Human Endogenous Retrovirus-H-Derived miR-4454 Inhibits the Expression of DNAJB4 and SASH1 in Non-Muscle-Invasive Bladder Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of miRNAs Derived from HERVs
2.2. Cell Culture
2.3. RNA Extraction and Complementary DNA Synthesis
2.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.5. Identification of the Putative Tumor Suppressor Targets of miR-4454 in Bladder Cancer Cells
2.6. Dual-Luciferase Reporter Assay
2.7. Statistical Analyses
3. Results
3.1. Identification of miRNAs Derived from HERVs in the Human Genome
3.2. Sequence Conservation of HERV-H-int-Derived miRNAs
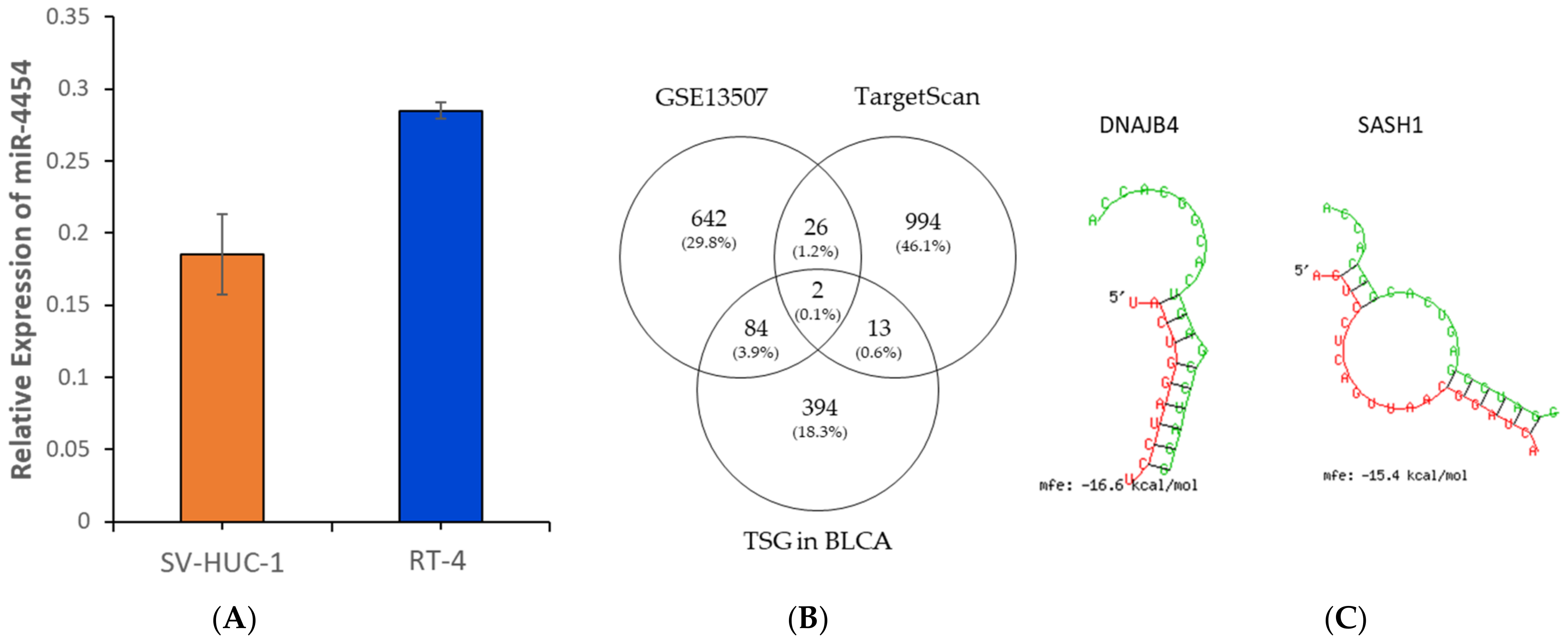
3.3. Expression of miR-4454 in Bladder Cancer Cell Lines
3.4. DNAJB4 and SASH1 Are Putative Tumor Suppressor Target Genes of miR-4454 in Bladder Cancer
3.5. The Negative Relationship between miR-4454 and DNAJB4/SASH1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Matylla-Kulinska, K.; Tafer, H.; Weiss, A.; Schroeder, R. Functional repeat-derived RNAs often originate from retrotransposon-propagated ncRNAs. Wiley Interdiscip. Rev. RNA 2014, 5, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-E.; Huh, J.-W.; Kim, H.-S. Bioinformatics analysis of evolution and human disease related transposable element-derived microRNAs. Life 2020, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Payer, L.M.; Burns, K.H. Transposable elements in human genetic disease. Nat. Rev. Genet. 2019, 20, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.H. Transposable elements in cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.R.; Greene, W.K. Transposable elements: Powerful facilitators of evolution. Bioessays 2009, 31, 703–714. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Lee, H.-E.; Park, S.-J.; Huh, J.-W.; Imai, H.; Kim, H.-S. Enhancer function of microRNA-3681 derived from long terminal repeats represses the activity of variable number tandem repeats in the 3′UTR of SHISA7. Mol. Cells 2020, 43, 607. [Google Scholar]
- Wu, K.L.; Tsai, Y.M.; Lien, C.T.; Kuo, P.L.; Hung, J.Y. The roles of MicroRNA in lung cancer. Int. J. Mol. Sci. 2019, 20, 1611. [Google Scholar] [CrossRef]
- Gebert, L.F.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Grandi, N.; Tramontano, E. HERV envelope proteins: Physiological role and pathogenic potential in cancer and autoimmunity. Front. Microbiol. 2018, 9, 462. [Google Scholar] [CrossRef]
- Seifarth, W.; Frank, O.; Zeilfelder, U.; Spiess, B.; Greenwood, A.D.; Hehlmann, R.; Leib-Mösch, C. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 2005, 79, 341–352. [Google Scholar] [CrossRef]
- Kim, H.-S.; Yi, J.-M.; Hirai, H.; Huh, J.-W.; Jeong, M.-S.; Jang, S.-B.; Kim, C.-G.; Saitou, N.; Hyun, B.-H.; Lee, W.-H. Human endogenous retrovirus (HERV)-R family in primates: Chromosomal location, gene expression, and evolution. Gene 2006, 370, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Bae, W.H.; Ha, H.; Park, E.G.; Lee, Y.J.; Kim, W.R.; Kim, H.-S. Z-DNA–Containing Long Terminal Repeats of Human Endogenous Retrovirus Families Provide Alternative Promoters for Human Functional Genes. Mol. Cells 2022, 45, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Bello-Morales, R.; Andreu, S.; Ripa, I.; López-Guerrero, J.A. Hsv-1 and endogenous retroviruses as risk factors in demyelination. Int. J. Mol. Sci. 2021, 22, 5738. [Google Scholar] [CrossRef]
- Wang, X.; Huang, J.; Zhu, F. Human endogenous retroviral envelope protein syncytin-1 and inflammatory abnormalities in neuropsychological diseases. Front. Psychiatry 2018, 9, 422. [Google Scholar] [CrossRef]
- Vargiu, L.; Rodriguez-Tomé, P.; Sperber, G.O.; Cadeddu, M.; Grandi, N.; Blikstad, V.; Tramontano, E.; Blomberg, J. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 2016, 13, 7. [Google Scholar] [CrossRef]
- Yi, J.-M.; Kim, H.-M.; Kim, H.-S. Human endogenous retrovirus HERV-H family in human tissues and cancer cells: Expression, identification, and phylogeny. Cancer Lett. 2006, 231, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Carter, T.A.; Singh, M.; Dumbović, G.; Chobirko, J.D.; Rinn, J.L.; Feschotte, C. Mosaic cis-regulatory evolution drives transcriptional partitioning of HERVH endogenous retrovirus in the human embryo. eLife 2022, 11, e76257. [Google Scholar] [CrossRef]
- Santoni, F.A.; Guerra, J.; Luban, J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology 2012, 9, 111. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, T.; Preissl, S.; Amaral, M.L.; Grinstein, J.D.; Farah, E.N.; Destici, E.; Qiu, Y.; Hu, R.; Lee, A.Y.; et al. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat. Genet. 2019, 51, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Sachs, F.; Ramsay, L.; Jacques, P.; Göke, J.; Bourque, G.; Ng, H.-H. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat. Struct. Mol. Biol. 2014, 21, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Ohnuki, M.; Tanabe, K.; Sutou, K.; Teramoto, I.; Sawamura, Y.; Narita, M.; Nakamura, M.; Tokunaga, Y.; Nakamura, M.; Watanabe, A.; et al. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc. Natl. Acad. Sci. USA 2014, 111, 12426–12431. [Google Scholar] [CrossRef] [PubMed]
- Mangeney, M.; De Parseval, N.; Thomas, G.; Heidmann, T. The full-length envelope of an HERV-H human endogenous retrovirus has immunosuppressive properties. J. Gen. Virol. 2001, 82, 2515–2518. [Google Scholar] [CrossRef] [PubMed]
- Kudo-Saito, C. Cancer-associated mesenchymal stem cells aggravate tumor progression. Front. Cell Dev. Biol. 2015, 3, 23. [Google Scholar] [CrossRef] [PubMed]
- Pérot, P.; Mullins, C.S.; Naville, M.; Bressan, C.; Hühns, M.; Gock, M.; Kühn, F.; Volff, J.-N.; Trillet-Lenoir, V.; Linnebacher, M.; et al. Expression of young HERV-H loci in the course of colorectal carcinoma and correlation with molecular subtypes. Oncotarget 2015, 6, 40095. [Google Scholar] [CrossRef]
- Wentzensen, N.; Coy, J.F.; Knaebel, H.-P.; Linnebacher, M.; Wilz, B.; Gebert, J.; Doeberitz, M.v.K. Expression of an endogenous retroviral sequence from the HERV-H group in gastrointestinal cancers. Int. J. Cancer 2007, 121, 1417–1423. [Google Scholar] [CrossRef]
- Manca, M.A.; Solinas, T.; Simula, E.R.; Noli, M.; Ruberto, S.; Madonia, M.; Sechi, L.A. HERV-K and HERV-H Env proteins induce a humoral response in prostate cancer patients. Pathogens 2022, 11, 95. [Google Scholar] [CrossRef]
- Park, E.G.; Ha, H.; Lee, D.H.; Kim, W.R.; Lee, Y.J.; Bae, W.H.; Kim, H.-S. Genomic Analyses of Non-Coding RNAs Overlapping Transposable Elements and Its Implication to Human Diseases. Int. J. Mol. Sci. 2022, 23, 8950. [Google Scholar] [CrossRef]
- Lin, H.; Zhang, R.; Wu, W.; Lei, L. miR-4454 promotes hepatic carcinoma progression by targeting Vps4A and Rab27A. Oxidative Med. Cell. Longev. 2021, 2021, 9230435. [Google Scholar] [CrossRef]
- Wang, H.; Hu, H.; Luo, Z.; Liu, S.; Wu, W.; Zhu, M.; Wang, J.; Liu, Y.; Lu, Z. miR-4454 up-regulated by HPV16 E6/E7 promotes invasion and migration by targeting ABHD2/NUDT21 in cervical cancer. Biosci. Rep. 2020, 40, BSR20200796. [Google Scholar] [CrossRef]
- Kannathasan, T.; Kuo, W.W.; Chen, M.C.; Viswanadha, V.P.; Shen, C.Y.; Tu, C.C.; Yeh, Y.L.; Bharath, M.; Shibu, M.A.; Huang, C.Y. Chemoresistance-associated silencing of miR-4454 promotes colorectal cancer aggression through the GNL3L and NF-κB pathway. Cancers 2020, 12, 1231. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Pandhiri, T.; Grassi, T.; Visscher, D.W.; Multinu, F.; Agarwal, K.; Mariani, A.; Shridhar, V.; Mitra, A.K. Signals from the Metastatic Niche Regulate Early and Advanced Ovarian Cancer Metastasis through miR-4454 Downregulation. Microenvironmental Regulation of OC Metastasis via p53 and FAK. Mol. Cancer Res. 2020, 18, 1202–1217. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.A.; Green, B.B.; Seigne, J.D.; Schned, A.R.; Marsit, C.J. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol. Cancer 2015, 14, 194. [Google Scholar] [CrossRef]
- Piriyapongsa, J.; Mariño-Ramirez, L.; Jordan, I.K. Origin and evolution of human microRNAs from transposable elements. Genetics 2007, 176, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.T.; Cardin, S.E.; Borchert, G.M. Burgeoning evidence indicates that microRNAs were initially formed from transposable element sequences. Mob. Genet. Elem. 2014, 4, e29255. [Google Scholar] [CrossRef]
- Qin, S.; Jin, P.; Zhou, X.; Chen, L.; Ma, F. The role of transposable elements in the origin and evolution of microRNAs in human. PLoS ONE 2015, 10, e0131365. [Google Scholar] [CrossRef]
- Ali, A.; Han, K.; Liang, P. Role of transposable elements in gene regulation in the human genome. Life 2021, 11, 118. [Google Scholar] [CrossRef]
- Liang, Q.; Xu, Z.; Xu, R.; Wu, L.; Zheng, S. Expression patterns of non-coding spliced transcripts from human endogenous retrovirus HERV-H elements in colon cancer. PLoS ONE 2012, 7, e29950. [Google Scholar] [CrossRef]
- Gibb, E.A.; Warren, R.L.; Wilson, G.W.; Brown, S.D.; Robertson, G.A.; Morin, G.B.; Holt, R.A. Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma. Genome Med. 2015, 7, 22. [Google Scholar] [CrossRef]
- Jin, X.; Xu, X.E.; Jiang, Y.Z.; Liu, Y.R.; Sun, W.; Guo, Y.J.; Ren, Y.X.; Zuo, W.J.; Hu, X.; Huang, S.L.; et al. The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci. Adv. 2019, 5, eaat9820. [Google Scholar] [CrossRef] [PubMed]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Zeller, C.; Hinzmann, B.; Seitz, S.; Prokoph, H.; Burkhard-Goettges, E.; Fischer, J.; Jandrig, B.; Schwarz, L.-E.; Rosenthal, A.; Scherneck, S. SASH1: A candidate tumor suppressor gene on chromosome 6q24. 3 is downregulated in breast cancer. Oncogene 2003, 22, 2972–2983. [Google Scholar] [PubMed]
- Rimkus, C.; Martini, M.; Friederichs, J.; Rosenberg, R.; Doll, D.; Siewert, J.R.; Holzmann, B.; Janssen, K.P. Prognostic significance of downregulated expression of the candidate tumour suppressor gene SASH1 in colon cancer. Br. J. Cancer 2006, 95, 1419–1423. [Google Scholar] [CrossRef] [PubMed]
- Burgess, J.T.; Bolderson, E.; Adams, M.N.; Duijf, P.H.G.; Zhang, S.-D.; Gray, S.G.; Wright, G.; Richard, D.J.; O’byrne, K.J. SASH1 is a prognostic indicator and potential therapeutic target in non-small cell lung cancer. Sci. Rep. 2020, 10, 18605. [Google Scholar] [CrossRef]
- Zhou, N.; Liu, C.; Wang, X.; Mao, Q.; Jin, Q.; Li, P. Downregulated SASH1 expression indicates poor clinical prognosis in gastric cancer. Hum. Pathol. 2018, 74, 83–91. [Google Scholar] [CrossRef]
- Xie, J.; Zhang, W.; Zhang, J.; Lv, Q.-Y.; Luan, Y.-F. Downregulation of SASH1 correlates with poor prognosis in cervical cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3781–3786. [Google Scholar]
- Martini, M.; Gnann, A.; Scheikl, D.; Holzmann, B.; Janssen, K.P. The candidate tumor suppressor SASH1 interacts with the actin cytoskeleton and stimulates cell–matrix adhesion. Int. J. Biochem. Cell Biol. 2011, 43, 1630–1640. [Google Scholar] [CrossRef]
- Chen, E.-G.; Chen, Y.; Dong, L.-L.; Zhang, J.-S. Effects of SASH1 on lung cancer cell proliferation, apoptosis, and invasion in vitro. Tumor Biol. 2012, 33, 1393–1401. [Google Scholar] [CrossRef]
- Zong, W.; Yu, C.; Wang, P.; Dong, L. Overexpression of SASH1 inhibits TGF-β1-induced EMT in gastric cancer cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2016, 24, 17–23. [Google Scholar] [CrossRef]
- Wang, C.C.; Tsai, M.F.; Hong, T.M.; Chang, G.C.; Chen, C.Y.; Yang, W.M.; Chen, J.J.; Yang, P.C. The transcriptional factor YY1 upregulates the novel invasion suppressor HLJ1 expression and inhibits cancer cell invasion. Oncogene 2005, 24, 4081–4093. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, J.; Zhang, C.; Fu, W.; Xiao, X.; Ruan, S.; Zhang, Y.; Luo, X.; Tang, M. HLJ1 is a novel biomarker for colorectal carcinoma progression and overall patient survival. Int. J. Clin. Exp. Pathol. 2014, 7, 969. [Google Scholar] [PubMed]
- Mo, L.; Liu, J.; Yang, Z.; Gong, X.; Meng, F.; Zou, R.; Hou, L.; Fang, F. DNAJB4 identified as a potential breast cancer marker: Evidence from bioinformatics analysis and basic experiments. Gland. Surg. 2020, 9, 1955. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.F.; Wang, C.C.; Chang, G.C.; Chen, C.Y.; Chen, H.Y.; Cheng, C.L.; Yang, Y.P.; Wu, C.Y.; Shih, F.Y.; Liu, C.C.; et al. A new tumor suppressor DnaJ-like heat shock protein, HLJ1, and survival of patients with non–small-cell lung carcinoma. J. Natl. Cancer Inst. 2006, 98, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-F.; Wang, C.-C.; Chen, J.J. Tumour suppressor HLJ1: A potential diagnostic, preventive and therapeutic target in non-small cell lung cancer. World J. Clin. Oncol. 2014, 5, 865. [Google Scholar] [CrossRef]



| Gene | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| SASH1 | GAATCCTGACCATGTCCATCCC | AATATGCAGCCTCACAGTGACC |
| DNAJB4 | AGTGAGAATGATGTTTTGAAGCA | AGGGTAAACTTTATGGCATGTAA |
| GAPDH | GAAATCCCATCACCATCTTCCAGG | GAGCCCCAGCCTTCTCCATG |
| Chr | Start | End | miRNA | Family | Subfamily |
|---|---|---|---|---|---|
| chr1 | 51,059,879 | 51,059,900 | hsa-miR-4421 | ERV1 | MER52A |
| chr1 | 51,060,031 | 51,060,054 | hsa-miR-6500-5p | ERV1 | MER52A |
| chr1 | 51,060,072 | 51,060,092 | hsa-miR-6500-3p | ERV1 | MER52A |
| chr1 | 237,471,164 | 237,471,185 | hsa-miR-4428 | ERV1 | LTR9A1 |
| chr3 | 32,506,342 | 32,506,362 | hsa-miR-548ay-5p | ERV1 | LTR43 |
| chr3 | 32,506,305 | 32,506,326 | hsa-miR-548ay-3p | ERV1 | LTR43 |
| chr3 | 164,171,523 | 164,171,544 | hsa-miR-1263 | ERV1 | MER39 |
| chr3 | 176,515,103 | 176,515,120 | hsa-miR-7977 | ERV1 | HUERS-P3-int |
| chr4 | 163,093,607 | 163,093,626 | hsa-miR-4454 | ERV1 | HERV-H-int |
| chr6 | 155,946,809 | 155,946,829 | hsa-miR-1202 | ERV1 | MER52A |
| chr7 | 64,679,069 | 64,679,090 | hsa-miR-6839-5p | ERV1 | LTR7C |
| chr8 | 1,801,133 | 1,801,154 | hsa-miR-3674 | ERV1 | LTR8A |
| chr12 | 122,695,964 | 122,695,985 | hsa-miR-9902 | ERV1 | HUERS-P1-int |
| chr15 | 85,825,655 | 85,825,673 | hsa-miR-548ap-5p | ERV1 | MER50 |
| chr15 | 85,825,692 | 85,825,710 | hsa-miR-548ap-3p | ERV1 | MER50 |
| chr17 | 82,668,281 | 82,668,301 | hsa-miR-4525 | ERV1 | MER52D |
| chr18 | 39,622,153 | 39,622,172 | hsa-miR-924 | ERV1 | MER101B |
| chr19 | 12,920,373 | 12,920,394 | hsa-miR-5695 | ERV1 | MER39B |
| chr19 | 55,123,225 | 55,123,242 | hsa-miR-7975 | ERV1 | HERV-H-int |
| chr3 | 183,886,860 | 183,886,879 | hsa-miR-4448 | ERVK | LTR13 |
| chr2 | 12,199,138 | 12,199,159 | hsa-miR-3681-5p | ERVL | LTR16D1 |
| chr2 | 12,199,174 | 12,199,195 | hsa-miR-3681-3p | ERVL | LTR16D1 |
| chr4 | 66,276,890 | 66,276,911 | hsa-miR-1269a | ERVL | MLT2B3 |
| chr8 | 28,505,126 | 28,505,142 | hsa-miR-4288 | ERVL | LTR86B1 |
| chr14 | 67,441,902 | 67,441,922 | hsa-miR-5694 | ERVL | MLT2B1 |
| chr16 | 14,907,756 | 14,907,779 | hsa-miR-3670 | ERVL | LTR16A1 |
| chr17 | 12,917,313 | 12,917,334 | hsa-miR-1269b | ERVL | MLT2B3 |
| chr20 | 60,308,534 | 60,308,552 | hsa-miR-646 | ERVL | LTR67B |
| chr22 | 30,731,569 | 30,731,590 | hsa-miR-3200-5p | ERVL | LTR16D2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, E.G.; Lee, D.H.; Kim, W.R.; Lee, Y.J.; Bae, W.H.; Kim, J.-m.; Shin, H.J.; Ha, H.; Yi, J.M.; Cho, S.G.; et al. Human Endogenous Retrovirus-H-Derived miR-4454 Inhibits the Expression of DNAJB4 and SASH1 in Non-Muscle-Invasive Bladder Cancer. Genes 2023, 14, 1410. https://doi.org/10.3390/genes14071410
Park EG, Lee DH, Kim WR, Lee YJ, Bae WH, Kim J-m, Shin HJ, Ha H, Yi JM, Cho SG, et al. Human Endogenous Retrovirus-H-Derived miR-4454 Inhibits the Expression of DNAJB4 and SASH1 in Non-Muscle-Invasive Bladder Cancer. Genes. 2023; 14(7):1410. https://doi.org/10.3390/genes14071410
Chicago/Turabian StylePark, Eun Gyung, Du Hyeong Lee, Woo Ryung Kim, Yun Ju Lee, Woo Hyeon Bae, Jung-min Kim, Hae Jin Shin, Hongseok Ha, Joo Mi Yi, Ssang Goo Cho, and et al. 2023. "Human Endogenous Retrovirus-H-Derived miR-4454 Inhibits the Expression of DNAJB4 and SASH1 in Non-Muscle-Invasive Bladder Cancer" Genes 14, no. 7: 1410. https://doi.org/10.3390/genes14071410
APA StylePark, E. G., Lee, D. H., Kim, W. R., Lee, Y. J., Bae, W. H., Kim, J.-m., Shin, H. J., Ha, H., Yi, J. M., Cho, S. G., Choi, Y. H., Leem, S. H., Cha, H. J., Kim, S. W., & Kim, H. S. (2023). Human Endogenous Retrovirus-H-Derived miR-4454 Inhibits the Expression of DNAJB4 and SASH1 in Non-Muscle-Invasive Bladder Cancer. Genes, 14(7), 1410. https://doi.org/10.3390/genes14071410











