The Dynamics of Gene Expression Unraveling the Immune Response of Macrobrachium rosenbergii Infected by Aeromonas veronii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Prawns and A. veronii Preparation
2.2. Sample Collection
2.3. Total RNA Extraction and Illumina Sequencing
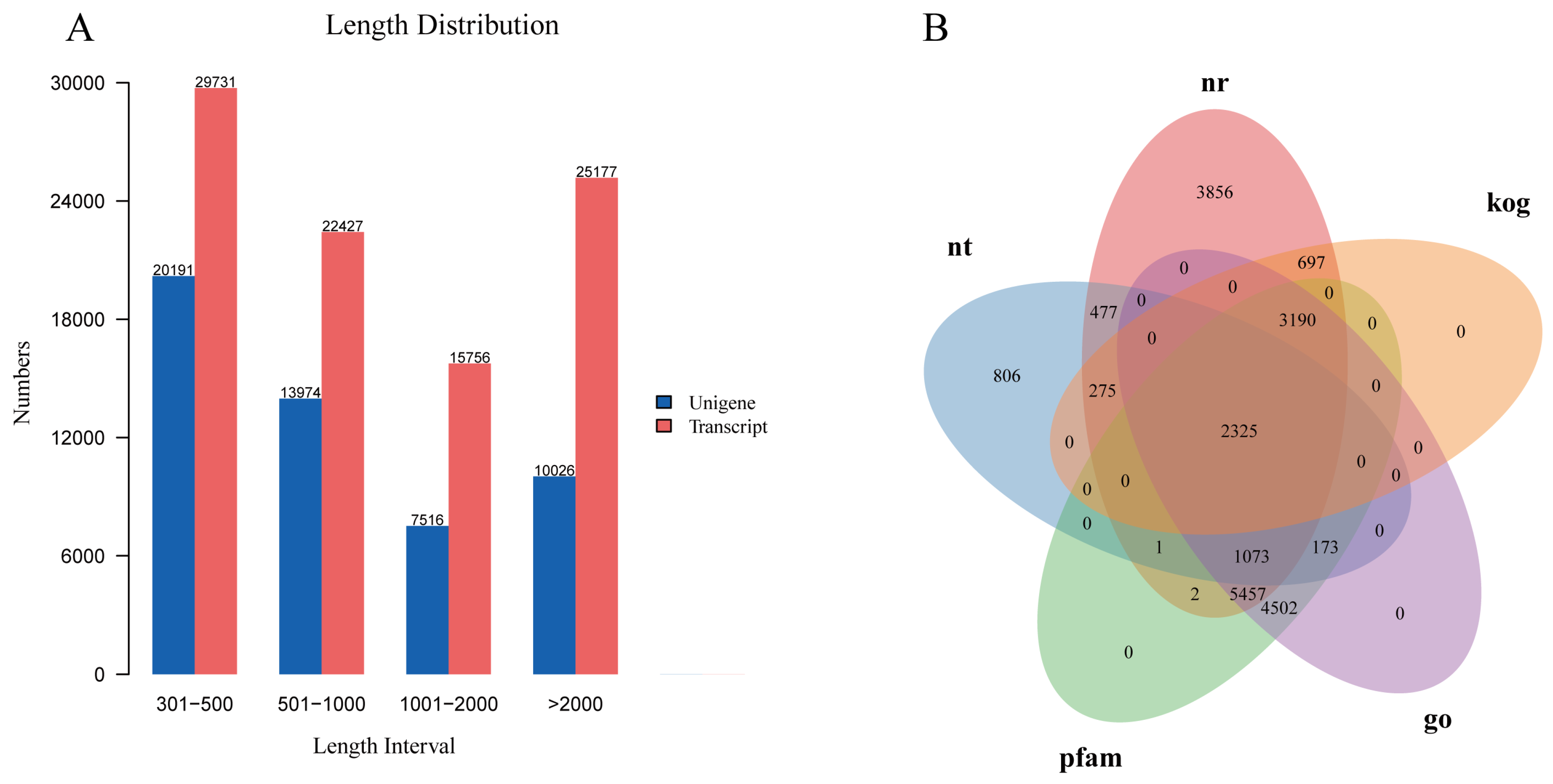
2.4. De Novo Assembly, Annotation, and Classification
2.5. DEGs, KEGG, and GO Enrichment Analysis
2.6. Validation of Immune-Related DEGs by RT-qPCR
2.7. Expression Patterns of Four Key Immune-Related DEGs in Different Tissues
2.8. Statistical Analysis
3. Results
3.1. Transcript Assembly, Gene Functional Annotation, GO, and KEGG Classification
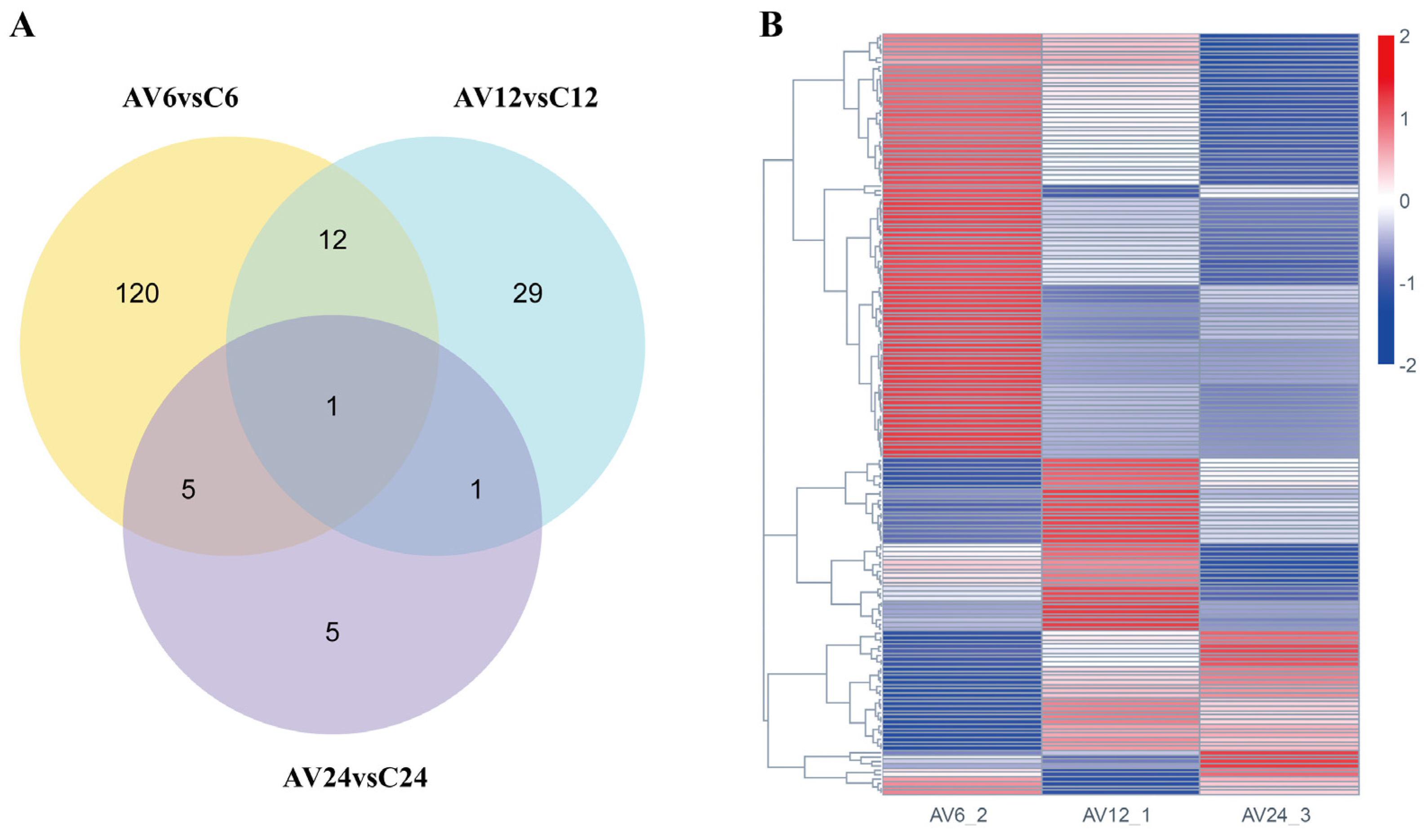
3.2. Identification of DEGs Related to A. veronii Infection
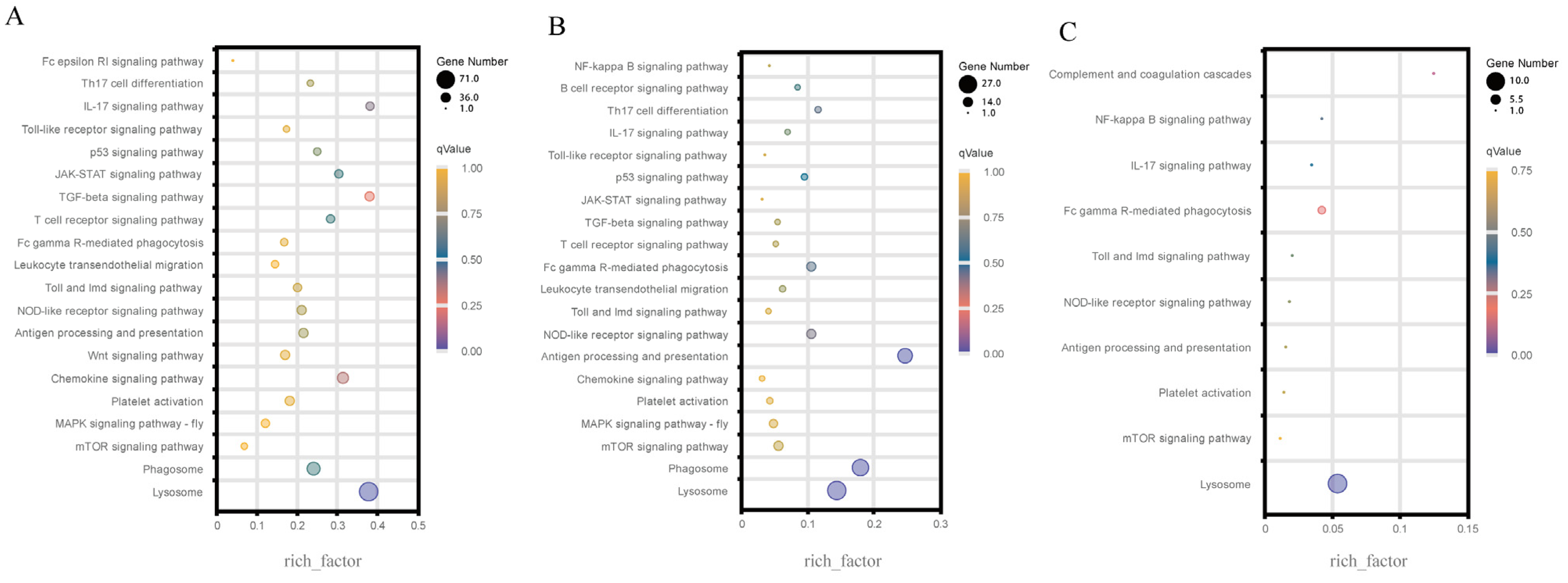
3.3. KEGG Enrichment of the Immune-Related DEGs
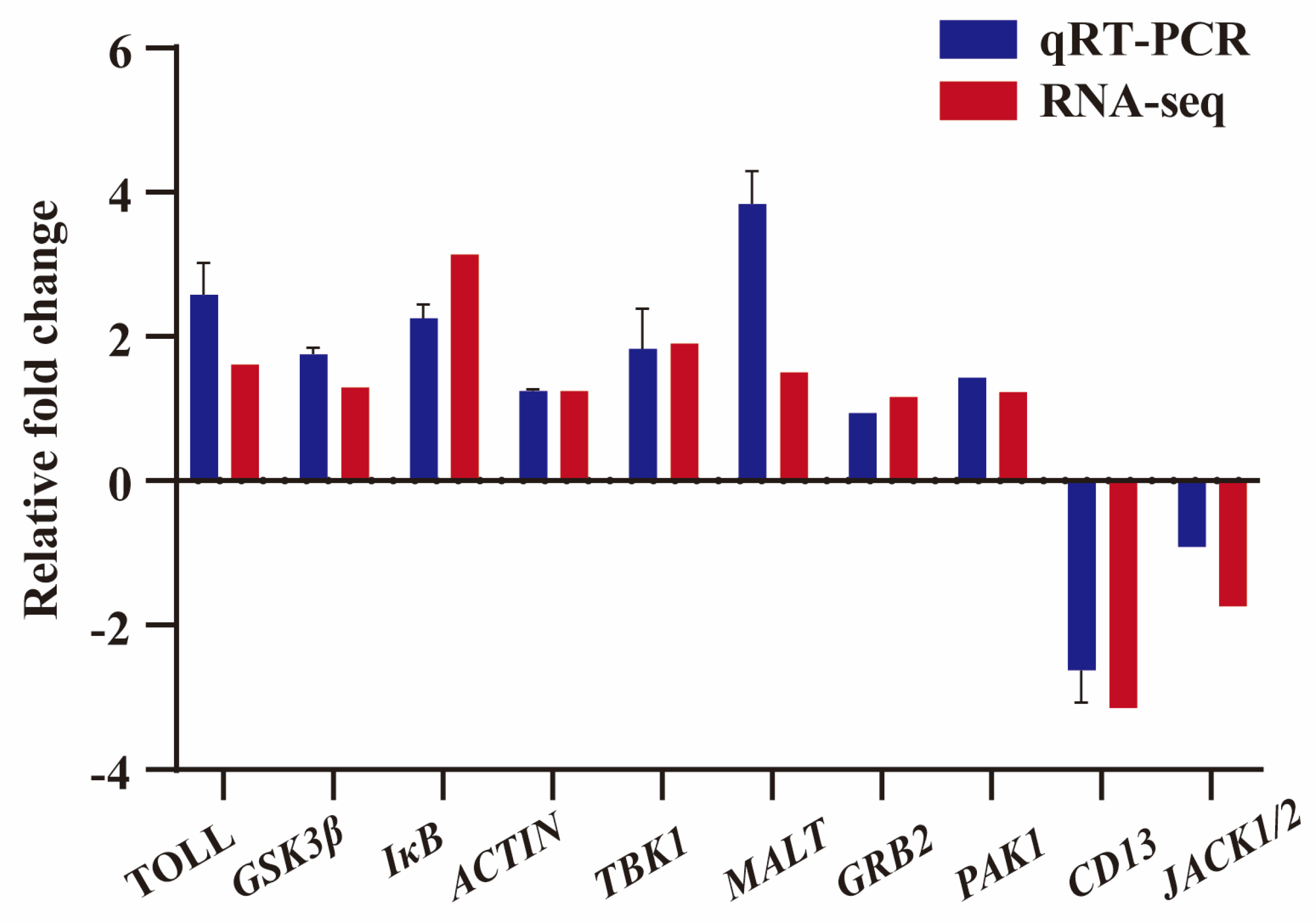
3.4. Validation of DEGs by qRT-PCR
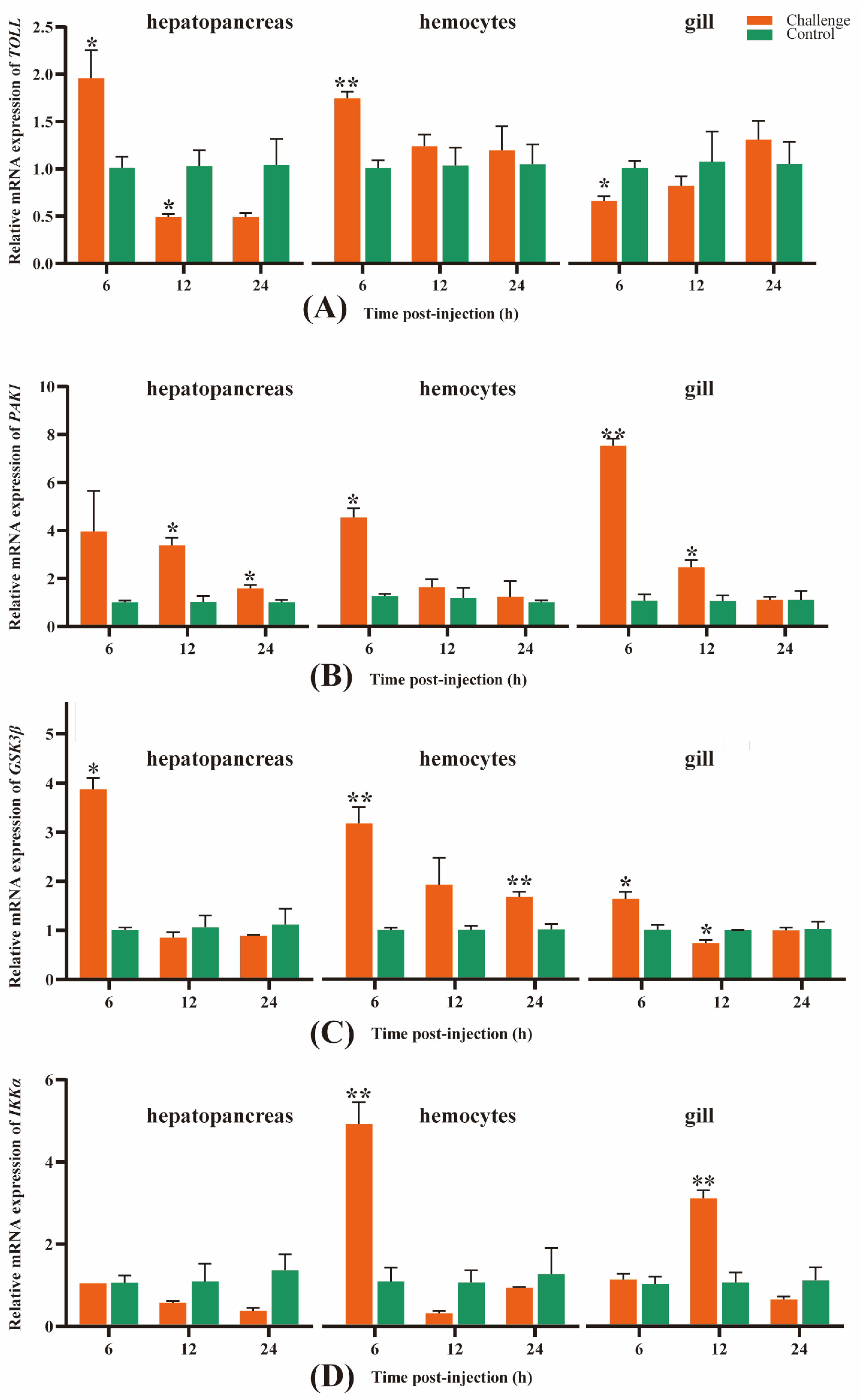
3.5. Temporal and Spatial Expression Levels of Four Key Immune-Related Genes in M. rosenbergii
4. Discussion
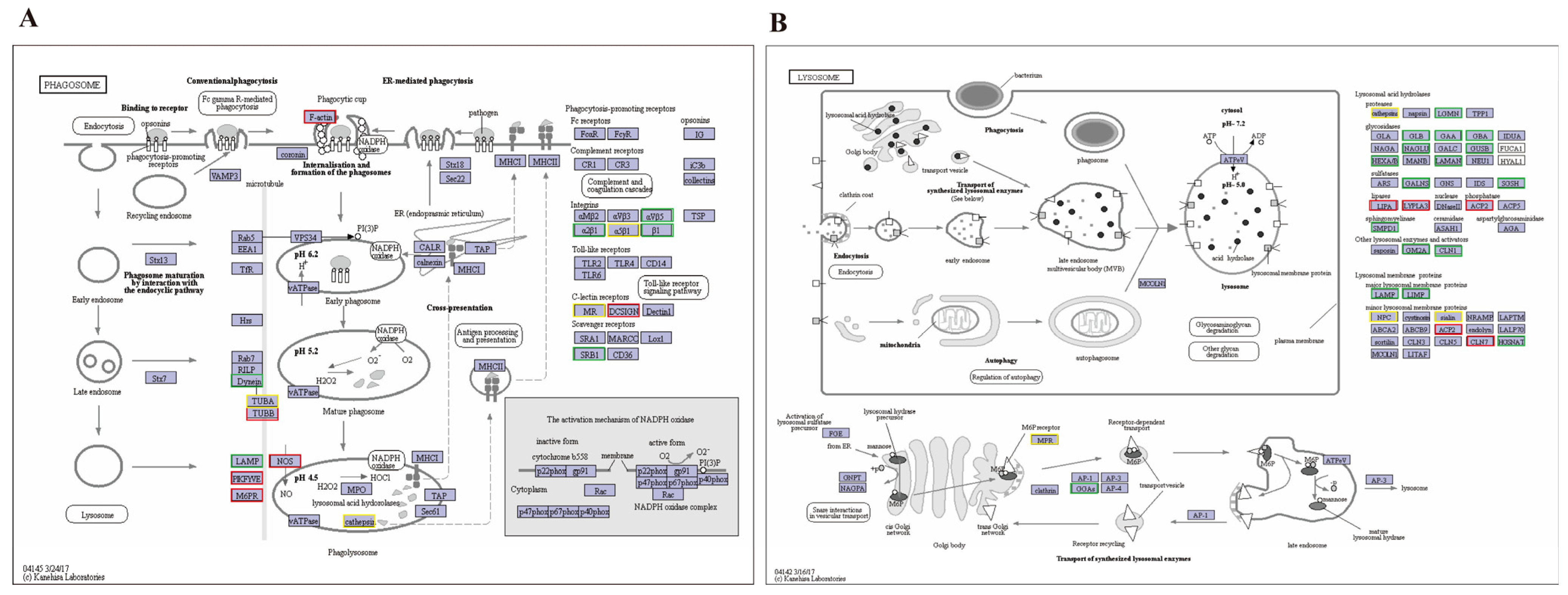
4.1. Phagosome and Lysosome Pathway Analysis
4.2. Important Immune-Related Genes Involved in the Immune Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fao, F.; Bondad-Reantaso, M.G.; Arthur, J.R. FAO Fisheries and Aquaculture Report NFIA/R1333 (En); FAO: Rome, Italy, 2021. [Google Scholar]
- Chen, K.F.; Maran, S.; Tan, W.S.; Ong, L.K.; Abidin, S.A.Z.; Othman, I.; Tey, B.T.; Lee, R.F.S. Meta-analysis of studies on pro tection provided by different prophylactic agents, their routes of administration and incubation times against nodavirus infection in Macrobrachium rosenbergii. Aquaculture 2023, 565, 739125. [Google Scholar] [CrossRef]
- Zhao, C.; Miu, Q.; Liu, S.; Zhou, D.; He, X.; Pang, J.; Weng, S.; He, J. Detection methods, epidemiological investigation, and host ranges of infectious precocity virus (IPV). Aquaculture 2023, 562, 738818. [Google Scholar] [CrossRef]
- Qian, Q.; Zhou, Y.; Chen, Z.; Zhu, Y.; Xu, J.; Gao, X.; Jiang, Q.; Wang, J.; Zhang, X. Pathogenesis and complete genome sequence of Decapod iridescent virus 1 (DIV1) associated with mass mortality in Macrobrachium rosenbergii. Aquaculture 2023, 566, 739220. [Google Scholar] [CrossRef]
- Zhao, C.; Wen, H.; Huang, S.; Weng, S.; He, J. A Novel Disease (Water Bubble Disease) of the Giant Freshwater Prawn Macrobrachium rosenbergii Caused by Citrobacter freundii: Antibiotic Treatment and Effects on the Antioxidant Enzyme Activity and Immune Responses. Antioxidants 2022, 11, 1491. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C. LPSN—List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef]
- Hickman-Brenner, F.W.; MacDonald, K.L.; Steigerwalt, A.G.; Fanning, G.R.; Brenner, D.J.; Farmer, J.J., 3rd. Aermonas veronii, a new ornithine decarboxylase-positive species that may cause diarrhea. J. Clin. Microbiol. 1987, 25, 900–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, H.T.; Techatanakitarnan, C.; Jindakittikul, P.; Thaiprayoon, A.; Taengphu, S.; Charoensapsri, W.; Khunrae, P.; Rattanarojpong, T.; Senapin, S. Aeromonas jandaei and Aeromonas veronii caused disease and mortality in Nile tilapia, Oreochromis niloticus (L.). J. Fish Dis. 2017, 40, 1395–1403. [Google Scholar] [CrossRef]
- Zhai, W.; Wang, Q.; Zhu, X.; Jia, X.; Chen, L. Pathogenic infection and microbial composition of yellow catfish (Pelteobagrus fulvidraco) challenged by Aeromonas veronii and Proteus mirabilis. Aquac. Fish 2023, 8, 166–173. [Google Scholar] [CrossRef]
- Liu, G.; Li, J.; Jiang, Z.; Zhu, X.; Gao, X.; Jiang, Q.; Wang, J.; Wei, W.; Zhang, X. Pathogenicity of Aeromonas veronii causing mass mortalities of Odontobutis potamophila and its induced host immune response. Fish Shellfish Immunol. 2022, 125, 180–189. [Google Scholar] [CrossRef]
- Peng, X.; Tu, H.H.; Luo, J.P.; Zhong, Z.X.; Lan, X.; Tang, Q.Y.; Yi, S.K.; Xia, Z.L.; Cai, M.Y.; Yang, G.L. Isolation, identification and virulence gene analysis of pathogenic Aeromonas veronii in Microbrachium rosenberdii and its histopathological observation. Acta Hydrobiol. Sin. 2023, 47, 874–882. [Google Scholar]
- Kumaresan, V.; Palanisamy, R.; Pasupuleti, M.; Arockiaraj, J. Impacts of environmental and biological stressors on immune system of Macrobrachium rosenbergii. Rev. Aquacult. 2017, 9, 283–307. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, B.; Liu, M.; Jiang, K.; Wang, M.; Wang, L. Comparative transcriptome analysis reveals the different roles between hepatopancreas and intestine of Litopenaeus vannamei in immune response to aflatoxin B1 (AFB1) challenge. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2019, 222, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, M.; Zheng, H.; Ye, H.; Zhang, X.; Li, S. Source of hemolymph microbiota and their roles in the immune system of mud crab. Dev. Comp. Immunol. 2020, 102, 103470. [Google Scholar] [CrossRef]
- Gao, X.; Jiang, Z.; Zhang, S.; Chen, Q.; Tong, S.; Liu, X.; Jiang, Q.; Yang, H.; Wei, W.; Zhang, X. Transcriptome analysis and immune-related genes expression reveals the immune responses of Macrobrachium rosenbergii infected by Enterobacter cloacae. Fish Shellfish Immunol. 2020, 101, 66–77. [Google Scholar] [CrossRef]
- Wang, D.-L.; Zuo, D.; Wang, L.-M.; Sun, T.; Wang, Q.; Zhao, Y.-L. Effects of white spot syndrome virus infection on immuno-enzyme activities and ultrastructure in gills of Cherax quadricarinatus. Fish Shellfish Immunol. 2012, 32, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhang, J.; Dong, H.; Wang, Y.; Liu, Q.; Li, H. Oxidative stress response of the black tiger shrimp Penaeus monodon to Vibrio parahaemolyticus challenge. Fish Shellfish Immunol. 2015, 46, 354–365. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome. Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Lv, Z.; Li, C.; Sun, Y.; Jiang, H.; Zhao, M.; Zhao, X.; Shao, Y.; Chang, Y. Transcriptome profiling reveals key roles of phagosome and NOD-like receptor pathway in spotting diseased Strongylocentrotus intermedius. Fish Shellfish Immunol. 2019, 84, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Torunn, T.E.; Løvdal, T.; Berg, T. Phagosome dynamics and function. Bioessays 2000, 22, 255–263. [Google Scholar]
- Swanson, J.A.; Baer, S.C. Phagocytosis by zippers and triggers. Trends. Cell Biol. 1995, 5, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Dambuza, I.M.; Brown, G.D. C-type lectins in immunity: Recent developments. Curr. Opin. Immunol. 2015, 32, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.B.; Yan, J.J.; Kang, J.S.; Kim, L.K.; Kim, Y.J. Macrophage C-type lectin is essential for phagosome maturation and acidification during Escherichia coli-induced peritonitis. Biochem. Biophys. Res. Commun. 2017, 493, 1491–1497. [Google Scholar] [CrossRef]
- Nishi, T.; Forgac, M. The vacuolar (H+)-ATPases--nature’s most versatile proton pumps. Nat. Rev. Mol. Cell. Biol. 2002, 3, 94–103. [Google Scholar] [CrossRef]
- Pu, J.; Guardia, C.M.; Keren-Kaplan, T.; Bonifacino, J.S. Mechanisms and functions of lysosome positioning. J. Cell. Sci. 2016, 129, 4329–4339. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Porto, A.F. Cholesteryl ester storage disease: Protean presentations of lysosomal acid lipase deficiency. J. Pediatr. Gastroenterol. Nutr. 2013, 56, 682–685. [Google Scholar] [CrossRef]
- Hiraoka, M.; Abe, A.; Shayman, J.A. Cloning and characterization of a lysosomal phospholipase A2, 1-O-acylceramide synthase. J. Biol. Chem. 2002, 277, 10090–10099. [Google Scholar] [CrossRef] [Green Version]
- Taniyama, Y.; Fuse, H.; Satomi, T.; Tozawa, R.; Yasuhara, Y.; Shimakawa, K.; Shibata, S.; Hattori, M.; Nakata, M.; Taketomi, S. Loss of lysophospholipase 3 increases atherosclerosis in apolipoprotein E-deficient mice. Biochem. Biophys. Res. Commun. 2005, 330, 104–110. [Google Scholar] [CrossRef]
- Du, J.; Zhu, H.; Liu, P.; Chen, J.; Xiu, Y.; Yao, W.; Wu, T.; Ren, Q.; Meng, Q.; Gu, W.; et al. Immune responses and gene expression in hepatopancreas from Macrobrachium rosenbergii challenged by a novel pathogen spiroplasma MR-1008. Fish Shellfish Immunol. 2013, 34, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Danyukova, T.; Ariunbat, K.; Thelen, M.; Brocke-Ahmadinejad, N.; Mole, S.E.; Storch, S. Loss of CLN7 results in depletion of soluble lysosomal proteins and impaired mTOR reactivation. Hum. Mol. Genet. 2018, 27, 1711–1722. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, E.J.A.; Li, L. Toll-Interacting Protein in Resolving and Non-Resolving Inflammation. Front. Immunol. 2017, 8, 511. [Google Scholar] [CrossRef] [Green Version]
- Ou, J.; Chen, H.; Liu, Q.; Bian, Y.; Luan, X.; Jiang, Q.; Ji, H.; Wang, Z.; Lv, L.; Dong, X.; et al. Integrated transcriptome analysis of immune-related mRNAs and microRNAs in Macrobrachium rosenbergii infected with Spiroplasma eriocheiris. Fish Shellfish Immunol. 2021, 119, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhao, L.; Jin, M.; Li, T.; Wu, L.; Chen, Y.; Ren, Q. Toll receptor response to white spot syndrome virus challenge in giant freshwater prawns (Macrobrachium rosenbergii). Fish Shellfish Immunol. 2016, 57, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Wang, X.; Jin, X.Y.; Solaro, R.J.; Lei, M. PAK1 is a novel cardiac protective signaling molecule. Front. Med. 2014, 8, 399–403. [Google Scholar] [CrossRef]
- Ren, L.J.; Li, K.Q.; Zhang, Y.Y.; Wang, y.; Yu, Y.; Cheng, Y.; Lin, K.; Song, J.; Chang, Y.Q. Isolation of a new PAK1 Gene from Sea Cucumber (Apostichopus japonicus) and its Expression analysis and function characterization. J. Ocean Univ. China 2019, 18, 1147–1157. [Google Scholar] [CrossRef]
- Beurel, E.; Grieco, S.F.; Jope, R.S. Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 2015, 148, 114–131. [Google Scholar] [CrossRef] [Green Version]
- Ruan, L.; Liu, H.; Shi, H. Characterization and function of GSK3β from Litopenaeus vannamei in WSSV infection. Fish Shellfish Immunol. 2018, 82, 220–228. [Google Scholar] [CrossRef]
- Zhang, S.; Zhu, L.; Hou, C.; Yuan, H.; Yang, S.; Dehwah, M.A.S.; Shi, L. GSK3β Plays a Negative Role During White Spot Syndrome Virus (WSSV) Infection by Regulating NF-κB Activity in Shrimp Litopenaeus vannamei. Front. Immunol. 2020, 11, 607543. [Google Scholar] [CrossRef]
- Solt, L.A.; May, M.J. The IκB kinase complex: Master regulator of NF-κB signaling. Immunol. Res. 2008, 42, 3–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viatour, P.; Merville, M.P.; Bours, V.; Chariot, A. Phosphorylation of NF-κB and IκB proteins: Implications in cancer and inflammation. Trends Biochem. Sci. 2005, 30, 43–45. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal. Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreiro, D.U.; Komives, E.A. Molecular mechanisms of system control of NF-κB signaling by IκBα. Biochemistry 2010, 49, 1560–1567. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Xia, Y.; Parker, A.S.; Verma, I.M. IKK biology. Immunol. Rev. 2012, 246, 239–253. [Google Scholar] [CrossRef] [Green Version]

| Primer | Gene Name or Usage | Sequence (5′→3′) | Annealing Temperature (°C) |
|---|---|---|---|
| 18S-F | Used as reference gene | TATACGCTAGTGGAGCTGGAA | 59 |
| 18S-R | GGGGAGGTAGTGACGAAAAAT | ||
| GSK3β-F | Glycogen synthase kinase 3 β | ACCCGTGAGCAGATTAGA | 59 |
| GSK3β-R | GCCTGAAGTGGCGTGATA | ||
| 1κB-F | NF-kappa-B inhibitor α | GCATAATGGCTATTGAACTG | 59 |
| 1κB-R | TCCCAAGATGGAACGCTA | ||
| PAK1-F | p21-activated kinase 1 | TTCGTCGGAAGGTAGAGG | 59 |
| PAK1-R | GAGGCTGGTCGGTGGTAT | ||
| TBK1-F | Tank-binding kinase 1 | AGAGGAGCAAGAAGGTCG | 59 |
| TBK1-R | CAGGCTTCAAGTCACGATGT | ||
| TOLL-F | Protein toll | CAAACCGTCGGAGGAACA | 59 |
| TOLL-R | CCTTGACTGCCACTGAAC | ||
| CD13-F | Aminopeptidase N | GAGTGCCGACTTCCAACC | 59 |
| CD13-R | CAAGACCTCCAGAACAATA | ||
| Actin-F | Actin β/γ 1 | ATGGTCGGTATGGGTCAGA | 59 |
| Actin-R | AGGTGCTACACGGAGTTCA | ||
| GRB2-F | Growth factor receptor-Bound protein 2 | GAAGGACTTATTCCCAGCAA | 59 |
| GRB2-R | ACCATCGCCACATTTAGG | ||
| IKKα-F | Inhibitor of nuclear factor kappa-B kinase subunit α | AATATCCCACTTGAAGCC | 59 |
| IKKα-R | CGTTGAAACAGGACGAAA | ||
| MALT-F | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | CGGAAGGACGGCGTTACAT | 59 |
| MALT-R | CACGGTCACGGGTCTGGTT | ||
| JAK1/2-F | Janus kinase 1 | AAAGAGCGGATGAGCAC | 59 |
| JAK1/2-R | CTGGCAAGTCCCGATGA |
| Genes | Fold Change | Up/Down Regulation | Pathway |
|---|---|---|---|
| Toll | 1.6164 | Up | Toll and Imd signaling pathway |
| PAK1 | 1.2329 | Up | Chemokine signaling pathway |
| Fc γ R-mediated phagocytosis | |||
| Natural killer cell mediated cytotoxicity | |||
| GSK3B | 1.2959 | Up | mTOR signaling pathway |
| Chemokine signaling pathway | |||
| Wnt signaling pathway | |||
| T cell receptor signaling pathway | |||
| IL-17 signaling pathway | |||
| B cell receptor signaling pathway | |||
| IKKα | −5.0217 | Down | mTOR signaling pathway |
| Chemokine signaling pathway | |||
| NOD-like receptor signaling pathway | |||
| T cell receptor signaling pathway | |||
| Toll-like receptor signaling pathway | |||
| IL-17 signaling pathway | |||
| Th17 cell differentiation | |||
| B cell receptor signaling pathway | |||
| NF-kappa B signaling pathway | |||
| RIG-I-like receptor signaling pathway | |||
| Cytosolic DNA-sensing pathway | |||
| Th1 and Th2 cell differentiation |
| Pathway | Genes | Fold Change | Up/Down Regulation | Genes | Fold Change | Up/Down Regulation |
|---|---|---|---|---|---|---|
| Phagosome | F-actin | 1.7692 | Up | Dvnein | −1.2674 | Down |
| TUBB | 2.9491 | Up | LAMP | −7.1505 | Down | |
| PIKFYVE | 1.1451 | Up | αVβ5 | −3.1955 | Down | |
| M6PR | 1.315 | Up | α2β1 | −2.9904 | Down | |
| NOS | 5.1266 | Up | β1 | −1.4217 | Down | |
| DCSIGN | 2.472 | Up | SRB1 | −2.3453 | Down | |
| Lysosome | LIPA | 1.7805 | Up | LGMN | −1.3984 | Down |
| LYPLA | 1.6242 | Up | GLB | −2.5314 | Down | |
| ACP2 | 1.7805 | Up | GAA | −1.8661 | Down | |
| CLN7 | 2.5625 | Up | GBA | −1.9262 | Down | |
| LAMAN | −5.2917 | Down | NAGLU | −2.5669 | Down | |
| GANLS | −2.0659 | Down | GUSB | −2.8311 | Down | |
| SGSH | −2.5976 | Down | HXA/P | −3.5577 | Down | |
| GM2A | −2.4086 | Down | LAMP | −7.1505 | Down | |
| CLN1 | −1.498 | Down | HGSNAT | −1.2058 | Down | |
| GGAS | −1.2001 | Down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, X.; Lan, X.; Zhong, Z.; Tu, H.; Yao, X.; Tang, Q.; Xia, Z.; Yang, G.; Yi, S. The Dynamics of Gene Expression Unraveling the Immune Response of Macrobrachium rosenbergii Infected by Aeromonas veronii. Genes 2023, 14, 1383. https://doi.org/10.3390/genes14071383
Peng X, Lan X, Zhong Z, Tu H, Yao X, Tang Q, Xia Z, Yang G, Yi S. The Dynamics of Gene Expression Unraveling the Immune Response of Macrobrachium rosenbergii Infected by Aeromonas veronii. Genes. 2023; 14(7):1383. https://doi.org/10.3390/genes14071383
Chicago/Turabian StylePeng, Xin, Xuan Lan, Zhenxiao Zhong, Haihui Tu, Xinyi Yao, Qiongying Tang, Zhenglong Xia, Guoliang Yang, and Shaokui Yi. 2023. "The Dynamics of Gene Expression Unraveling the Immune Response of Macrobrachium rosenbergii Infected by Aeromonas veronii" Genes 14, no. 7: 1383. https://doi.org/10.3390/genes14071383





