Genetic Diversity of Durum Wheat (Triticum turgidum L. ssp. durum, Desf) Germplasm as Revealed by Morphological and SSR Markers
Abstract
:1. Introduction
2. Materials and Methods
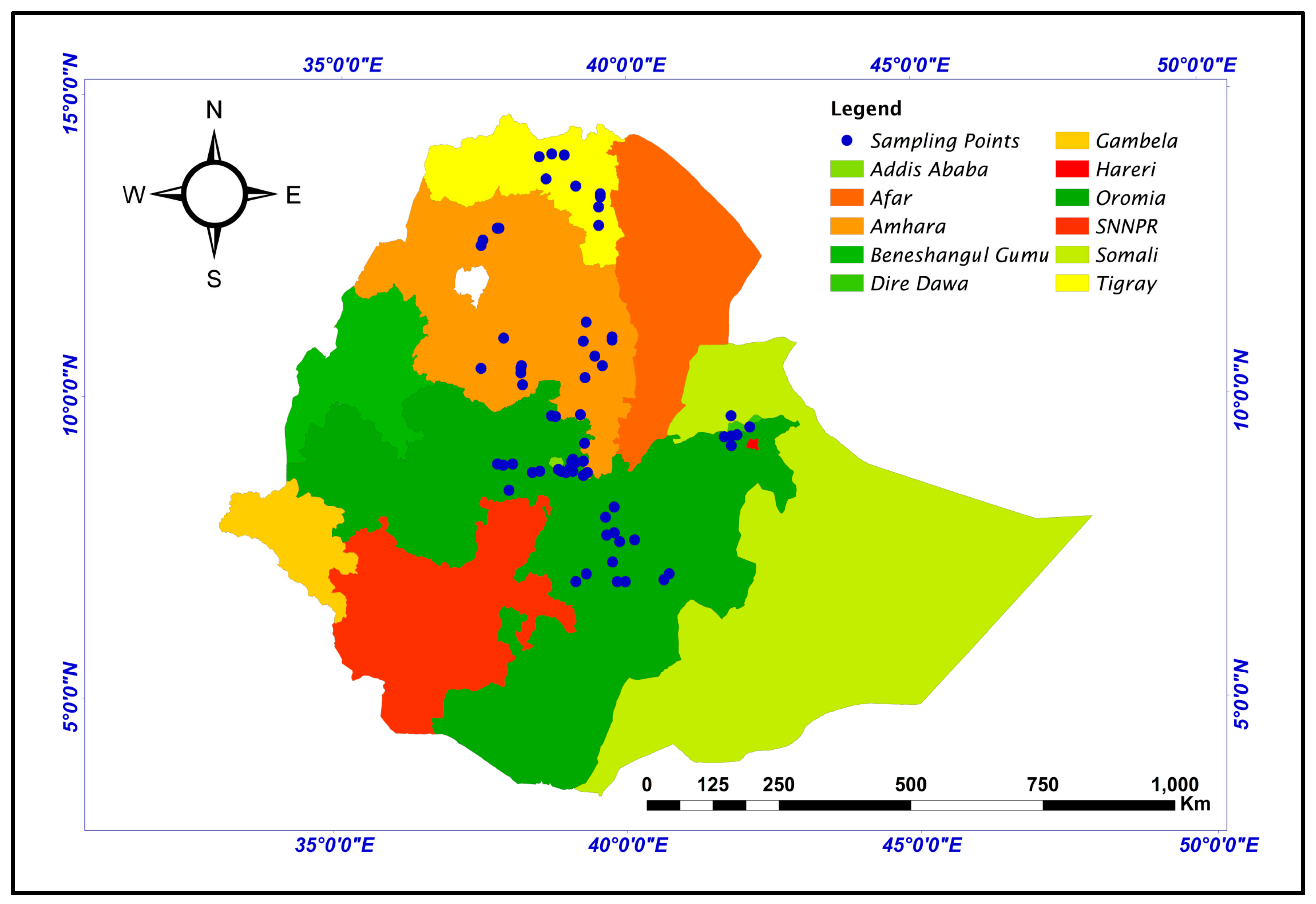
2.1. Plant Material and Field Experiment
2.2. Measurement of Morphological and Grain Quality Traits
2.3. SSR-Based Molecular Characterization
2.3.1. Genomic DNA Extraction
2.3.2. SSR Marker Selection
2.3.3. PCR Amplification
2.3.4. Gel-Electrophoresis and Data Scoring
2.4. Statistical Analysis
2.4.1. Morphological Data Analysis
2.4.2. SSR Data Analysis
3. Results
3.1. Morphological Diversity
3.1.1. Shannon Diversity Index
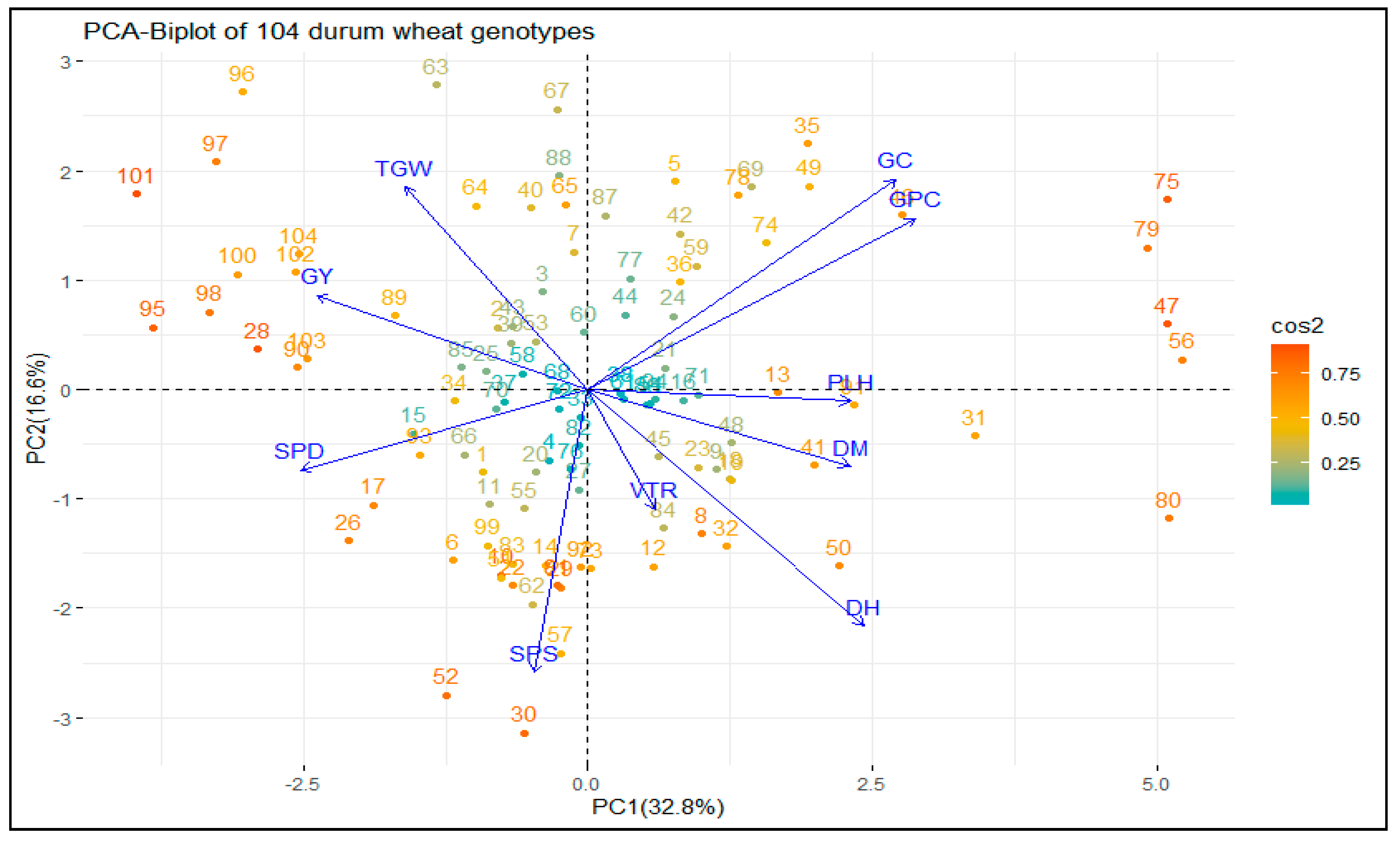
3.1.2. Principal Component Analysis (PCA)
3.2. SSR Diversity
3.2.1. Level of Polymorphism of the SSR Loci
3.2.2. Genetic Diversity across Populations
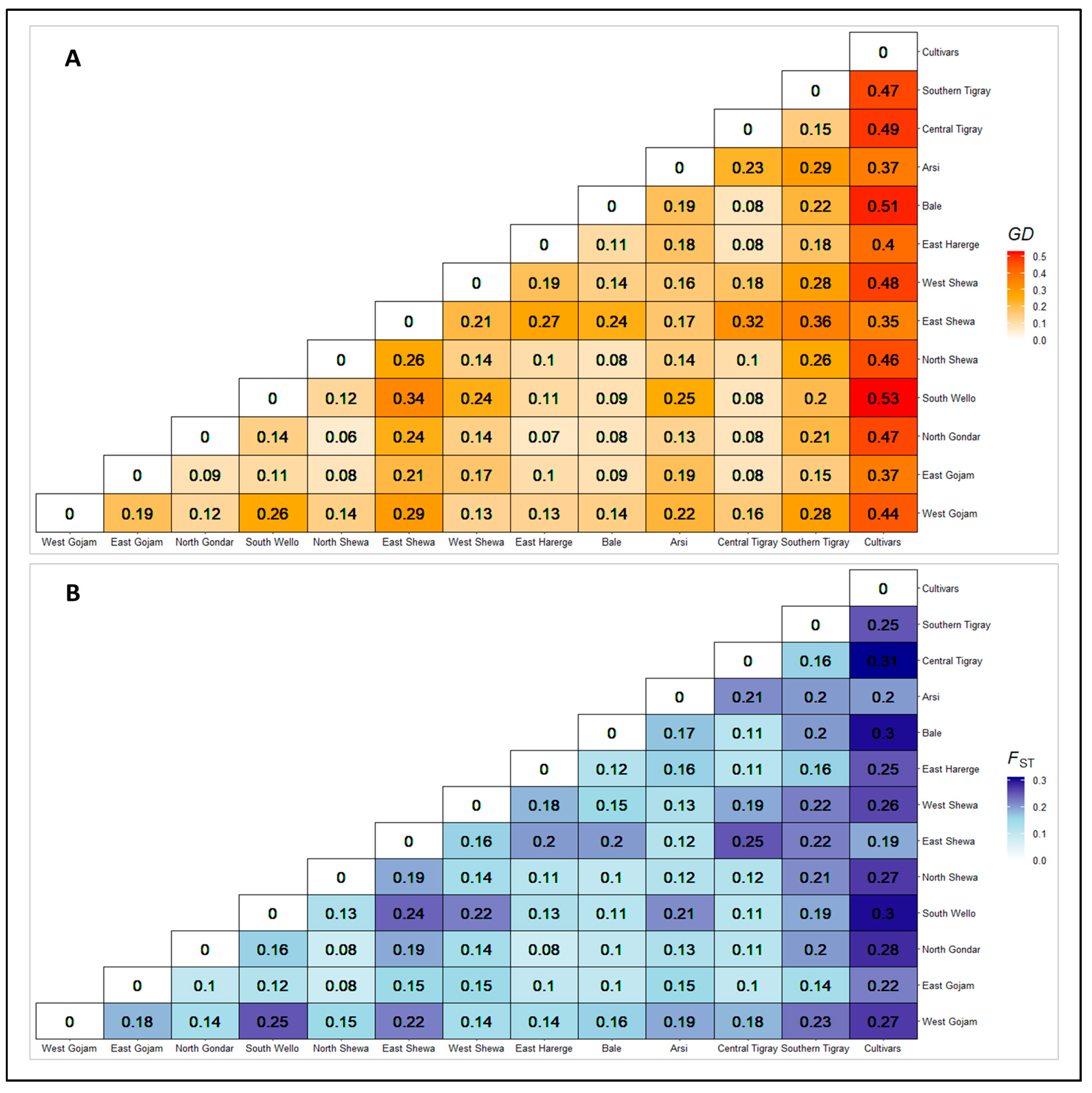
3.2.3. Genetic Distance and Differentiation between Populations
3.2.4. Analysis of Molecular Variance (AMOVA)
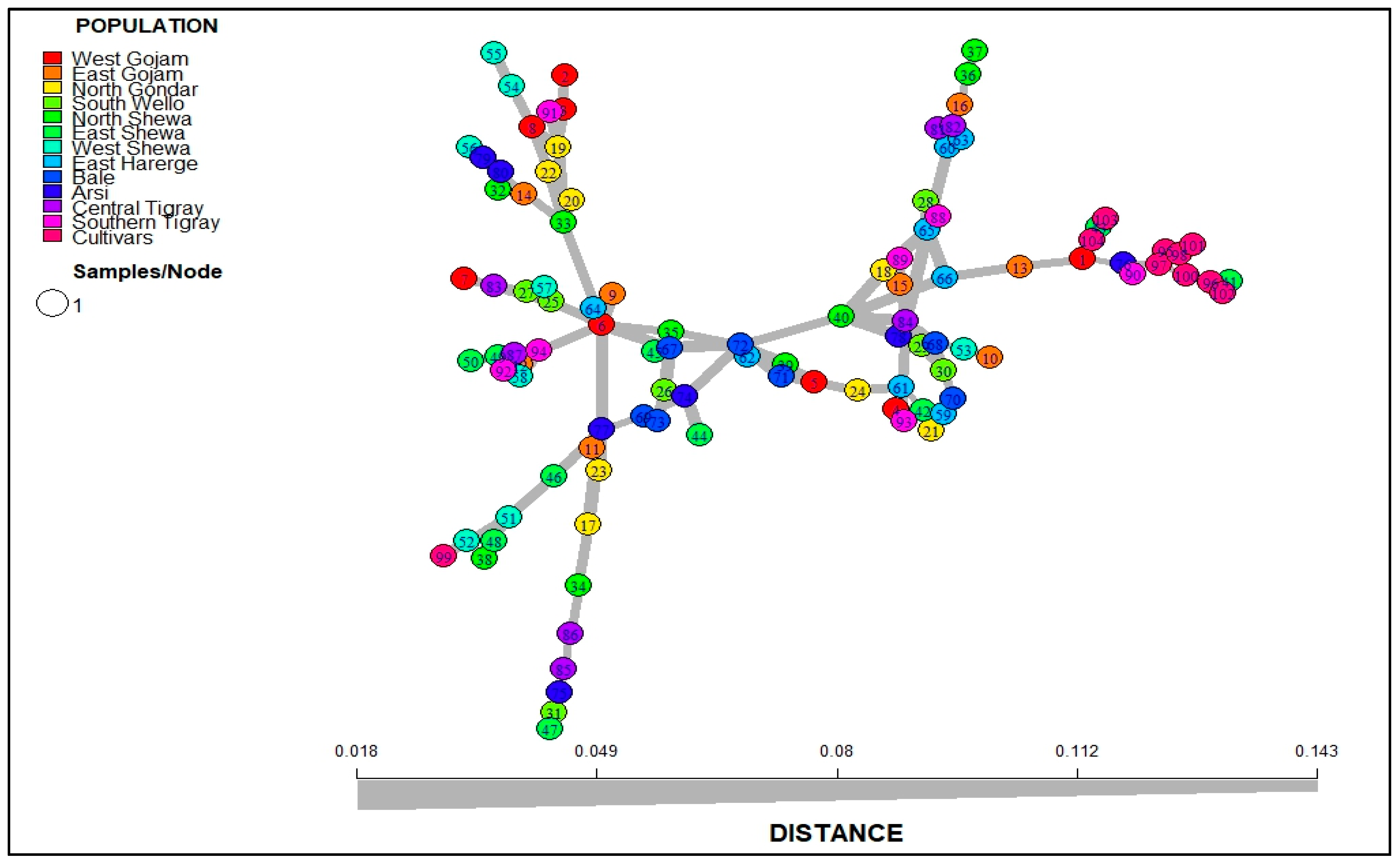
3.2.5. Minimum Spanning Network (MSN) and Discriminant Analysis of Principal Component (DAPC)
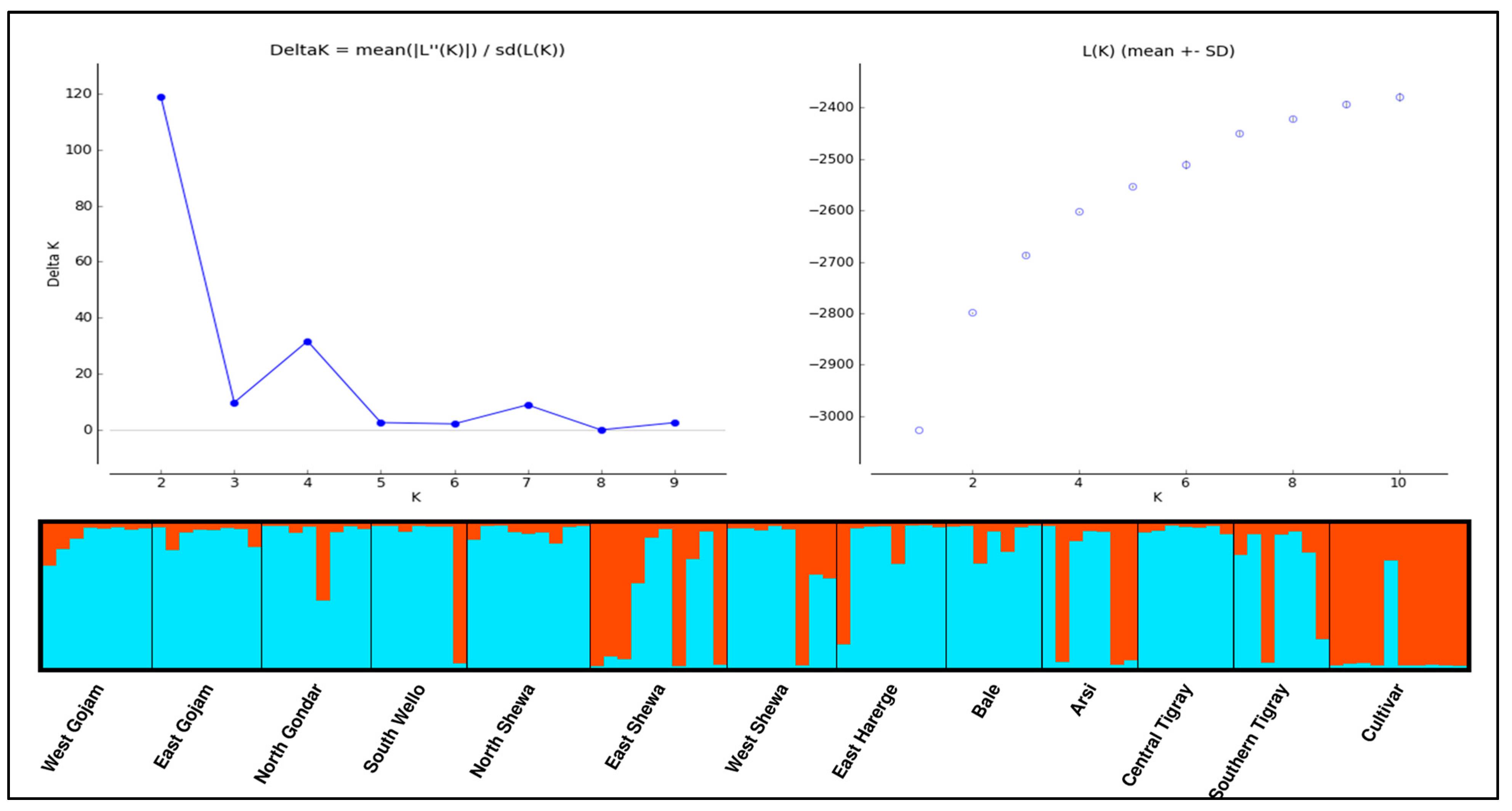
3.2.6. Population Structure
3.2.7. Relationship between Markers and Associated Traits
4. Discussion
4.1. Magnitudes of SSR Markers-Based Diversity
4.2. Genetic and Morphological Diversity within Populations
4.3. Genetic Differentiation across Populations
4.4. Genetic Relationship, Population Structure and Association with Phenotypic Traits
4.5. Relationship between SSR Markers and Associated Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hancock, J.F. Plant Evolution and the Origin of Crop Species, 2nd ed.; CABI Publishing: Cambridge, UK, 2004; Volume 123, ISBN 085199685X. [Google Scholar]
- Özkan, H.; Willcox, G.; Graner, A.; Salamini, F.; Kilian, B. Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides). Genet. Resour. Crop. Evol. 2011, 58, 11–53. [Google Scholar] [CrossRef]
- Pecetti, L.; Annicchiarico, P.; Damania, A.B. Biodiversity in a germplasm collection of durum wheat. Euphytica 1992, 60, 229–238. [Google Scholar] [CrossRef]
- Kabbaj, H.; Sall, A.T.; Al-Abdallat, A.; Geleta, M.; Amri, A.; Filali-Maltouf, A.; Belkadi, B.; Ortiz, R.; Bassi, F.M. Genetic Diversity within a Global Panel of Durum Wheat (Triticum durum) Landraces and Modern Germplasm Reveals the History of Alleles Exchange. Front. Plant Sci. 2017, 8, 1277. [Google Scholar] [CrossRef] [PubMed]
- Teklu, Y.; Hammer, K.; Huang, X.; Röder, M. Analysis of Microsatellite Diversity in Ethiopian Tetraploid Wheat Landraces. Genet. Resour. Crop Evol. 2006, 53, 1115–1126. [Google Scholar] [CrossRef]
- Asmamaw, M.; Keneni, G.; Tesfaye, K. Genetic Diversity of Ethiopian Durum Wheat (Triticum durum Desf.) Landrace Collections as Reveled by SSR Markers. Adv. Crop Sci. Technol. 2019, 7, 413. [Google Scholar] [CrossRef]
- Alemu, Y.A.; Anley, A.M.; Abebe, T.D. Genetic variability and association of traits in Ethiopian durum wheat (Triticum turgidium L. var. durum) landraces at Dabat Research Station, North Gondar. Cogent Food Agric. 2020, 6, 1778604. [Google Scholar] [CrossRef]
- Negisho, K.; Shibru, S.; Pillen, K.; Ordon, F.; Wehner, G. Genetic diversity of Ethiopian durum wheat landraces. PLoS ONE 2021, 16, e0247016. [Google Scholar] [CrossRef]
- Dagnaw, T.; Mulugeta, B.; Haileselassie, T.; Geleta, M.; Tesfaye, K. Phenotypic Variability, Heritability and Associations of Agronomic and Quality Traits in Cultivated Ethiopian Durum Wheat (Triticum turgidum L. ssp. Durum, Desf.). Agronomy 2022, 12, 1714. [Google Scholar] [CrossRef]
- Brasesco, F.; Asgedom, D.; Casari, G. Strategic Analysis and Intervention Plan for Fresh and Industrial Tomato in the AgroCommodities Procurement Zone of the Pilot Integrated AgroIndustrial Park in Central-Eastern Oromia, Ethiopia; Food and Agriculture Organization of the United Nations (FAO): Addis Ababa, Ethiopia, 2019; Volume 53, ISBN 9788578110796. [Google Scholar]
- Tsegaye, B.; Berg, T. Utilization of durum wheat landraces in East Shewa, central Ethiopia: Are home uses an incentive for on-farm conservation? Agric. Hum. Values 2007, 24, 219–230. [Google Scholar] [CrossRef]
- Uddin, M.; Boerner, A. Genetic Diversity in Hexaploid and Tetraploid Wheat Genotypes Using Microsatellite Markers. Plant Tissue Cult. Biotechnol. 2009, 18, 65–73. [Google Scholar] [CrossRef]
- Negassa, A.; Shiferaw, B.; Koo, J.; Sonder, K.; Smale, M.; Braun, H.J.; Gbegbelegbe, S.; Guo, Z.; Hodson, D.; Wood, S.; et al. The Potential for Wheat Production in Africa: Analysis of Biophysical Suitability and Economic Profitability; CIMMYT: Mexico City, Mexico, 2013; Volume 53. [Google Scholar]
- Wheat Atlas—Ethioian Wheat Varieties. Available online: http://wheatatlas.org/(X(1)S(vczqlbkabx0vcgd4mlpdtvxo))/country/varieties/ETH/0?AspxAutoDetectCookieSupport=1 (accessed on 19 December 2022).
- Tehseen, M.M.; Tonk, F.A.; Tosun, M.; Istipliler, D.; Amri, A.; Sansaloni, C.P.; Kurtulus, E.; Mubarik, M.S.; Nazari, K. Exploring the Genetic Diversity and Population Structure of Wheat Landrace Population Conserved at ICARDA Genebank. Front. Genet. 2022, 13, 900572. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.K.; Pandey, A.; Thomas, G.; Akkaya, M.S.; Kayis, S.A.; Ozsensoy, Y.; Hamurcu, M.; Gezgin, S.; Topal, A.; Hakki, E.E. Genetic Diversity and Population Structure of Wheat in India and Turkey. AoB Plants 2015, 7, plv083. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Langridge, P. Breeding Technologies to Increase Crop Production in a Changing World. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Broccanello, C.; Bellin, D.; DalCorso, G.; Furini, A.; Taranto, F. Genetic Approaches to Exploit Landraces for Improvement of Triticum turgidum ssp. durum in the Age of Climate Change. Front. Plant Sci. 2023, 14, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Tan, B.; Liu, H.; Zhu, W.; Xu, L.; Wang, Y.; Fan, X.; Sha, L.; Zhang, H.; Zeng, J.; et al. Genetic Diversity and Population Structure of Asian and European Common Wheat Accessions Based on Genotyping-By-Sequencing. Front. Genet. 2020, 11, 580782. [Google Scholar] [CrossRef]
- Hinge, V.R.; Shaikh, I.M.; Chavhan, R.L.; Deshmukh, A.S.; Shelake, R.M.; Ghuge, S.A.; Dethe, A.M.; Suprasanna, P.; Kadam, U.S. Assessment of Genetic Diversity and Volatile Content of Commercially Grown Banana (Musa spp.) Cultivars. Sci. Rep. 2022, 12, 7979. [Google Scholar] [CrossRef]
- Upadhyay, A.; Kadam, U.S.; Chacko, P.M.; Aher, L.; Karibasappa, G.S. Microsatellite Analysis to Differentiate Clones of Thompson Seedless Grapevine. Indian J. Hortic. 2010, 67, 260–263. [Google Scholar]
- Upadhyay, A.; Kadam, U.S.; Chacko, P.; Karibasappa, G.S. Microsatellite and RAPD Analysis of Grape (Vitis spp.) Accessions and Identification of Duplicates/Misnomers in Germplasm Collection. Indian J. Hortic. 2010, 67, 8–15. [Google Scholar]
- Paux, E.; Sourdille, P.; Mackay, I.; Feuillet, C. Sequence-Based Marker Development in Wheat: Advances and Applications to Breeding. Biotechnol. Adv. 2012, 30, 1071–1088. [Google Scholar] [CrossRef]
- Teshome, A.; Bryngelsson, T.; Dagne, K.; Geleta, M. Assessment of Genetic Diversity in Ethiopian Field Pea (Pisum sativum L.) Accessions with Newly Developed EST-SSR Markers. BMC Genet. 2015, 16, 102. [Google Scholar] [CrossRef]
- Röder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixier, M.H.; Leroy, P.; Ganal, M.W. A Microsatellite Map of Wheat. Genetics 1998, 149, 2007–2023. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Gupta, V.K.; Misra, A.K.; Modi, D.R.; Pandey, B.K. Potential of Molecular Markers in Plant Biotechnology. Plant Omics J. 2009, 2, 141–162. [Google Scholar]
- Alamerew, S.; Chebotar, S.; Huang, X.; Röder, M.; Börner, A. Genetic Diversity in Ethiopian Hexaploid and Tetraploid Wheat Germplasm Assessed by Microsatellite Markers. Genet. Resour. Crop Evol. 2004, 51, 559–567. [Google Scholar] [CrossRef]
- Mondini, L.; Farina, A.; Porceddu, E.; Pagnotta, M.A. Analysis of Durum Wheat Germplasm Adapted to Different Climatic Conditions. Ann. Appl. Biol. 2010, 156, 211–219. [Google Scholar] [CrossRef]
- Haile, J.K.; Hammer, K.; Badebo, A.; Nachit, M.M.; Röder, M.S. Genetic Diversity Assessment of Ethiopian Tetraploid Wheat Landraces and Improved Durum Wheat Varieties Using Microsatellites and Markers Linked with Stem Rust Resistance. Genet. Resour. Crop Evol. 2013, 60, 513–527. [Google Scholar] [CrossRef]
- International Board for Plant Genetic Resources (IBPGR). Descriptors for Wheat (Revised); IBPGR: Rome, Italy, 1985. [Google Scholar]
- Hailu, F.; Johansson, E.; Merker, A. Patterns of Phenotypic Diversity for Phenologic and Qualitative Traits in Ethiopian Tetraploid Wheat Germplasm. Genet. Resour. Crop Evol. 2010, 57, 781–790. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of AOAC International, 20th ed.; George, W., Latimer, J., Eds.; AOAC International: Rockville, MD, USA, 2016; ISBN 0-935584-87-0. [Google Scholar]
- Abdie, O.; Dagne, K.; Feyissa, T.; Tesfaye, K.; Durai, J.; Hyder, M.Z. Genetic Diversity, Population Structure, and Gene Flow Analysis of Lowland Bamboo [Oxytenanthera abyssinica (A. Rich.) Munro] in Ethiopia. Ecol. Evol. 2020, 10, 11217–11236. [Google Scholar] [CrossRef]
- Zhang, W.; Chao, S.; Manthey, F.; Chicaiza, O.; Brevis, J.C.; Echenique, V.; Dubcovsky, J. QTL Analysis of Pasta Quality Using a Composite Microsatellite and SNP Map of Durum Wheat. Theor. Appl. Genet. 2008, 117, 1361–1377. [Google Scholar] [CrossRef]
- Ramya, P.; Chaubal, A.; Kulkarni, K.; Gupta, L.; Kadoo, N.; Dhaliwal, H.S.; Chhuneja, P.; Lagu, M.; Gupta, V. QTL Mapping of 1000-Kernel Weight, Kernel Length, and Kernel Width in Bread Wheat (Triticum aestivum L.). J. Appl. Genet. 2010, 51, 421–429. [Google Scholar] [CrossRef]
- MacCaferri, M.; Sanguineti, M.C.; Demontis, A.; El-Ahmed, A.; Del Moral, L.G.; Maalouf, F.; Nachit, M.; Nserallah, N.; Ouabbou, H.; Rhouma, S.; et al. Association Mapping in Durum Wheat Grown across a Broad Range of Water Regimes. J. Exp. Bot. 2011, 62, 409–438. [Google Scholar] [CrossRef]
- Blanco, A.; Mangini, G.; Giancaspro, A.; Giove, S.; Colasuonno, P.; Simeone, R.; Signorile, A.; De Vita, P.; Mastrangelo, A.M.; Cattivelli, L.; et al. Relationships between Grain Protein Content and Grain Yield Components through Quantitative Trait Locus Analyses in a Recombinant Inbred Line Population Derived from Two Elite Durum Wheat Cultivars. Mol. Breed. 2012, 30, 79–92. [Google Scholar] [CrossRef]
- Moradi, N.; Badakhshan, H.; Mohammadzadeh, H.; Zakeri, M.R.; Mirzaghaderi, G. Assessment of Genetic Diversity and Identification of SSR Markers Associated with Grain Iron Content in Iranian Prevalent Wheat Genotypes. J. Plant Mol. Breed. 2014, 2, 64–73. [Google Scholar] [CrossRef]
- Amallah, L.; Taghouti, M.; Rhrib, K.; Gaboun, F.; Arahou, M.; Hassikou, R.; Diria, G. Validation of Simple Sequence Repeats Associated with Quality Traits in Durum Wheat. J. Crop Sci. Biotechnol. 2016, 19, 137–150. [Google Scholar] [CrossRef]
- Prasad, M.; Kumar, N.; Kulwal, P.L.; Röder, M.S.; Balyan, H.S.; Dhaliwal, H.S.; Gupta, P.K. QTL Analysis for Grain Protein Content Using SSR Markers and Validation Studies Using NILs in Bread Wheat. Theor. Appl. Genet. 2003, 106, 659–667. [Google Scholar] [CrossRef]
- Eticha, F.; Bekele, E.; Belay, G.; Börner, A. Phenotypic Diversity in Tetraploid Wheats Collected from Bale and Weallo Regions of Ethiopia. Plant Genet. Resour. 2005, 3, 35–43. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses, R Package v. 1.0.7. 2020. Available online: https://cran.r-project.org/web/packages/factoextra/index.html (accessed on 4 May 2023).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-7. 2020. Available online: https://www.researchgate.net/publication/346579465_vegan_community_ecology_package_version_25-7_November_2020 (accessed on 4 May 2023).
- Paradis, E. Pegas: An R Package for Population Genetics with an Integrated-Modular Approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, R. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMaker: An Integrated Analysis Environment for Genetic Maker Analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Goudet, J.; Jombart, T.; Kamvar, Z.N.; Archer, E.; Hardy, O. Hierfstat: Estimation and Tests of Hierarchical F-Statistics. R Package Version 0.5-11. 2022. Available online: https://cran.r-project.org/web/packages/hierfstat/hierfstat.pdf (accessed on 4 May 2023).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Volume 35, ISBN 978-0-387-98140-6. [Google Scholar]
- Bruvo, R.; Michiels, N.K.; D’Souza, T.G.; Schulenburg, H. A Simple Method for the Calculation of Microsatellite Genotype Distances Irrespective of Ploidy Level. Mol. Ecol. 2004, 13, 2101–2106. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R Package for Genetic Analysis of Populations with Clonal, Partially Clonal, and/or Sexual Reproduction. PeerJ 2014, 2014, e281. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Wen, X.; Falush, D. Documentation for Structure Software: Version 2.3; University of Chicago: Chicago, IL, USA, 2010; pp. 1–37. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. Structure Harvester: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A Program for Identifying Clustering Modes and Packaging Population Structure Inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Sicilia, A.; Anastasi, U.; Bizzini, M.; Montemagno, S.; Nicotra, C.; Blangiforti, S.; Spina, A.; Cosentino, S.L.; Lo Piero, A.R. Genetic and Morpho-Agronomic Characterization of Sicilian Tetraploid Wheat Germplasm. Plants 2022, 11, 130. [Google Scholar] [CrossRef]
- Christov, N.K.; Tsonev, S.; Dragov, R.; Taneva, K.; Bozhanova, V.; Todorovska, E.G. Genetic Diversity and Population Structure of Modern Bulgarian and Foreign Durum Wheat Based on Microsatellite and Agronomic Data. Biotechnol. Biotechnol. Equip. 2022, 36, 637–652. [Google Scholar] [CrossRef]
- Marzario, S.; Logozzo, G.; David, J.L.; Zeuli, P.S.; Gioia, T. Molecular Genotyping (SSR) and Agronomic Phenotyping for Utilization of Durum Wheat (Triticum durum Desf.) Ex Situ Collection from Southern Italy: A Combined Approach Including Pedigreed Varieties. Genes 2018, 9, 465. [Google Scholar] [CrossRef]
- Ouaja, M.; Bahri, B.A.; Aouini, L.; Ferjaoui, S.; Medini, M.; Marcel, T.C.; Hamza, S. Morphological Characterization and Genetic Diversity Analysis of Tunisian Durum Wheat (Triticum Turgidum var. durum) Accessions. BMC Genom. Data 2021, 22, 3. [Google Scholar] [CrossRef]
- Soriano, J.M.; Villegas, D.; Aranzana, M.J.; García Del Moral, L.F.; Royo, C. Genetic Structure of Modern Durum Wheat Cultivars and Mediterranean Landraces Matches with Their Agronomic Performance. PLoS ONE 2016, 11, e0160983. [Google Scholar] [CrossRef]
- Mengistu, D.K.; Kiros, A.Y.; Pè, M.E. Phenotypic Diversity in Ethiopian Durum Wheat (Triticum Turgidum var. durum) Landraces. Crop J. 2015, 3, 190–199. [Google Scholar] [CrossRef]
- Arora, A.; Kundu, S.; Dilbaghi, N.; Sharma, I.; Tiwari, R. Population Structure and Genetic Diversity among Indian Wheat Varieties Using Microsatellite (SSR) Markers. Aust. J. Crop Sci. 2014, 8, 1281–1289. [Google Scholar]
- Slim, A.; Piarulli, L.; Kourda, H.C.; Rouaissi, M.; Robbana, C.; Chaabane, R.; Pignone, D.; Montemurro, C. Genetic Structure Analysis of a Collection of Tunisian Durum Wheat Germplasm. Int. J. Mol. Sci. 2019, 20, 3362. [Google Scholar] [CrossRef] [PubMed]
- Quinn, G.P.; Keough, M. Experimental Design and Data Analysis for Biologists Anessential; Cambridge University Press: New York, NY, USA, 2002. [Google Scholar]
- Kirouani, A.; Henkrar, F.; Udupa, S.M.; Boukhalfoun, L.; Bouzerzour, H. Genetic Diversity in Algerian Durum Wheat Varieties (Triticum Turgidum L. var. durum) Using Microsatellite Markers. Biosci. J. 2018, 34, 1575–1583. [Google Scholar] [CrossRef]
- Gesesse, C.A.; Nigir, B.; de Sousa, K.; Gianfranceschi, L.; Gallo, G.R.; Poland, J.; Kidane, Y.G.; Abate Desta, E.; Fadda, C.; Pè, M.E.; et al. Genomics-Driven Breeding for Local Adaptation of Durum Wheat Is Enhanced by Farmers’ Traditional Knowledge. Proc. Natl. Acad. Sci. USA 2023, 120, e2205774119. [Google Scholar] [CrossRef]
- Branković, G.; Dodig, D.; Pajić, V.; Kandić, V.; Knežević, D.; Đurić, N.; Živanović, T. Triticum aestivum Ir T. durum Genetiniai Parametrai Kokybės Technologinėms Savybėms Serbijoje. Zemdirbyste 2018, 105, 39–48. [Google Scholar] [CrossRef]
- Fiore, M.C.; Mercati, F.; Spina, A.; Blangiforti, S.; Venora, G.; Dell’Acqua, M.; Lupini, A.; Preiti, G.; Monti, M.; Pè, M.E.; et al. High-Throughput Genotype, Morphology, and Quality Traits Evaluation for the Assessment of Genetic Diversity of Wheat Landraces from Sicily. Plants 2019, 8, 116. [Google Scholar] [CrossRef]
- Mengistu, D.; Pè, M. Revisiting the Ignored Ethiopian Durum Wheat (Triticum Turgidum var. durum) Landraces for Genetic Diversity Exploitation in Future Wheat Breeding Programs. J. Plant Breed. Crop Sci. 2016, 8, 45–59. [Google Scholar] [CrossRef]
- Kehel, Z.; Garcia-Ferrer, A.; Nachit, M.M. Using Bayesian and Eigen Approaches to Study Spatial Genetic Structure of Moroccan and Syrian Durum Wheat Landraces. Am. J. Mol. Biol. 2013, 3, 17–31. [Google Scholar] [CrossRef]


| SSR | Chr. Position (cM) | Ta | Primer Sequence (5′-3′) | Associated Trait | References | |
|---|---|---|---|---|---|---|
| Xwmc24 | 1AS (35.9) | 52 | Fw | GTGAGCAATTTTGATTATACTG | TKW | [34] |
| Rv | TACCCTGATGCTGTAATATGTG | TW, GY | [36] | |||
| Xbarc240 | 1BL (46.4) | 60 | Fw | AGAGGACGCTGAGAACTTTAGAGAA | TW, YP | [34] |
| Rv | GCGATCTTTGTAATGCATGGTGAAC | |||||
| Xgwm294 | 2AL (118.3) | 61 | Fw | GGATTGGAGTTAAGAGAGAACCG | DH, PLH, TKW | [36] |
| Rv | GCAGAGTGATCAATGCCAGA | |||||
| Xgwm47.1 | 2BL (116.8) | 61 | Fw | TTGCTACCATGCATGACCAT | PLH, TKW | [36] |
| Rv | TTCACCTCGATTGAGGTCCT | |||||
| Xbarc12 | 3AS (12.8) | 56 | Fw | CGACAGAGTGATCACCCAAATATAA | TKW | [36] |
| Rv | CATCGGTCTAATTGTCAATGTA | |||||
| Xgwm493 | 3BS (10.7) | 60 | Fw | TTCCCATAACTAAAACCGCG | DH, PLH, TW, GY | [36] |
| Rv | GGAACATCATTTCTGGACTTTG | GPC | [37] | |||
| Xbarc155 | 4AL (18.2) | 59 | Fw | GCGAGTATTGACGTCTTATTTTTGAA | PLH, TW | [36] |
| Rv | GCGTCATGAATTCTAACAATGTGCATA | |||||
| Xwmc617 | 4BS (6.0) | 59 | Fw | CCACTAGGAAGAAGGGGAAACT | TKW | [35,38] |
| Rv | ATCTGGATTACTGGCCAACTGT | GNS | [37] | |||
| Xgwm513 | 4BL (38.1) | 58 | Fw | ATCCGTAGCACCTACTGGTCA | GPC | [40] |
| Rv | GGTCTGTTCATGCCACATTG | |||||
| Xgwm120 | 5AS (26.0) | 58 | Fw | GATCCACCTTCCTCTCTCTC | GPC, GL | [34] |
| Rv | GATTATACTGGTGCCGAAAC | |||||
| Xgwm371 | 5BL (58.0) | 63 | Fw | GACCAAGATATTCAAACTGGCC | TKW, TW | [35,39] |
| Rv | AGCTCAGCTTGCTTGGTACC | |||||
| Xwmc256 | 6AL (65.5) | 61 | Fw | CCAAATCTTCGAACAAGAACCC | DH, GY | [36] |
| Rv | ACCGATCGATGGTGTATACTGA | |||||
| Xbarc178 | 6BL (50.0) | 59 | Fw | GCGTATTAGCAAAACAGAAGTGAG | TKW | [35] |
| Rv | GCGACTAGTACGAACACCACAAAA | |||||
| Xgwm46 | 7BS (73.9) | 60 | Fw | GCACGTGAATGGATTGGAC | GPC | [39] |
| Rv | TGACCCAATAGTGGTGGTCA | GY | [36] | |||
| Grouping Methods | No. | DH | DM | SPS | PLH | TKW | GY | GL | GPC | SPD | VTR | Mean | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All genotypes as a single group | 104 | 0.96 | 0.42 | 0.84 | 0.77 | 0.84 | 0.8 | 0.83 | 0.87 | 0.93 | 0.54 | 0.78 | ||
| Population | 1 | West Gojam | 8 | 0.91 | 0.41 | 0.87 | 0.82 | 0.71 | 0.84 | 0.77 | 0.91 | 0.82 | 0.00 | 0.71 |
| 2 | East Gojam | 8 | 0.89 | 0.21 | 0.89 | 0.56 | 0.68 | 0.83 | 0.83 | 0.80 | 0.97 | 0.67 | 0.73 | |
| 3 | North Gondar | 8 | 0.92 | 0.44 | 0.70 | 0.63 | 0.63 | 0.75 | 0.67 | 0.73 | 0.94 | 0.64 | 0.71 | |
| 4 | South Wello | 7 | 0.80 | 0.48 | 0.84 | 0.58 | 0.71 | 0.67 | 0.86 | 0.77 | 0.54 | 0.55 | 0.68 | |
| 5 | North Shewa | 9 | 0.98 | 0.57 | 0.76 | 0.71 | 0.96 | 0.58 | 0.85 | 0.90 | 0.70 | 0.55 | 0.76 | |
| 6 | East Shewa | 10 | 0.92 | 0.28 | 0.79 | 0.76 | 0.64 | 0.76 | 0.68 | 0.80 | 0.76 | 0.57 | 0.70 | |
| 7 | West Shewa | 8 | 0.97 | 0.54 | 0.82 | 0.75 | 0.59 | 0.43 | 0.97 | 0.94 | 0.77 | 0.54 | 0.73 | |
| 8 | East Harerge | 8 | 0.94 | 0.31 | 0.98 | 0.76 | 0.83 | 0.79 | 0.73 | 0.82 | 0.84 | 0.00 | 0.70 | |
| 9 | Bale | 7 | 0.97 | 0.23 | 0.65 | 0.83 | 0.63 | 0.88 | 0.70 | 0.76 | 0.96 | 0.00 | 0.66 | |
| 10 | Arsi | 7 | 0.74 | 0.37 | 0.78 | 0.69 | 0.75 | 0.78 | 0.69 | 0.82 | 0.81 | 0.37 | 0.68 | |
| 11 | Central Tigray | 7 | 0.97 | 0.51 | 0.92 | 0.59 | 0.61 | 0.58 | 0.60 | 0.87 | 0.77 | 0.80 | 0.72 | |
| 12 | Southern Tigray | 7 | 0.65 | 0.51 | 0.58 | 0.83 | 0.76 | 0.84 | 0.69 | 0.63 | 0.24 | 0.73 | 0.65 | |
| 13 | Cultivars | 10 | 0.78 | 0.30 | 0.90 | 0.64 | 0.79 | 0.74 | 0.84 | 0.92 | 0.82 | 0.38 | 0.71 | |
| Mean | 0.89 | 0.40 | 0.81 | 0.71 | 0.72 | 0.73 | 0.77 | 0.82 | 0.78 | 0.45 | 0.71 | |||
| Region | 1 | Amhara | 34 | 0.93 | 0.38 | 0.78 | 0.74 | 0.67 | 0.80 | 0.75 | 0.80 | 0.83 | 0.53 | 0.72 |
| 2 | Oromia | 46 | 0.94 | 0.41 | 0.90 | 0.75 | 0.84 | 0.77 | 0.89 | 0.92 | 0.90 | 0.46 | 0.78 | |
| 3 | Tigray | 14 | 0.89 | 0.45 | 0.87 | 0.65 | 0.69 | 0.72 | 0.69 | 0.85 | 0.79 | 0.64 | 0.72 | |
| Mean | 0.89 | 0.39 | 0.86 | 0.70 | 0.75 | 0.76 | 0.79 | 0.87 | 0.84 | 0.5 | 0.74 | |||
| Altitudinal class (m.a.s.l) | 1 | <2000 | 10 | 0.89 | 0.49 | 0.94 | 0.75 | 0.81 | 0.69 | 0.86 | 0.91 | 0.83 | 0.38 | 0.76 |
| 2 | 2001–2400 | 42 | 0.95 | 0.39 | 0.80 | 0.67 | 0.75 | 0.78 | 0.83 | 0.88 | 0.84 | 0.64 | 0.75 | |
| 3 | 2401–2800 | 31 | 0.91 | 0.38 | 0.88 | 0.75 | 0.76 | 0.81 | 0.81 | 0.82 | 0.85 | 0.36 | 0.73 | |
| 4 | >2800 | 11 | 0.94 | 0.46 | 0.86 | 0.79 | 0.72 | 0.73 | 0.83 | 0.95 | 0.91 | 0.43 | 0.76 | |
| Mean | 0.92 | 0.43 | 0.87 | 0.74 | 0.76 | 0.75 | 0.83 | 0.89 | 0.86 | 0.45 | 0.75 | |||
| SSR | Expected Size (bp) | Range of Fragments (bp) | Na | MAF | h | I | Ho | He | Fst | Fis | Fit | Nm | PIC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xwmc24 | 122 | 116–152 | 4.00 | 0.70 | 0.48 | 0.68 | 0.36 | 0.39 | 0.17 | 0.06 | 0.23 | 1.19 | 0.45 |
| Xbarc240 | 267 | 247–292 | 6.00 | 0.40 | 0.73 | 1.08 | 0.08 | 0.61 | 0.15 | 0.87 | 0.89 | 1.37 | 0.69 |
| Xgwm294 | 102 | 88–118 | 5.00 | 0.37 | 0.74 | 1.06 | 0.25 | 0.60 | 0.19 | 0.58 | 0.65 | 1.08 | 0.70 |
| Xgwm47.1 | 166 | 128–178 | 5.00 | 0.70 | 0.48 | 0.64 | 0.13 | 0.36 | 0.21 | 0.63 | 0.71 | 0.94 | 0.45 |
| Xbarc12 | 200 | 212–257 | 5.00 | 0.61 | 0.57 | 0.85 | 0.25 | 0.49 | 0.14 | 0.49 | 0.56 | 1.50 | 0.53 |
| Xgwm493 | 179 | 145–179 | 4.00 | 0.55 | 0.58 | 0.86 | 0.66 | 0.53 | 0.08 | −0.24 | −0.14 | 3.03 | 0.51 |
| Xbarc155 | 182 | 180–204 | 4.00 | 0.65 | 0.52 | 0.61 | 0.02 | 0.37 | 0.26 | 0.95 | 0.97 | 0.73 | 0.48 |
| Xwmc617 | 199 | 196–220 | 4.00 | 0.60 | 0.56 | 0.72 | 0.05 | 0.44 | 0.20 | 0.88 | 0.90 | 1.01 | 0.51 |
| Xgwm513 | 152 | 122–144 | 2.00 | 0.65 | 0.45 | 0.63 | 0.69 | 0.44 | 0.04 | −0.59 | −0.53 | 6.36 | 0.35 |
| Xgwm120 | 162 | 128–153 | 3.00 | 0.61 | 0.56 | 0.73 | 0.17 | 0.44 | 0.19 | 0.62 | 0.69 | 1.10 | 0.50 |
| Xgwm371 | 191 | 126–179 | 5.00 | 0.49 | 0.69 | 0.95 | 0.45 | 0.54 | 0.21 | 0.17 | 0.34 | 0.95 | 0.65 |
| Xwmc256 | 117 | 111–117 | 2.00 | 0.89 | 0.19 | 0.27 | 0.10 | 0.17 | 0.12 | 0.37 | 0.44 | 1.91 | 0.17 |
| Xbarc178 | 266 | 274–296 | 3.00 | 0.55 | 0.56 | 0.77 | 0.00 | 0.48 | 0.14 | 1.00 | 1.00 | 1.54 | 0.48 |
| Xgwm46 | 187 | 152–179 | 4.00 | 0.38 | 0.68 | 0.89 | 0.02 | 0.54 | 0.20 | 0.97 | 0.97 | 0.99 | 0.62 |
| Mean | 4.00 | 0.58 | 0.56 | 0.77 | 0.23 | 0.46 | 0.16 | 0.48 | 0.55 | 1.69 | 0.50 | ||
| Popuation | N | Na | Ne | I | Ho | He | F | PPL (%) |
|---|---|---|---|---|---|---|---|---|
| West Gojam | 8.00 | 2.29 | 1.87 | 0.65 | 0.21 | 0.42 | 0.48 | 93 |
| East Gojam | 8.00 | 2.71 | 2.14 | 0.79 | 0.21 | 0.48 | 0.52 | 93 |
| North Gondar | 8.00 | 2.36 | 1.80 | 0.65 | 0.24 | 0.41 | 0.39 | 100 |
| South Wello | 7.00 | 2.57 | 1.86 | 0.68 | 0.27 | 0.41 | 0.32 | 93 |
| North Shewa | 9.00 | 2.57 | 1.97 | 0.72 | 0.25 | 0.44 | 0.42 | 93 |
| East Shewa | 10.00 | 3.29 | 2.39 | 0.95 | 0.22 | 0.55 | 0.58 | 100 |
| West Shewa | 8.00 | 2.86 | 2.02 | 0.79 | 0.21 | 0.46 | 0.56 | 100 |
| East Harerge | 8.00 | 2.71 | 1.95 | 0.73 | 0.15 | 0.43 | 0.65 | 93 |
| Bale | 7.00 | 2.57 | 1.86 | 0.68 | 0.20 | 0.40 | 0.43 | 93 |
| Arsi | 7.00 | 2.86 | 2.28 | 0.87 | 0.27 | 0.52 | 0.42 | 100 |
| Central Tigray | 7.00 | 2.36 | 1.79 | 0.62 | 0.17 | 0.38 | 0.51 | 93 |
| Southern Tigray | 7.00 | 2.79 | 2.18 | 0.81 | 0.23 | 0.48 | 0.51 | 86 |
| Cultivars | 10.00 | 3.57 | 2.68 | 1.04 | 0.37 | 0.58 | 0.36 | 93 |
| Mean | 8.00 | 2.73 | 2.06 | 0.77 | 0.23 | 0.46 | 0.47 | 95 |
| ± SE | 0.08 | 0.07 | 0.05 | 0.03 | 0.02 | 0.01 | 0.04 | 1 |
| Grouping Units | Source | df | SS | MS | Estimated Variance | Percent Variance | Fst | p | Nm |
|---|---|---|---|---|---|---|---|---|---|
| Populations | Among | 12 | 276.62 | 23.05 | 1.51 | 12% | 0.12 | 0.001 | 1.83 |
| Within | 91 | 1002.97 | 11.02 | 11.02 | 88% | ||||
| Total | 103 | 1279.59 | 12.53 | 100% | |||||
| Regions | Among | 2 | 43.06 | 21.53 | 0.35 | 3% | 0.03 | 0.003 | 8.37 |
| Within | 91 | 1057.39 | 11.62 | 11.62 | 97% | ||||
| Total | 93 | 1100.45 | 11.97 | 100% | |||||
| Altitudes | Among | 3 | 58.29 | 19.43 | 0.38 | 3% | 0.03 | 0.008 | 7.70 |
| Within | 90 | 1042.16 | 11.58 | 11.58 | 97% | ||||
| Total | 93 | 1100.45 | 11.96 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dagnaw, T.; Mulugeta, B.; Haileselassie, T.; Geleta, M.; Ortiz, R.; Tesfaye, K. Genetic Diversity of Durum Wheat (Triticum turgidum L. ssp. durum, Desf) Germplasm as Revealed by Morphological and SSR Markers. Genes 2023, 14, 1155. https://doi.org/10.3390/genes14061155
Dagnaw T, Mulugeta B, Haileselassie T, Geleta M, Ortiz R, Tesfaye K. Genetic Diversity of Durum Wheat (Triticum turgidum L. ssp. durum, Desf) Germplasm as Revealed by Morphological and SSR Markers. Genes. 2023; 14(6):1155. https://doi.org/10.3390/genes14061155
Chicago/Turabian StyleDagnaw, Temesgen, Behailu Mulugeta, Teklehaimanot Haileselassie, Mulatu Geleta, Rodomiro Ortiz, and Kassahun Tesfaye. 2023. "Genetic Diversity of Durum Wheat (Triticum turgidum L. ssp. durum, Desf) Germplasm as Revealed by Morphological and SSR Markers" Genes 14, no. 6: 1155. https://doi.org/10.3390/genes14061155








