Localization of Insertion Sequences in Plasmids for L-Cysteine Production in E. coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Deep Sequencing
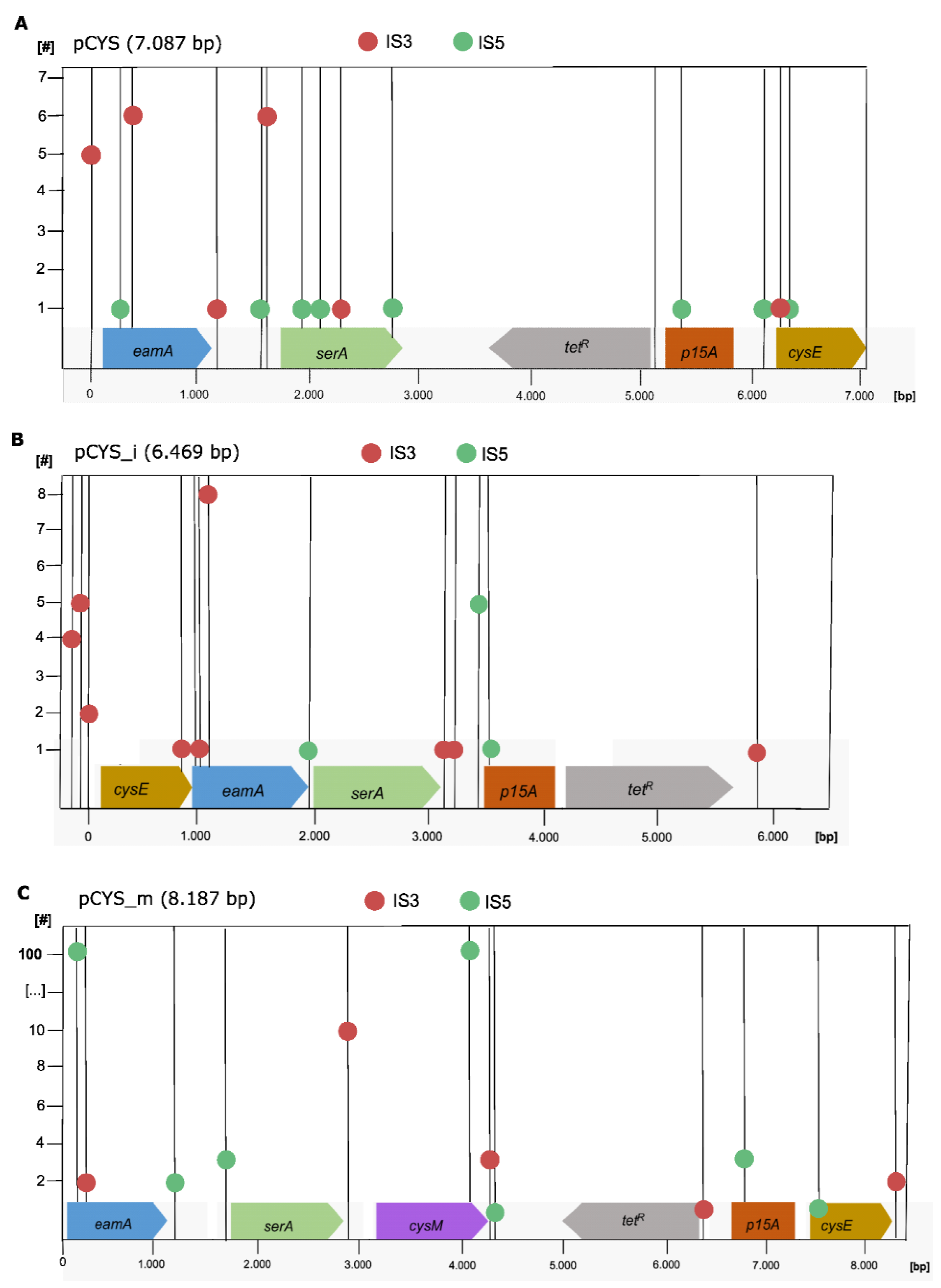
2.2. Localization of IS Target Site Duplications within Plasmids
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Aziz, R.K.; Breitbart, M.; Edwards, R.A. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010, 38, 4207–4217. [Google Scholar] [CrossRef]
- Hickman, A.B.; Dyda, F. Mechanisms of DNA Transposition. Microbiol. Spectr. 2015, 3, MDNA3-0034-2014. [Google Scholar] [CrossRef]
- Lee, H.; Doak, T.G.; Popodi, E.; Foster, P.L.; Tang, H. Insertion sequence-caused large-scale rearrangements in the genome of Escherichia coli. Nucleic Acids Res. 2016, 44, 7109–7119. [Google Scholar] [CrossRef]
- Rugbjerg, P.; Myling-Petersen, N.; Porse, A.; Sarup-Lytzen, K.; Sommer, M.O.A. Diverse genetic error modes constrain large-scale bio-based production. Nat. Commun. 2018, 9, 787. [Google Scholar] [CrossRef]
- Heieck, K.; Arnold, N.D.; Bruck, T.B. Metabolic stress constrains microbial L-cysteine production in Escherichia coli by accelerating transposition through mobile genetic elements. Microb. Cell. Fact. 2023, 22, 10. [Google Scholar] [CrossRef]
- Drake, J.W. Rates of Spontaneous Mutation. Genetics 1998, 148, 1667–1686. [Google Scholar] [CrossRef]
- Porse, A.; Gumpert, H.; Kubicek-Sutherland, J.Z.; Karami, N.; Adlerberth, I.; Wold, A.E.; Andersson, D.I.; Sommer, M.O.A. Genome Dynamics of Escherichia coli during Antibiotic Treatment: Transfer, Loss, and Persistence of Genetic Elements In situ of the Infant Gut. Front. Cell. Infect. Microbiol. 2017, 7, 126. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, G.A.L. Evidence that the insertion events of IS2 transposition are biased towards abrupt compositional shifts in target DNA and modulated by a diverse set of culture parameters. Biotechnol. Prod. A Proc. Eng. 2014, 98, 6609–6619. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Ecovoiu, A.A.; Ghionoiu, I.C.; Ciuca, A.M.; Ratiu, A.C. Genome ARTIST: A robust, high-accuracy aligner tool for mapping transposon insertions and self-insertions. Mob. DNA 2016, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.; Fayet, O.; Rousseau, P.; Ton Hoang, B.; Duval-Valentin, G. Copy-out-Paste-in Transposition of IS911: A Major Transposition Pathway. Microbiol. Spectr. 2015, 3, MDNA3-0031-2014. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.; Fayet, O. Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol. 1993, 7, 497–503. [Google Scholar] [CrossRef]
- Rousseau, P.; Gueguen, E.; Duval-Valentin, G.; Chandler, M. The helix-turn-helix motif of bacterial insertion sequence IS911 transposase is required for DNA binding. Nucleic Acids Res. 2004, 32, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Haren, L.; Normand, C.; Polard, P.; Alazard, R.; Chandler, M. IS911 transposition is regulated by protein-protein interactions via a leucine zipper motif. J. Mol. Biol. 2000, 296, 757–768. [Google Scholar] [CrossRef]
- Haren, L. Integrating DNA: Transposases and Retroviral Integrases. Annu. Rev. Microbiol. 1999, 53, 245–281. [Google Scholar] [CrossRef]
- Sharma, V.; Firth, A.E.; Antonov, I.; Fayet, O.; Atkins, J.F.; Borodovsky, M.; Baranov, P.V. A pilot study of bacterial genes with disrupted ORFs reveals a surprising profusion of protein sequence recoding mediated by ribosomal frameshifting and transcriptional realignment. Mol. Biol. Evol. 2011, 28, 3195–3211. [Google Scholar] [CrossRef]
- Sharma, V.; Prere, M.F.; Canal, I.; Firth, A.E.; Atkins, J.F.; Baranov, P.V.; Fayet, O. Analysis of tetra- and hepta-nucleotides motifs promoting −1 ribosomal frameshifting in Escherichia coli. Nucleic Acids Res. 2014, 42, 7210–7225. [Google Scholar] [CrossRef]
- Vögele, K. High-level ribosomal frameshifting directs the synthesis of IS150 gene products. Nucleic Acid. Res. 1991, 19, 4377–4385. [Google Scholar] [CrossRef] [PubMed]
- Mahillon, J. Insertion Sequences. Microbiol. Mol. Biol. Rev. 1998, 62, 725–774. [Google Scholar] [CrossRef]
- Humayun, M.Z.; Zhang, Z.; Butcher, A.M.; Moshayedi, A.; Saier, M.H., Jr. Hopping into a hot seat: Role of DNA structural features on IS5-mediated gene activation and inactivation under stress. PLoS ONE 2017, 12, e0180156. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef]
- Zhang, Z.; Saier, M.H., Jr. A mechanism of transposon-mediated directed mutation. Mol. Microbiol. 2009, 74, 29–43. [Google Scholar] [CrossRef]
- Zhang, Z.; Saier, M.H., Jr. A novel mechanism of transposon-mediated gene activation. PLoS Genet. 2009, 5, e1000689. [Google Scholar] [CrossRef] [PubMed]
- Schnetz, K. IS5: A mobile enhancer of transcription in Escherichia coli. Proc. Natl. Acad. Sci. USA 1992, 89, 1244–1248. [Google Scholar] [CrossRef] [PubMed]
- Fye, R.M. Exact method for numerically analyzing a model of local denaturation in superhelically stressed DNA. Phys. Rev. E 1999, 59, 3408. [Google Scholar] [CrossRef]
- Sheridan, S.D.; Benham, C.J.; Hatfield, G.W. Activation of gene expression by a novel DNA structural transmission mechanism that requires supercoiling-induced DNA duplex destabilization in an upstream activating sequence. J. Biol. Chem. 1998, 273, 21298–21308. [Google Scholar] [CrossRef] [PubMed]
- Umenhoffer, K.; Feher, T.; Baliko, G.; Ayaydin, F.; Posfai, J.; Blattner, F.R.; Posfai, G. Reduced evolvability of Escherichia coli MDS42, an IS-less cellular chassis for molecular and synthetic biology applications. Microb. Cell. Fact. 2010, 9, 38. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heieck, K.; Brück, T. Localization of Insertion Sequences in Plasmids for L-Cysteine Production in E. coli. Genes 2023, 14, 1317. https://doi.org/10.3390/genes14071317
Heieck K, Brück T. Localization of Insertion Sequences in Plasmids for L-Cysteine Production in E. coli. Genes. 2023; 14(7):1317. https://doi.org/10.3390/genes14071317
Chicago/Turabian StyleHeieck, Kevin, and Thomas Brück. 2023. "Localization of Insertion Sequences in Plasmids for L-Cysteine Production in E. coli" Genes 14, no. 7: 1317. https://doi.org/10.3390/genes14071317
APA StyleHeieck, K., & Brück, T. (2023). Localization of Insertion Sequences in Plasmids for L-Cysteine Production in E. coli. Genes, 14(7), 1317. https://doi.org/10.3390/genes14071317





