Transcriptomic Analysis of the Developing Testis and Spermatogenesis in Qianbei Ma Goats
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animal Handling and Sample Collection
2.3. RNA Quantitation and Quality
2.4. Transcriptome Sequencing
2.5. Quantification of Gene Expression
2.6. Differential Expression Analysis
2.7. GO and KEGG Enrichment Analyses of DEGs
2.8. Prediction of New Transcripts and Alternative Splicing Analysis
2.9. qRT-PCR for RNA-Seq Data Validation
3. Results
3.1. Gene Expression Profiling during Testicular Development in Qianbei Ma Goats
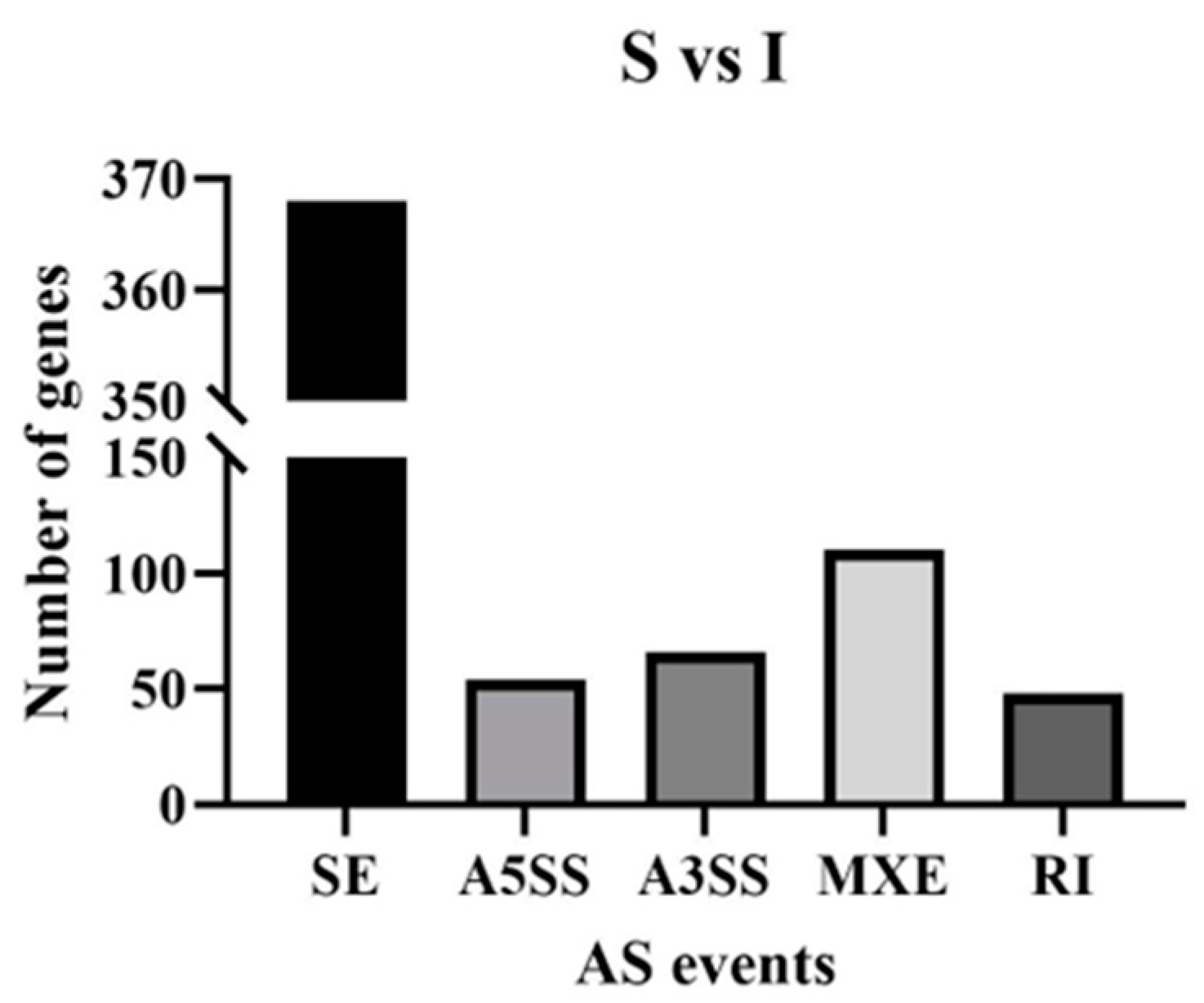
3.2. Alternative Splicing Data
3.3. Analysis of DEGs
3.4. GO Analysis of DEGs
3.5. KEGG Pathway Analysis of DEGs
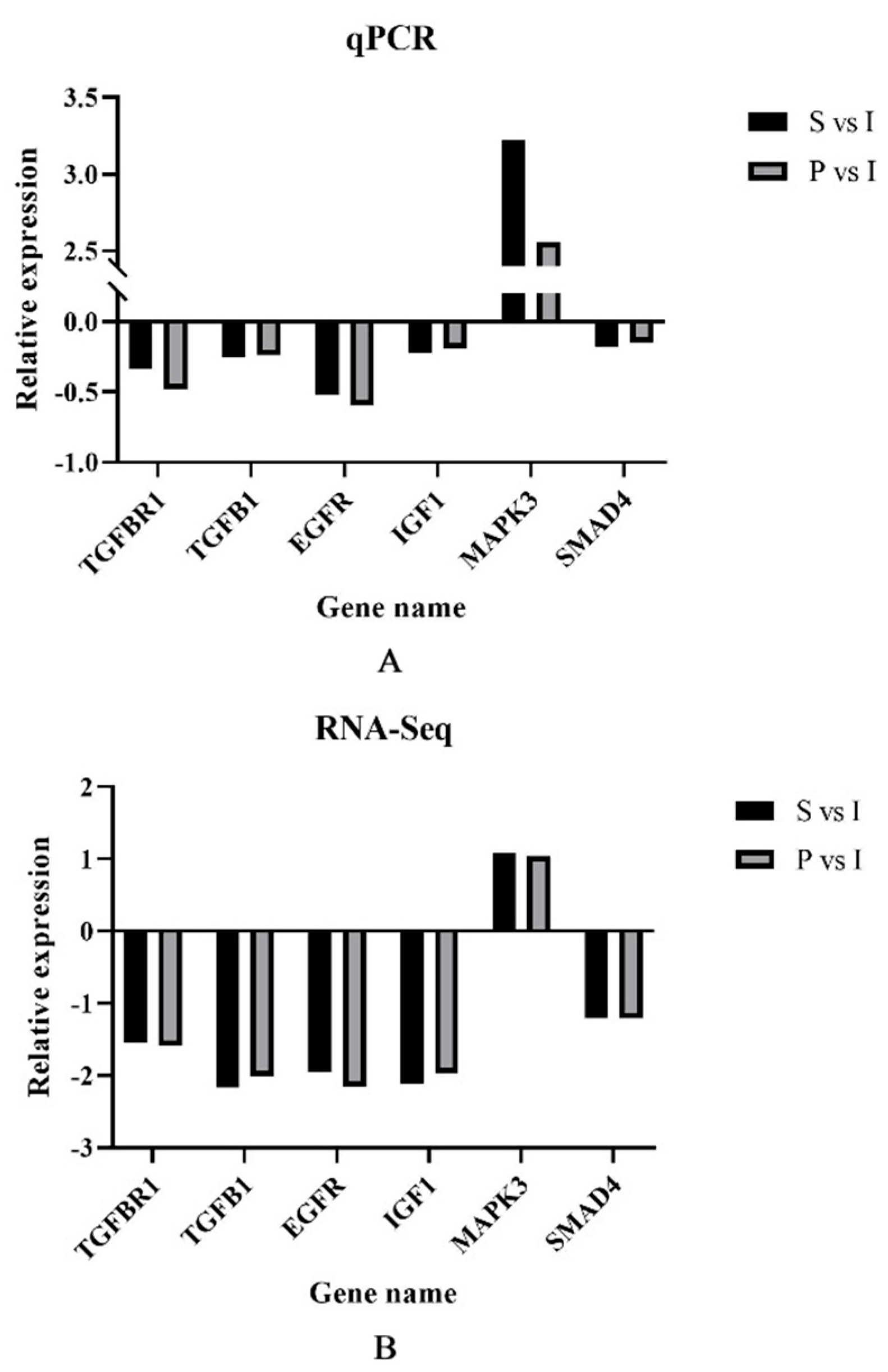
3.6. qRT-PCR Validation of DEGs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hecht, N.B. Molecular mechanisms of male germ cell differentiation. BioEssays 1998, 20, 555–561. [Google Scholar] [CrossRef]
- Eddy, E.M.; O’Brien, D.A. Gene expression during mammalian meiosis. Curr. Top. Dev. Biol. 1998, 37, 141–200. [Google Scholar] [PubMed]
- Hess, R.A.; De Franca, L.R. Spermatogenesis and cycle of the seminiferous epithelium. Adv. Exp. Med. Biol. 2008, 636, 1–15. [Google Scholar] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Marioni, J.C.; Mason, C.E.; Mane, S.M.; Stephens, M.; Gilad, Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008, 18, 1509–1517. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Reviews. Genet. 2010, 10, 57–63. [Google Scholar] [CrossRef]
- Ramsköld, D.; Wang, E.T.; Burge, C.B.; Sandberg, R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009, 5, e1000598. [Google Scholar] [CrossRef]
- Djureinovic, D.; Fagerberg, L.; Hallstrom, B.; Danielsson, A.; Lindskog, C.; Uhlen, M.; Ponten, F. The human testis-specific proteome defined by transcriptomics and antibody-based profiling. Mol. Hum. Reprod. 2014, 20, 476–488. [Google Scholar] [CrossRef]
- Anand, M.; Prasad, B.V. The computational analysis of human testis transcriptome reveals closer ties to pluripotency. J. Hum. Reprod. Sci. 2012, 5, 266–273. [Google Scholar]
- Fu, K.; Chen, X.; Guo, W.; Zhou, Z.; Zhang, Y.; Ji, T.; Yang, P.; Tian, X.; Wang, W.; Zou, Y. Effects of N Acetylcysteine on the Expression of Genes Associated with Reproductive Performance in the Goat Uterus during Early Gestation. Animals 2022, 12, 2431. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based Genome Alignment and Genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Erratum: Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 888. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Shumate, A.; Wong, B.; Pertea, G.; Pertea, M. Improved Transcriptome Assembly Using a Hybrid of Long and Short Reads with StringTie. PLoS Comput. Biol. 2022, 18, e1009730. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-seq Reads. Nat. Biotechnol. 2015, 33, 290–296. [Google Scholar] [CrossRef]
- Moraveji, S.F.; Esfandiari, F.; Taleahmad, S.; Nikeghbalian, S.; Sayahpour, F.A.; Masoudi, N.S.; Shahverdi, A.; Baharvand, H. Suppression of transforming growth factor-beta signaling enhances spermatogonial proliferation and spermatogenesis recovery following chemotherapy. Hum. Reprod. 2019, 34, 2430–2442. [Google Scholar] [CrossRef]
- Memon, M.A.; Anway, M.D.; Covert, T.R.; Uzumcu, M.; Skinner, M.K. Transforming growth factor beta (TGF beta 1, TGF beta 2 and TGF beta 3) null-mutant phenotypes in embryonic gonadal development. Mol. Cell. Endocrinol. 2008, 294, 70–80. [Google Scholar] [CrossRef]
- He, J.; Dong, C.; You, R.; Zhu, Z.; Lv, L.; Smith, G.W. Localization of epidermal growth factor (EGF) and its receptor (EGFR) during postnatal testis development in the alpaca (Lama pacos). Anim. Reprod. Sci. 2009, 116, 155–161. [Google Scholar] [CrossRef]
- Baker, J.; Hardy, M.P. Effects of an Igf1 gene null mutation on mouse reproduction. Mol. Endocrinol. 1996, 10, 903–918. [Google Scholar]
- Manesh, K.P.S.; Saradha, B.; Suresh, C.S. Telomere Signaling and Maintenance Pathways in Spermatozoa of Infertile Men Treated with Antioxidants: An in silico Approach Using Bioinformatic Analysis. Front. Cell Dev. Biol. 2021, 9, 768510. [Google Scholar]
- He, J.; Zhang, X.; Wen, X.; Zhao, L. Expression and localization of Smad2 and Smad4 in domestic cat testis during testicular development and spermatogenesis. Acta Anat. Sin. 2010, 41, 598–602. [Google Scholar]
- Zhou, C.; Zhang, L.; Xu, D.; Ding, H.; Zheng, S.; Liu, M. MeDIP-seq and RNA-seq Analysis during Porcine Testis Development Reveals Functional DMR at the Promoter of LDHC. Genomics 2022, 114, 110467. [Google Scholar] [CrossRef]
- Zhang, L.; Li, F.; Lei, P.; Guo, M.; Liu, R.; Wang, L.; Yu, T.; Lv, Y.; Zhang, T.; Zeng, W.; et al. Single-cell RNA-sequencing Reveals the Dynamic Process and Novel Markers in Porcine Spermatogenesis. J. Anim. Sci. Biotechnol. 2021, 12, 122. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.; Yang, Y.; Retzel, E.F.; Liu, W. Male-specific Region of the Bovine Y Chromosome Is Gene Rich with a High Transcriptomic Activity in Testis Development. Proc. Natl. Acad. Sci. USA 2013, 110, 12373–12378. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, M.; Fan, Y.; Li, S.; Lai, Z.; Huang, Y.; Lan, X.; Lei, C.; Chen, H.; Dang, R. Identification and Characterization of Circular RNAs in Qinchuan Cattle Testis. R. Soc. Open Sci. 2018, 5, 180413. [Google Scholar] [CrossRef]
- Fu, Y.; Wu, P.; Beane, T.; Zamore, P.D.; Weng, Z. Elimination of PCR Duplicates in RNA-seq and Small RNA-seq Using Unique Molecular Identifiers. BMC Genom. 2018, 19, 531. [Google Scholar] [CrossRef] [PubMed]
- Green, C.D.; Ma, Q.; Manske, G.L.; Shami, A.N.; Zheng, X.; Marini, S.; Moritz, L.; Sultan, C.; Gurczynski, S.J.; Moore, B.B.; et al. A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev. Cell 2018, 46, 651–667. [Google Scholar] [CrossRef]
- Zou, X.; Lu, T.; Zhao, Z.; Liu, G.; Lian, Z.; Guo, Y.; Sun, B.; Liu, D.; Li, Y. Comprehensive Analysis of MRNAs and MiRNAs in the Ovarian Follicles of Uniparous and Multiple Goats at Estrus Phase. BMC Genom. 2020, 21, 267. [Google Scholar] [CrossRef]
- Zi, X.; Lu, J.; Zhou, H.; Ma, L.; Xia, W.; Xiong, X.; Lan, D.; Wu, X. Comparative Analysis of Ovarian Transcriptomes between Prolific and Non-prolific Goat Breeds via High-throughput Sequencing. Reprod. Domest. Anim. 2018, 53, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Hu, Q.; Zang, X.; Xie, Y.; Zhou, C.; Zou, X.; Li, Y.; Deng, M.; Guo, Y.; Liu, G.; et al. Analysis and Screening of Reproductive Long Non-coding RNAs Through Genome-Wide Analyses of Goat Endometrium During the Pre-attachment Phase. Front. Genet. 2020, 11, 568017. [Google Scholar] [CrossRef]
- Liu, H.; Wang, C.; Li, Z.; Shang, C.; Zhang, X.; Zhang, R.; Wang, A.; Jin, Y.; Lin, P. Transcriptomic Analysis of STAT1/3 in the Goat Endometrium During Embryo Implantation. Front. Vet. Sci. 2021, 8, 757759. [Google Scholar] [CrossRef]
- Bo, D.; Jiang, X.; Liu, G.; Hu, R.; Chong, Y. RNA-Seq Implies Divergent Regulation Patterns of LincRNA on Spermatogenesis and Testis Growth in Goats. Animals 2021, 11, 625. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, P.; Yang, H.; Wang, Z.; El-Samahy, M.A.; Wang, F.; Zhang, Y. Dietary Supplementation with Metformin Improves Testis Function and Semen Quality and Increases Antioxidants and Autophagy Capacity in Goats. Theriogenology 2022, 188, 79–89. [Google Scholar] [CrossRef]
- Gong, W.; Pan, L.; Lin, Q.; Zhou, Y.; Xin, C.; Yu, X.; Cui, P.; Hu, S.; Yu, J. Transcriptome Profiling of the Developing Postnatal Mouse Testis Using Next-generation Sequencing. Sci. China 2013, 56, 1–12. [Google Scholar] [CrossRef][Green Version]
- Shen, F.; Geng, Y.; Zhang, L.; Luo, L.; Yan, G.; Hou, R.; Yue, B.; Zhang, X. Transcriptome Analysis Reveals the Alternative Splicing Changes in the Immune-Related Genes of the Giant Panda (Ailuropoda melanoleuca), in Response to the Canine Distemper Vaccine. Zool. Sci. 2022, 39, 275. [Google Scholar] [CrossRef]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, M.; Kuroyanagi, H. 2SE01 Regulatory Mechanisms of Alternative Splicing in Metazoans. Seibutsu Butsuri 2005, 45, S23. [Google Scholar] [CrossRef][Green Version]
- Liu, X.; He, P.; Zhang, Z.; Gong, P.; Niu, Y.; Bao, Z.; Yang, Y.; Gan, L.; Muhuyati. Candidate Genes and Their Alternative Splicing May Be Potential Biomarkers of Acute Myocardial Infarction: A Study of Mouse Model. BMC Cardiovasc. Disord. 2022, 22, 505. [Google Scholar] [CrossRef]
- Zhu, Z.; Ni, S.; Zhang, J.; Yuan, Y.; Bai, Y.; Yin, X.; Zhu, Z. Genome-wide Analysis of Dysregulated RNA-binding Proteins and Alternative Splicing Genes in Keloid. Front. Genet. 2023, 14, 1118999. [Google Scholar] [CrossRef]
- Schomburg, L.; Liao, X.; Majed, F.A.; Abdullah, M.S.Y.; Refetoff, S.; Boran, G.; Dumitrescu, A.M.; Lado-Abeal, J.; Moeller, L.C.; Weiss, R.E. Mutations in SECISBP2 Result in Abnormal Thyroid Hormone Metabolism. Nat. Genet. 2005, 37, 1247–1252. [Google Scholar]
- Schoenmakers, E.; Chatterjee, K. Identification of Genetic Disorders Causing Disruption of Selenoprotein Biosynthesis. Methods Mol. Biol. 2018, 1661, 325–335. [Google Scholar]
- Mohammadabadi, T.; Tabatbaei, S.; Ghezi, Z.; Swelum, A.A. Effect of Dietary Palm Kernel on Semen Quality, Reproductive and Thyroid Hormones and Blood Chemistry Parameters of Arabi Rams. Anim. Nutr. Feed Technol. 2020, 20, 93. [Google Scholar] [CrossRef]
- Mogheiseh, A.; Vara, N.; Ayaseh, M.; Jalali, P. Effects of Cabergoline on Thyroid Hormones and Semen Quality of Dog. Top. Companion Anim. Med. 2017, 32, 13–15. [Google Scholar] [CrossRef]
- Quartuccio, M.; Fazio, E.; Medica, P.; Cristarella, S.; Emmanuele, G.; Sinagra, L.; Liotta, L. Correlation between Sperm Parameters and Circulating Thyroid Hormones and Testosterone Concentrations in Labrador Retriever Dog. Ital. J. Anim. Sci. 2021, 20, 947–954. [Google Scholar] [CrossRef]
- Majdic, G.; Snoj, T.; Horvat, A.; Mrkun, J.; Kosec, M.; Cestnik, V. Higher Thyroid Hormone Levels in Neonatal Life Result in Reduced Testis Volume in Postpubertal Bulls. Int. J. Androl. 1998, 21, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Ghasemzadeh-Hasankolaei, M.; Sedighi-Gilani, M.A.; Eslaminejad, M.B. Induction of Ram Bone Marrow Mesenchymal Stem Cells into Germ Cell Lineage Using Transforming Growth Factor-β Superfamily Growth Factors. Reprod. Domest. Anim. 2014, 49, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Rojas-García, P.P.; Recabarren, M.P.; Sir-Petermann, T.; Rey, R.; Palma, S.; Carrasco, A.; Perez-Marin, C.C.; Padmanabhan, V.; Recabarren, S.E. Altered Testicular Development as a Consequence of Increase Number of Sertoli Cell in Male Lambs Exposed Prenatally to Excess Testosterone. Endocrine 2013, 43, 705–713. [Google Scholar] [CrossRef]
- Ingman, W.V.; Robertson, S.A. The Essential Roles of TGFB1 in Reproduction. Cytokine Growth Factor Rev. 2009, 20, 233–239. [Google Scholar] [CrossRef]
- McGrath, L.J.; Ingman, W.V.; Robker, R.L.; Robertson, S.A. Exogenous Transforming Growth Factor Beta1 Replacement and Fertility in Male Tgfb1 Null Mutant Mice. Reprod. Fertil. Dev. 2009, 21, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Cui, Y.; Yu, S.; Zhang, Q.; Fan, J.; Abdul, R.B.; Yang, K. The Expression of Epidermal Growth Factor (EGF) and Its Receptor (EGFR) During Post-Natal Testes Development in the Yak. Reprod. Domest. Anim. 2014, 49, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Kassab, M.; Abd-Elmaksoud, A.; Ali, M.A. Localization of the Epidermal Growth Factor (EGF) and Epidermal Growth Factor Receptor (EGFR) in the Bovine Testis. J. Mol. Histol. 2007, 38, 207–214. [Google Scholar] [CrossRef]
- Cannarella, R.; Condorelli, R.A.; La Vignera, S.; Calogero, A.E. Effects of the Insulin-like Growth Factor System on Testicular Differentiation and Function: A Review of the Literature. Andrology 2018, 6, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Pitetti, J.; Calvel, P.; Zimmermann, C.; Conne, B.; Papaioannou, M.D.; Aubry, F.; Cederroth, C.R.; Urner, F.; Fumel, B.; Crausaz, M.; et al. An Essential Role for Insulin and IGF1 Receptors in Regulating Sertoli Cell Proliferation, Testis Size, and FSH Action in Mice. Mol. Endocrinol. 2013, 27, 814–827. [Google Scholar] [CrossRef]
- Cannarella, R.; La Vignera, S.; Condorelli, R.A.; Calogero, A.E. The IGF1/FSH Ratio Correlates with Sperm Count and Testicular Volume. Endocrines 2022, 3, 624–632. [Google Scholar] [CrossRef]
- Müller, L.; Kowalewski, M.P.; Reichler, I.M.; Kollár, E.; Balogh, O. Different Expression of Leptin and IGF1 in the Adult and Prepubertal Testis in Dogs. Reprod. Domest. Anim. 2017, 52, 187–192. [Google Scholar] [CrossRef]
- Radovic, S.M.; Starovlah, I.M.; Capo, I.; Miljkovic, D.; Nef, S.; Kostic, T.S.; Andric, S.A. Insulin/IGF1 Signaling Regulates the Mitochondrial Biogenesis Markers in Steroidogenic Cells of Prepubertal Testis, but Not Ovary. Biol. Reprod. 2019, 100, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Neirijnck, Y.; Calvel, P.; Kilcoyne, K.R.; Kühne, F.; Stévant, I.; Griffeth, R.J.; Pitetti, J.; Andric, S.A.; Hu, M.; Pralong, F.; et al. Insulin and IGF1 Receptors Are Essential for the Development and Steroidogenic Function of Adult Leydig Cells. FASEB J. 2018, 32, 3321–3335. [Google Scholar] [CrossRef]
- Rojas, M.; Conei, D.; Bustos-Obregon, E. Epithelial-Mesenchymal Transitions in the Development of Testis/Interacciones Epitelio-Mesenquimaticas En El Desarrollo Testicular. Int. J. Morphol. 2017, 35, 1444. [Google Scholar] [CrossRef]
- Stratikopoulos, E.; Szabolcs, M.; Dragatsis, I.; Klinakis, A.; Efstratiadis, A. Hormonal Action of IGF1 in Postnatal Mouse Growth. Proc. Natl. Acad. Sci. USA 2008, 105, 19378–19383. [Google Scholar] [CrossRef]
- Pitetti, J.; Calvel, P.; Romero, Y.; Conne, B.; Vy, T.; Papaioannou, M.D.; Schaad, O.; Docquier, M.; Herrera, P.L.; Wilhelm, D.; et al. Insulin and IGF1 Receptors Are Essential for XX and XY Gonadal Differentiation and Adrenal Development in Mice. PLoS Genet. 2013, 9, E1003160. [Google Scholar] [CrossRef] [PubMed]
- Warr, N.; Carre, G.; Siggers, P.; Faleato, J.V.; Brixey, R.; Pope, M.; Bogani, D.; Childers, M.; Wells, S.; Scudamore, C.L.; et al. Gadd45γ and Map3k4 Interactions Regulate Mouse Testis Determination via P38 MAPK-Mediated Control of Sry Expression. Dev. Cell 2012, 23, 1020–1031. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ren, L.; Wei, J.; Zhang, J.; Zhu, Y.; Li, X.; Jing, L.; Duan, J.; Zhou, X.; Sun, Z. Fine Particle Matter Disrupts the Blood–testis Barrier by Activating TGF-β3/p38 MAPK Pathway and Decreasing Testosterone Secretion in Rat. Environ. Toxicol. 2018, 33, 711–719. [Google Scholar] [CrossRef]
- Shen, L.; Tang, X.; Wei, Y.; Long, C.; Tan, B.; Wu, S.; Sun, M.; Zhou, Y.; Cao, X.; Wei, G. Vitamin E and Vitamin C Attenuate Di-(2-ethylhexyl) Phthalate-induced Blood-testis Barrier Disruption by P38 MAPK in Immature SD Rats. Reprod. Toxicol. 2018, 81, 17–27. [Google Scholar] [CrossRef]
- Luo, D.; He, Z.; Yu, C.; Guan, Q. Role of P38 MAPK Signalling in Testis Development and Male Fertility. Oxidative Med. Cell. Longev. 2022, 2022, 6891897. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Sun, K.; Wang, S.; Gong, D. Polystyrene Microplastics Induce Apoptosis and Necroptosis in Swine Testis Cells via ROS/MAPK/HIF1α Pathway. Environ. Toxicol. 2022, 37, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.R.; Dorfman, V.B.; Vitullo, A.D. IGF1 Regulation of BOULE and CDC25A Transcripts via a Testosterone-independent Pathway in Spermatogenesis of Adult Mice. Reprod. Biol. 2015, 15, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shan, B.; Duan, Y.; Zhu, J.; Jiang, L.; Liu, Y.; Zhang, Y.; Qi, F.; Niu, S. Effects of Heshouwuyin on Gene Expression of the Insulin/IGF Signalling Pathway in Rat Testis and Spermatogenic Cells. Pharm. Biol. 2020, 58, 1208–1219. [Google Scholar] [CrossRef]



| Primer Name | Gene ID | Primer Sequence | Fragment Length | Annealing Temperature |
|---|---|---|---|---|
| GAPDH-F GAPDH-R | XM_005680968.3 | ATGTTTGTGATGGGCGTGAA GGCGTGGACAGTGGTCATAAGT | 153 bp | 60 °C |
| TGFBR1-F TGFBR1-R | XM_018052233.1 | TTCAAACGTGCTGACATCTATGC ACTGATGGATCGGAAGGTACAAG | 128 bp | 60 °C |
| SMAD4-F SMAD4-R | XM_018039535.1 | CATAACAGCACTACCACCTGGACT GGATGATTAGAAATAGGAGGCTGG | 173 bp | 60 °C |
| TGFB1-F TGFB1-R | NM_001314142.1 | CAACAATTCCTGGCGCTACCT ATGTCCACTTGAAGCGTGTTATCC | 183 bp | 60 °C |
| EGFR-F EGFR-R | XM_018067044.1 | CCGTGCGATTCAGTAACAACC GGTCAATTTCTGGCAGTTCTCCTC | 194 bp | 60 °C |
| IGF1-F IGF1-R | NM_001285697.1 | AATCAGCAGTCTTCCAACCCAA AGCAAGCACAGGGCCAGATA | 114 bp | 60 °C |
| MAPK3-F MAPK3-R | XM_018040780.1 | CTGGACCGGATGTTGACCTTTA CTCCTTCAGTCGTTCCTTGGG | 138 bp | 60 °C |
| Sample Name | Library Number | Raw Reads (n) | Clean Reads (n) | Error Rate | Q20 | Q30 | GC pct |
|---|---|---|---|---|---|---|---|
| I1 | 1 | 40,405,582 | 39,128,916 | 0.03 | 97.36 | 93.09 | 50.99 |
| I2 | 2 | 43,841,646 | 42,366,502 | 0.03 | 97.41 | 93.2 | 51.16 |
| I3 | 3 | 39,433,208 | 38,237,518 | 0.03 | 97.42 | 93.25 | 51.42 |
| I4 | 4 | 42,396,474 | 40,926,952 | 0.03 | 97.43 | 93.23 | 51.26 |
| I5 | 5 | 45,463,988 | 43,854,898 | 0.03 | 97.41 | 93.19 | 51.1 |
| I6 | 6 | 39,351,212 | 38,000,364 | 0.03 | 97.48 | 93.36 | 50.91 |
| S1 | 7 | 44,934,642 | 43,739,660 | 0.03 | 97.51 | 93.41 | 52.09 |
| S2 | 8 | 43,025,142 | 41,894,994 | 0.03 | 97.39 | 93.17 | 52.36 |
| S3 | 9 | 45,845,898 | 44,647,422 | 0.03 | 97.59 | 93.55 | 52.15 |
| S4 | 10 | 41,237,336 | 40,020,062 | 0.03 | 97.37 | 93.11 | 52.29 |
| S5 | 11 | 41,130,946 | 39,901,252 | 0.03 | 97.36 | 93.09 | 51.09 |
| S6 | 12 | 41,242,240 | 40,019,490 | 0.03 | 97.51 | 93.4 | 51.9 |
| P1 | 13 | 45,377,178 | 44,131,516 | 0.03 | 97.48 | 93.34 | 52.27 |
| P2 | 14 | 44,664,918 | 43,577,078 | 0.03 | 97.49 | 93.36 | 52.06 |
| P3 | 15 | 43,792,502 | 42,601,198 | 0.03 | 97.47 | 93.32 | 51.98 |
| P4 | 16 | 43,921,138 | 42,702,250 | 0.03 | 97.5 | 93.39 | 52.17 |
| P5 | 17 | 43,223,452 | 42,134,186 | 0.03 | 97.44 | 93.27 | 52.14 |
| P6 | 18 | 45,571,906 | 44,622,618 | 0.03 | 97.3 | 92.91 | 50.37 |
| Sample | Total Reads | Total Map | Unique Map | Multi Map | Positive Map | Negative Map |
|---|---|---|---|---|---|---|
| I1 | 39,128,916 | 37,563,635 (96.00%) | 35,626,251 (91.05%) | 1,937,384 (4.95%) | 17,793,049 (45.47%) | 17,833,202 (45.58%) |
| I2 | 42,366,502 | 40,625,554 (95.89%) | 38,612,044 (91.14%) | 2,013,510 (4.75%) | 19,294,015 (45.54%) | 19,318,029 (45.60%) |
| I3 | 38,237,518 | 36,735,876 (96.07%) | 34,685,761 (90.71%) | 2,050,115 (5.36%) | 17,323,488 (45.30%) | 17,362,273 (45.41%) |
| I4 | 40,926,952 | 39,282,398 (95.98%) | 37,389,786 (91.36%) | 1,892,612 (4.62%) | 18,672,350 (45.62%) | 18,717,436 (45.73%) |
| I5 | 43,854,898 | 42,092,675 (95.98%) | 40,255,938 (91.79%) | 1,836,737 (4.19%) | 20,105,549 (45.85%) | 20,150,389 (45.95%) |
| I6 | 38,000,364 | 36,500,392 (96.05%) | 34,501,227 (90.79%) | 1,999,165 (5.26%) | 17,230,278 (45.34%) | 17,270,949 (45.45%) |
| S1 | 43,739,660 | 42,076,928 (96.20%) | 40,551,689 (92.71%) | 1,525,239 (3.49%) | 20,259,699 (46.32%) | 20,291,990 (46.39%) |
| S2 | 41,894,994 | 40,318,684 (96.24%) | 39,006,285 (93.10%) | 1,312,399 (3.13%) | 19,484,346 (46.51%) | 19,521,939 (46.60%) |
| S3 | 44,647,422 | 43,061,457 (96.45%) | 41,514,183 (92.98%) | 1,547,274 (3.47%) | 20,739,259 (46.45%) | 20,774,924 (46.53%) |
| S4 | 40,020,062 | 38,465,949 (96.12%) | 36,829,538 (92.03%) | 1,636,411 (4.09%) | 18,396,451 (45.97%) | 18,433,087 (46.06%) |
| S5 | 39,901,252 | 38,334,062 (96.07%) | 36,942,595 (92.59%) | 1,391,467 (3.49%) | 18,452,256 (46.24%) | 18,490,339 (46.34%) |
| S6 | 40,019,490 | 38,522,907 (96.26%) | 36,912,252 (92.24%) | 1,610,655 (4.02%) | 18,439,977 (46.08%) | 18,472,275 (46.16%) |
| P1 | 44,131,516 | 42,514,601 (96.34%) | 41,073,585 (93.07%) | 1,441,016 (3.27%) | 20,518,750 (46.49%) | 20,554,835 (46.58%) |
| P2 | 43,577,078 | 41,944,011 (96.25%) | 40,303,516 (92.49%) | 1,640,495 (3.76%) | 20,133,389 (46.20%) | 20,170,127 (46.29%) |
| P3 | 42,601,198 | 41,016,519 (96.28%) | 39,440,892 (92.58%) | 1,575,627 (3.70%) | 19,701,050 (46.25%) | 19,739,842 (46.34%) |
| P4 | 42,702,250 | 41,098,071 (96.24%) | 39,528,322 (92.57%) | 1,569,749 (3.68%) | 19,745,068 (46.24%) | 19,783,254 (46.33%) |
| P5 | 42,134,186 | 40,510,371 (96.15%) | 38,849,502 (92.20%) | 1,660,869 (3.94%) | 19,405,731 (46.06%) | 19,443,771 (46.15%) |
| P6 | 44,622,618 | 38,550,615 (86.39%) | 37,036,723 (83.00%) | 1,513,892 (3.39%) | 18,501,103 (41.46%) | 18,535,620 (41.54%) |
| Sample Name | Exonic Region | Intronic Region | Intergenic Region |
|---|---|---|---|
| I1 | 4,466,151,533 (79.48%) | 611,822,545 (10.89%) | 541,085,383 (9.63%) |
| I2 | 4,779,622,398 (78.65%) | 761,383,464 (12.53%) | 535,946,498 (8.82%) |
| I3 | 4,432,093,997 (80.66%) | 570,346,947 (10.38%) | 492,580,965 (8.96%) |
| I4 | 4,614,052,978 (78.53%) | 738,532,433 (12.57%) | 523,236,881 (8.90%) |
| I5 | 5,160,159,063 (81.96%) | 605,739,527 (9.62%) | 530,252,297 (8.42%) |
| I6 | 4,298,336,210 (78.73%) | 599,791,632 (10.99%) | 561,800,986 (10.29%) |
| S1 | 5,004,805,293 (79.49%) | 606,545,683 (9.63%) | 684,973,145 (10.88%) |
| S2 | 4,891,821,345 (81.09%) | 533,206,687 (8.84%) | 607,543,338 (10.07%) |
| S3 | 5,173,907,638 (80.30%) | 600,212,860 (9.32%) | 668,983,341 (10.38%) |
| S4 | 4,579,030,919 (79.56%) | 552,189,052 (9.59%) | 623,968,138 (10.84%) |
| S5 | 4,488,498,885 (78.27%) | 613,561,446 (10.70%) | 632,649,316 (11.03%) |
| S6 | 4,595,667,229 (79.73%) | 543,550,434 (9.43%) | 624,459,696 (10.83%) |
| P1 | 5,058,268,703 (79.52%) | 636,322,977 (10.00%) | 666,220,762 (10.47%) |
| P2 | 4,921,081,484 (78.42%) | 644,761,157 (10.27%) | 709,616,381 (11.31%) |
| P3 | 4,871,901,450 (79.39%) | 593,069,117 (9.66%) | 671,905,236 (10.95%) |
| P4 | 4,918,914,052 (80.00%) | 601,562,419 (9.78%) | 628,332,353 (10.22%) |
| P5 | 4,886,718,972 (80.63%) | 554,456,620 (9.15%) | 619,529,425 (10.22%) |
| P6 | 4,658,554,613 (80.75%) | 537,138,992 (9.31%) | 573,327,628 (9.94%) |
| S vs. I | P vs. S | |
|---|---|---|
| BP | protein phosphorylation | sodium ion transport |
| ion transport | metal ion transport | |
| transmembrane transport | monovalent inorganic cation transport | |
| reproduction | immune response | |
| reproductive process | immune system process | |
| MF | ion channel activity | voltage-gated sodium channel activity |
| channel activity | voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential | |
| passive transmembrane transporter activity | sodium channel activity | |
| substrate-specific channel activity | chemokine activity | |
| scavenger receptor activity | chemokine receptor activity | |
| CC | myosin complex | voltage-gated sodium channel complex |
| extracellular matrix | sodium channel complex | |
| plasma membrane | ion channel complex | |
| plasma membrane part | cation channel complex | |
| cell periphery | integral component of plasma membrane |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, Y.; Chen, X.; Tian, X.; Guo, W.; Ruan, Y.; Tang, W.; Fu, K.; Ji, T. Transcriptomic Analysis of the Developing Testis and Spermatogenesis in Qianbei Ma Goats. Genes 2023, 14, 1334. https://doi.org/10.3390/genes14071334
Zou Y, Chen X, Tian X, Guo W, Ruan Y, Tang W, Fu K, Ji T. Transcriptomic Analysis of the Developing Testis and Spermatogenesis in Qianbei Ma Goats. Genes. 2023; 14(7):1334. https://doi.org/10.3390/genes14071334
Chicago/Turabian StyleZou, Yue, Xiang Chen, Xingzhou Tian, Wei Guo, Yong Ruan, Wen Tang, Kaibin Fu, and Taotao Ji. 2023. "Transcriptomic Analysis of the Developing Testis and Spermatogenesis in Qianbei Ma Goats" Genes 14, no. 7: 1334. https://doi.org/10.3390/genes14071334
APA StyleZou, Y., Chen, X., Tian, X., Guo, W., Ruan, Y., Tang, W., Fu, K., & Ji, T. (2023). Transcriptomic Analysis of the Developing Testis and Spermatogenesis in Qianbei Ma Goats. Genes, 14(7), 1334. https://doi.org/10.3390/genes14071334






