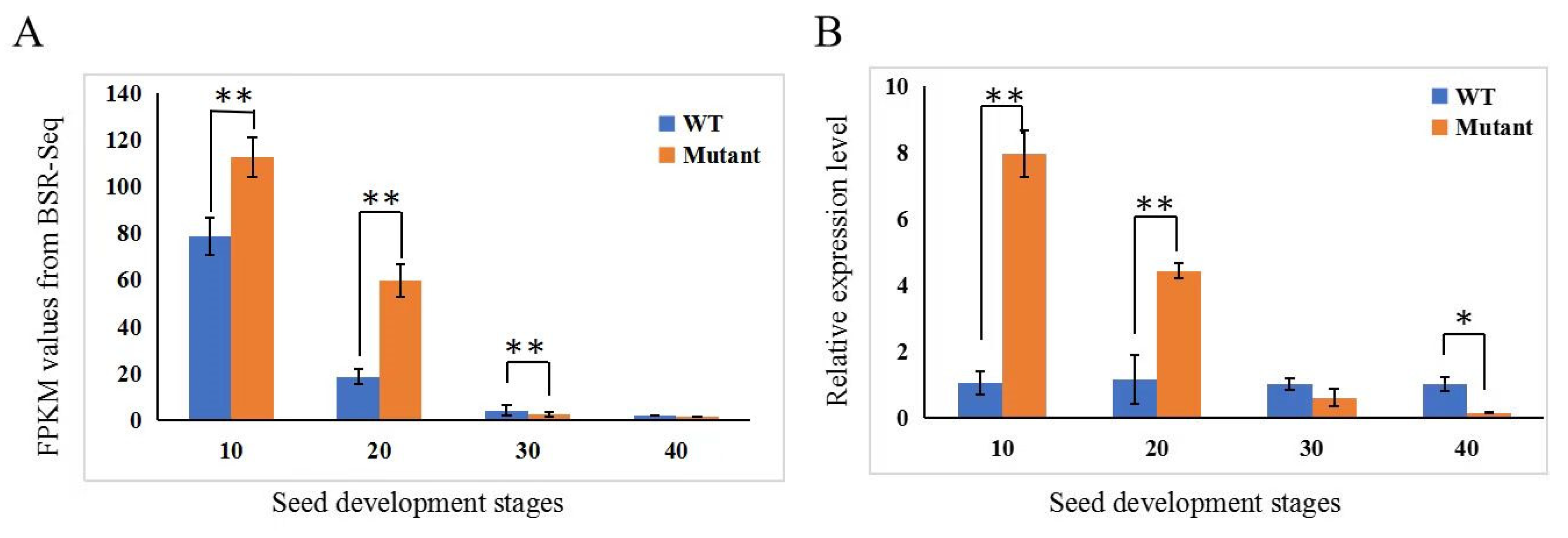
3.4. Sequence Analysis Suggests Alternative Splicing in the CmNST1 Transcript May Contribute to Defective Seed Coat Development in the Mutant
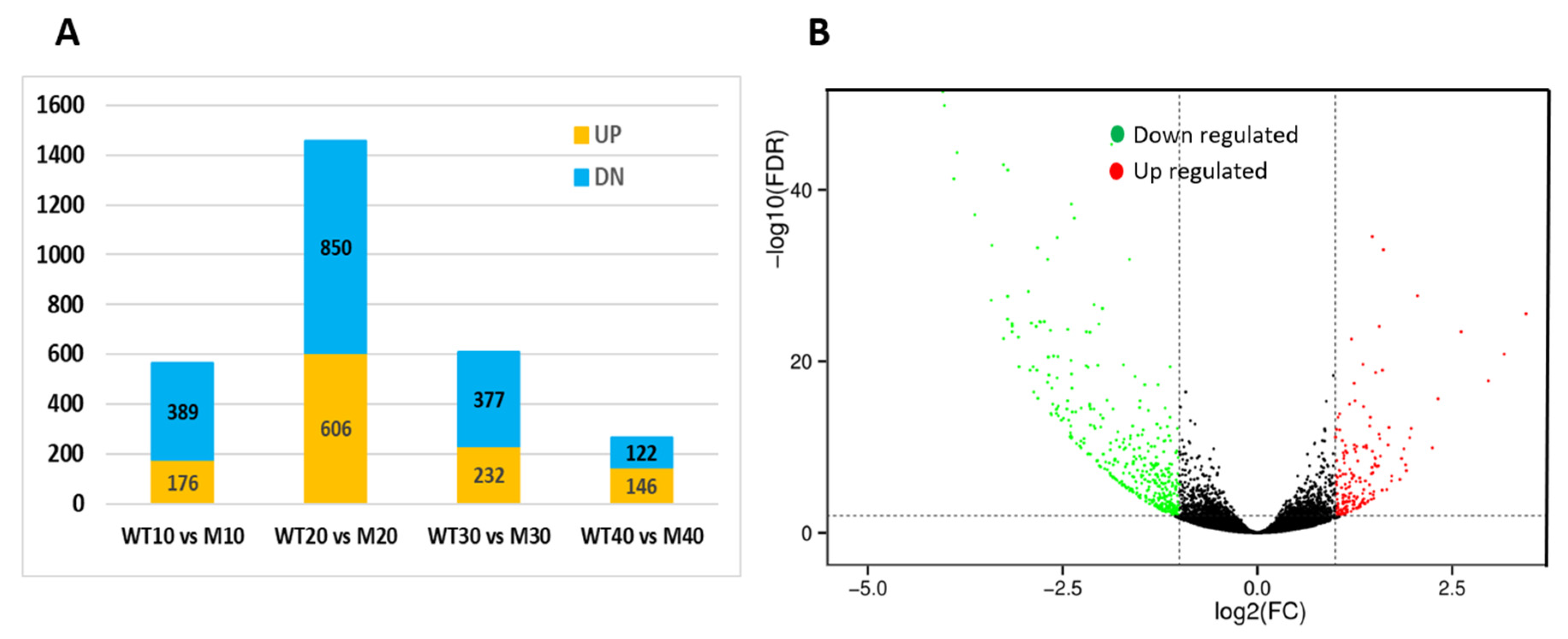
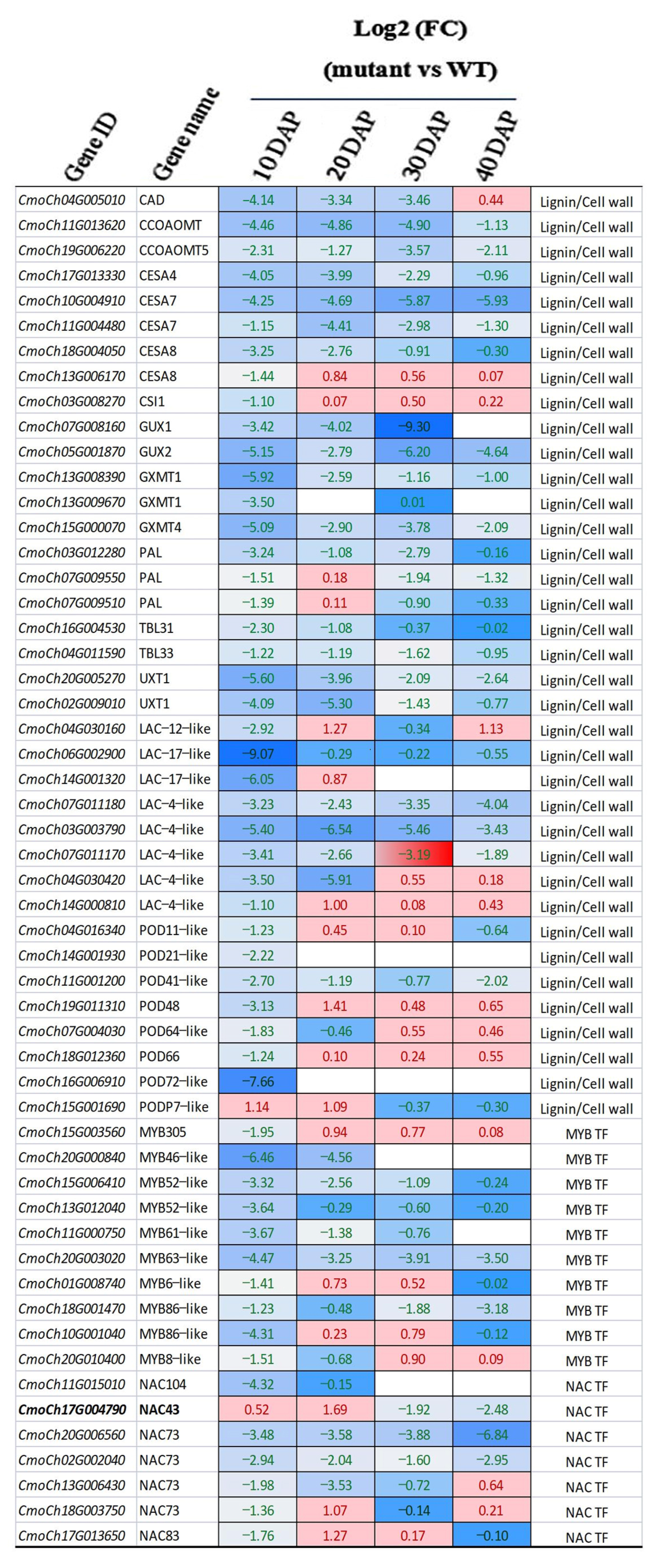
3.5. Transcriptome Profiling Reveals Regulatory Gene Network for Seed Coat Development in C. moschata
4. Discussion
4.1. CmNST1 Is a Candidate Gene for the Naked Seed (n) Locus
4.2. Alternative Splicing Contributes to Naked Seed Mutation in C. moschata
4.3. CmNST1-Regulated Seed Coat Development Involves a Complex Regulatory Network for Lignin Biosynthesis and Secondary Cell Wall Formation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferriol, M.; Picó, B. Pumpkin and winter squash. In Vegetables I: Asteraceae, Brassicaceae, Chenopodicaceae, and Cucurbitaceae; Springer: Berlin/Heidelberg, Germany, 2008; pp. 317–349. [Google Scholar]
- Paris, H.S. Germplasm enhancement of Cucurbita pepo (pumpkin, squash, gourd: Cucurbitaceae): Progress and challenges. Euphytica 2016, 208, 415–438. [Google Scholar] [CrossRef]
- Lelley, T.; Loy, B.; Murkovic, M. Hull-less oil seed pumpkin. Oil Crops 2010, 4, 469–492. [Google Scholar]
- Stevenson, D.G.; Eller, F.J.; Wang, L.; Jane, J.L.; Wang, T.; Inglett, G.E. Oil and tocopherol content and composition of pumpkin seed oil in 12 cultivars. J. Agric. Food Chem. 2007, 55, 4005–4013. [Google Scholar] [CrossRef]
- Fruhwirth, G.O.; Hermetter, A. Seeds and oil of the Styrian oil pumpkin: Components and biological activities. Eur. J. Lipid Sci. Technol. 2007, 109, 1128–1140. [Google Scholar] [CrossRef]
- Bergantin, C.; Maietti, A.; Tedeschi, P.; Font, G.; Manyes, L.; Marchetti, N. HPLC–UV/Vis–APCI–MS/MS determination of major carotenoids and their bioaccessibility from “Delica”(Cucurbita maxima) and “Violina”(Cucurbita moschata) pumpkins as food traceability markers. Molecules 2018, 23, 2791. [Google Scholar] [CrossRef]
- Kim, E.Y.; Kwon, C.W.; Chang, P.S. Purification and characterization of a novel acid-tolerant and heterodimeric β-glucosidase from pumpkin (Cucurbita moschata) seed. J. Biosci. Bioeng. 2021, 132, 125–131. [Google Scholar] [CrossRef]
- Vinayashree, S.; Vasu, P. Biochemical, nutritional and functional properties of protein isolate and fractions from pumpkin (Cucurbita moschata var. Kashi Harit) seeds. Food Chem. 2021, 340, 128177. [Google Scholar] [CrossRef]
- Baxter, G.; Murphy, K.; Paech, A. The Potential to Produce Pumpkim Seed for Processing in North East Victoria; Rural Industries Development Corporation: Kingston, Australia, 2012; Volume 11. [Google Scholar]
- Loy, J.B. Morpho-physiological aspects of productivity and quality in squash and pumpkins (Cucurbita spp.). Crit. Rev. Plant Sci. 2004, 23, 337–363. [Google Scholar] [CrossRef]
- Wenzl, T.; Prettner, E.; Schweiger, K.; Wagner, F.S. An improved method to discover adulteration of Styrian pumpkin seed oil. J. Biochem. Biophys. Methods 2002, 53, 193–202. [Google Scholar] [CrossRef]
- Zraidi, A.; Pachner, M.; Lelley, T.; Obermayer, R. On the genetics and histology of the hull-less character of Styrian oil-pumpkin (Cucurbita pepo L.). Mol. Breed. 2003, 26, 57–61. [Google Scholar]
- Zhou, X.L. Study on the breeding of naked kernel pumpkin and its genetic behavior. Acta Hortic. Sin. 1987, 14, 115–118. [Google Scholar]
- Paris, H.S.; Brown, R.N. The genes of pumpkin and squash. HortScience 2005, 40, 1620–1630. [Google Scholar] [CrossRef]
- Gong, L.; Stift, G.; Kofler, R.; Pachner, M.; Lelley, T. Microsatellites for the genus Cucurbita and an SSR-based genetic linkage map of Cucurbita pepo L. Theor. Appl. Genet. 2008, 117, 37–48. [Google Scholar] [CrossRef]
- Zraidi, A.; Stift, G.; Pachner, M.; Shojaeiyan, A.; Gong, L.; Lelley, T. A consensus map for Cucurbita pepo. Mol. Breed. 2007, 20, 375–388. [Google Scholar] [CrossRef]
- Inan, N.; Yildiz, M.; Sensoy, S.; Kafkas, S.; Abak, K. Efficacy of ISSR and SRAP techniques for molecular characterization of some Cucurbita genotypes including naked (hull-less) seed pumpkin. J. Anim. Plant Sci. 2012, 22, 126–136. [Google Scholar] [CrossRef]
- Chahal, G.K.; Kaur, A.; Dhatt, A.S. A single-gene mutation changed the architecture of pumpkin seed: A review. J. Plant Growth Regul. 2022, 41, 113–118. [Google Scholar] [CrossRef]
- Lv, X.; Shi, L.; Zhao, M.; Li, Z.; Liao, N.; Meng, Y.; Ma, Y.; Zhou, Y.; Xue, Q.; Hu, Z. A natural mutation of the NST1 gene arrests secondary cell wall biosynthesis in the seed coat of a hull-less pumpkin accession. Hortic. Res. 2022, 9, uhac136. [Google Scholar] [CrossRef]
- Meru, G.; Fu, Y.; Shrestha, S.; Michael, V.N.E.; Dorval, M.; Mainviel, R. Genomic position and markers associated with the hull-less seed trait in pumpkin. Plants 2022, 11, 1238. [Google Scholar] [CrossRef]
- Stuart, S.G.; Loy, J.B. Comparison of testa development in normal and hull-less seeded strains of Cucurbita pepo L. Bot. Gaz. 1983, 144, 491–500. [Google Scholar] [CrossRef]
- Bezold, T.N.; Loy, J.B.; Minocha, S.C. Changes in the cellular content of polyamines in different tissues of seed and fruit of a normal and a hull-less seed variety of pumpkin during development. Plant Sci. 2003, 164, 743–752. [Google Scholar] [CrossRef]
- Kreft, M.; Zorec, R.; Janeš, D.; Kreft, S. Histolocalisation of the oil and pigments in the pumpkin seed. Ann. Appl. Biol. 2009, 154, 413–418. [Google Scholar] [CrossRef]
- Xue, Y.; Shen, Z.; Tao, F.; Zhou, J.; Xu, B. Transcriptomic analysis reveal the molecular mechanisms of seed coat development in Cucurbita pepo L. Front. Plant Sci. 2022, 13, 51. [Google Scholar] [CrossRef]
- Shen, Q.; Wu, L.; Zhao, X.; Li, Y. Comparative analysis of electron microscopic structure of seed testa, lignin and biosynthesis-related enzyme activities in hulled and hull-less seeds of Cucurbita moschata. Sci. Hortic. 2019, 245, 137–143. [Google Scholar] [CrossRef]
- Bezold, T.N.; Mathews, D.; Loy, J.B.; Minocha, S.C. Molecular analysis of the hull-less seed trait in pumpkin: Expression profiles of genes related to seed coat development. Seed Sci. Res. 2005, 15, 205–217. [Google Scholar] [CrossRef]
- Zhou, X.L. Study on the breeding and genetic rules of naked pumpkin. Shanxi Agric. Sci. 1983, 12, 11–12. [Google Scholar]
- Wu, J.X.; Chang, Z.J. Study on seed characteristics of two naked kernel Cucurbita moschata. Shanxi Agric. Sci. 2011, 39, 230–231. [Google Scholar] [CrossRef]
- Li, Q.; Li, L.; Yang, X.; Warburton, M.L.; Bai, G.H.; Dai, J.R.; Li, J.S.; Yan, J.B. Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 2010, 10, 143. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods Cell Sci. 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Paris, H.S.; Kabelka, E. Gene list for Cucurbita species, 2009. Cucurbit Genet. Coop. Rep. 2009, 31, 44–69. [Google Scholar]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ye, Z. Complexity of the transcriptional network controlling secondary wall biosynthesis. Plant Sci. 2014, 229, 193–207. [Google Scholar] [CrossRef]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Xie, M.; Zhang, J.; Tschaplinski, T.J.; Tuskan, G.A.; Chen, J.; Muchero, W. Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front. Plant Sci. 2018, 9, 1427. [Google Scholar] [CrossRef]
- Pratyusha, D.S.; Sarada, D.V. MYB transcription factors–master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses. Plant Cell Rep. 2022, 41, 2245–2260. [Google Scholar] [CrossRef]
- Kaur, B.; Garcha, K.S.; Bhatia, D.; Khosa, J.S.; Sharma, M.; Mittal, A.; Verma, N.; Dhatt, A.S. Identification of single major QTL and candidate gene(s) governing hull-less seed trait in pumpkin. Front. Plant Sci. 2022, 13, 948106. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, Q.; Chen, F.; Wang, M.; Dixon, R.A. NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 2011, 68, 1104–1114. [Google Scholar] [CrossRef]
- Yu, H.; Tian, C.; Yu, Y.; Jiao, Y. Transcriptome survey of the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana. Mol. Plant Pathol. 2016, 9, 749–752. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Priest, H.D.; Givan, S.A.; Shen, R.; Bryant, D.W.; Fox, S.E.; Wong, W.K.; Mockler, T.C. Genome–wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010, 20, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Lu, G.; Fan, D.; Zhu, C.; Li, W.; Zhao, Q.; Feng, Q.; Zhao, Y.; Guo, Y.; Li, W. Function annotation of the rice transcriptome at single–nucleotide resolution by RNA–seq. Genome Res. 2010, 20, 1238–1249. [Google Scholar] [CrossRef]
- Huang, J.; Lu, X.; Wu, H.; Xie, Y.; Peng, Q.; Gu, L.; Wu, J.; Wang, Y.; Reddy, A.S.; Dong, S. Phytophthora effectors modulate genome–wide alternative splicing of host mRNAs to reprogram plant immunity. Mol. Plant Pathol. 2020, 13, 1470–1484. [Google Scholar] [CrossRef]
- Martín, G.; Márquez, Y.; Mantica, F.; Duque, P.; Irimia, M. Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol. 2021, 22, 35. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W. Alternative splicing in plants–coming of age. Trends Plant Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef]
- Tognacca, R.S.; Rodríguez, F.S.; Aballay, F.E.; Cartagena, C.M.; Servi, L.; Petrillo, E. Alternative splicing in plants: Current knowledge and future directions for assessing the biological relevance of splice variants. J. Exp. Bot. 2022, 74, 2251–2272. [Google Scholar] [CrossRef]
- Black, D.L. Mechanisms of alternative pre–messenger RNA splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef]
- Barbazuk, W.B.; Fu, Y.; McGinnis, K.M. Genome–wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 2008, 18, 1381–1392. [Google Scholar] [CrossRef]
- Ule, J.; Blencowe, B.J. Alternative splicing regulatory networks: Functions, mechanisms, and evolution. Mol. Cell 2019, 76, 329–345. [Google Scholar] [CrossRef]
- Zhong, R.; Demura, T.; Ye, Z. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Dixon, R.A. On–off switches for secondary cell wall biosynthesis. Mol. Plant Pathol. 2012, 5, 297–303. [Google Scholar] [CrossRef]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Choi, H.; An, G. Roles of lignin biosynthesis and regulatory genes in plant development. J. Integr. Plant Biol. 2015, 57, 902–912. [Google Scholar] [CrossRef]
- Kim, W.C.; Kim, J.Y.; Ko, J.H.; Kang, H.; Han, K.H. Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol. Biol. Rep. 2014, 85, 589–599. [Google Scholar] [CrossRef]
- Rao, X.; Dixon, R.A. Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front. Plant Sci. 2018, 9, 399. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, Q.; Weng, Y. Alternative Splicing of NAC Transcription Factor Gene CmNST1 Is Associated with Naked Seed Mutation in Pumpkin, Cucurbita moschata. Genes 2023, 14, 962. https://doi.org/10.3390/genes14050962
Shen Q, Weng Y. Alternative Splicing of NAC Transcription Factor Gene CmNST1 Is Associated with Naked Seed Mutation in Pumpkin, Cucurbita moschata. Genes. 2023; 14(5):962. https://doi.org/10.3390/genes14050962
Chicago/Turabian StyleShen, Qiong, and Yiqun Weng. 2023. "Alternative Splicing of NAC Transcription Factor Gene CmNST1 Is Associated with Naked Seed Mutation in Pumpkin, Cucurbita moschata" Genes 14, no. 5: 962. https://doi.org/10.3390/genes14050962
APA StyleShen, Q., & Weng, Y. (2023). Alternative Splicing of NAC Transcription Factor Gene CmNST1 Is Associated with Naked Seed Mutation in Pumpkin, Cucurbita moschata. Genes, 14(5), 962. https://doi.org/10.3390/genes14050962





