Association of Single Nucleotide Polymorphisms of Cytokine Genes with Depression, Schizophrenia and Bipolar Disorder
Abstract
:1. Introduction
2. Materials and Methods
2.1. Inclusion and Exclusion Criteria
2.2. Search Strategy
2.3. Assessment of Studies
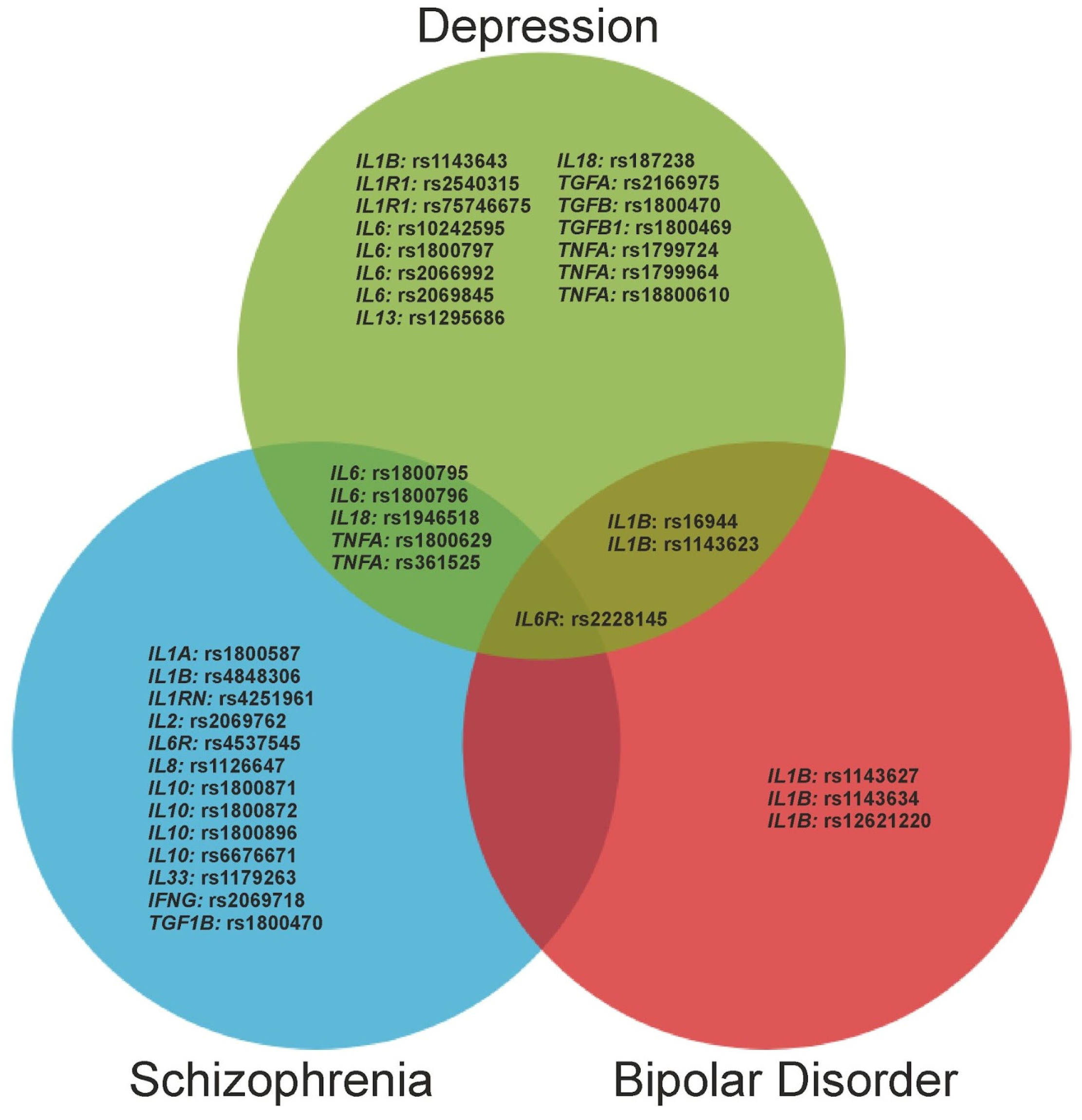
3. Results
3.1. Depressive Disorders
3.2. Schizophrenia
3.3. Bipolar Disorder
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stein, D.J.; Palk, A.C.; Kendler, K.S. What Is a Mental Disorder? An Exemplar-Focused Approach. Psychol. Med. 2021, 51, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Beurel, E.; Toups, M.; Nemeroff, C.B. The Bidirectional Relationship of Depression and Inflammation: Double Trouble. Neuron 2020, 107, 234–256. [Google Scholar] [CrossRef]
- Pedraz-Petrozzi, B.; Elyamany, O.; Rummel, C.; Mulert, C. Effects of Inflammation on the Kynurenine Pathway in Schizophrenia—A Systematic Review. J. Neuroinflammation 2020, 17, 56. [Google Scholar] [CrossRef] [Green Version]
- Troubat, R.; Barone, P.; Leman, S.; Desmidt, T.; Cressant, A.; Atanasova, B.; Brizard, B.; El Hage, W.; Surget, A.; Belzung, C.; et al. Neuroinflammation and Depression: A Review. Eur. J. Neurosci. 2021, 53, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Yuan, N.; Chen, Y.; Xia, Y.; Dai, J.; Liu, C. Inflammation-Related Biomarkers in Major Psychiatric Disorders: A Cross-Disorder Assessment of Reproducibility and Specificity in 43 Meta-Analyses. Transl. Psychiatry 2019, 9, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becher, B.; Spath, S.; Goverman, J. Cytokine Networks in Neuroinflammation. Nat. Rev. Immunol. 2017, 17, 49–59. [Google Scholar] [CrossRef]
- Chakrabarty, T.; Torres, I.J.; Bond, D.J.; Yatham, L.N. Inflammatory Cytokines and Cognitive Functioning in Early-Stage Bipolar I Disorder. J. Affect. Disord. 2019, 245, 679–685. [Google Scholar] [CrossRef]
- Strawbridge, R.; Hodsoll, J.; Powell, T.R.; Hotopf, M.; Hatch, S.L.; Breen, G.; Cleare, A.J. Inflammatory Profiles of Severe Treatment-Resistant Depression. J. Affect. Disord. 2019, 246, 42–51. [Google Scholar] [CrossRef] [Green Version]
- Wilhelm, C.J.; Fuller, B.E.; Huckans, M.; Loftis, J.M. Peripheral Immune Factors Are Elevated in Women with Current or Recent Alcohol Dependence and Associated with Altered Mood and Memory. Drug Alcohol Depend. 2017, 176, 71–78. [Google Scholar] [CrossRef]
- Comer, A.L.; Carrier, M.; Tremblay, M.-È.; Cruz-Martín, A. The Inflamed Brain in Schizophrenia: The Convergence of Genetic and Environmental Risk Factors That Lead to Uncontrolled Neuroinflammation. Front. Cell. Neurosci. 2020, 14, 274. [Google Scholar] [CrossRef]
- Ermakov, E.A.; Mednova, I.A.; Boiko, A.S.; Buneva, V.N.; Ivanova, S.A. Chemokine Dysregulation and Neuroinflammation in Schizophrenia: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 2215. [Google Scholar] [CrossRef] [PubMed]
- Ermakov, E.A.; Melamud, M.M.; Buneva, V.N.; Ivanova, S.A. Immune System Abnormalities in Schizophrenia: An Integrative View and Translational Perspectives. Front. Psychiatry 2022, 13, 880568. [Google Scholar] [CrossRef] [PubMed]
- Imai, R.; Hori, H.; Itoh, M.; Lin, M.; Niwa, M.; Ino, K.; Ogawa, S.; Ishida, M.; Sekiguchi, A.; Matsui, M.; et al. Inflammatory Markers and Their Possible Effects on Cognitive Function in Women with Posttraumatic Stress Disorder. J. Psychiatr. Res. 2018, 102, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.; Mondelli, V.; Pariante, C.M. Genetic Contributions of Inflammation to Depression. Neuropsychopharmacology 2017, 42, 81–98. [Google Scholar] [CrossRef] [Green Version]
- Dunn, G.A.; Loftis, J.M.; Sullivan, E.L. Neuroinflammation in Psychiatric Disorders: An Introductory Primer. Pharmacol. Biochem. Behav. 2020, 196, 172981. [Google Scholar] [CrossRef]
- Chen, X.; Yao, T.; Cai, J.; Fu, X.; Li, H.; Wu, J. Systemic Inflammatory Regulators and 7 Major Psychiatric Disorders: A Two-Sample Mendelian Randomization Study. Prog. Neuropsychopharmacol. Biol. Psychiatry 2022, 116, 110534. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Oskooei, V.K.; Omrani, M.D.; Taheri, M. Dysregulation of Cytokine Coding Genes in Peripheral Blood of Bipolar Patients. J. Affect. Disord. 2019, 256, 578–583. [Google Scholar] [CrossRef]
- Bialek, K.; Czarny, P.; Watala, C.; Synowiec, E.; Wigner, P.; Bijak, M.; Talarowska, M.; Galecki, P.; Szemraj, J.; Sliwinski, T. Preliminary Study of the Impact of Single-Nucleotide Polymorphisms of IL-1α, IL-1β and TNF-α Genes on the Occurrence, Severity and Treatment Effectiveness of the Major Depressive Disorder. Cell. Mol. Neurobiol. 2020, 40, 1049–1056. [Google Scholar] [CrossRef]
- Mednova, I.A.; Levchuk, L.A.; Boiko, A.S.; Roschina, O.V.; Simutkin, G.G.; Bokhan, N.A.; Loonen, A.J.M.; Ivanova, S.A. Cytokine Level in Patients with Mood Disorder, Alcohol Use Disorder and Their Comorbidity. World J. Biol. Psychiatry 2023, 24, 243–253. [Google Scholar] [CrossRef]
- Çakici, N.; Sutterland, A.L.; Penninx, B.W.J.H.; Dalm, V.A.; de Haan, L.; van Beveren, N.J.M. Altered Peripheral Blood Compounds in Drug-Naïve First-Episode Patients with Either Schizophrenia or Major Depressive Disorder: A Meta-Analysis. Brain Behav. Immun. 2020, 88, 547–558. [Google Scholar] [CrossRef]
- Kaufmann, F.N.; Costa, A.P.; Ghisleni, G.; Diaz, A.P.; Rodrigues, A.L.S.; Peluffo, H.; Kaster, M.P. NLRP3 Inflammasome-Driven Pathways in Depression: Clinical and Preclinical Findings. Brain Behav. Immun. 2017, 64, 367–383. [Google Scholar] [CrossRef]
- Poletti, S.; Vai, B.; Mazza, M.G.; Zanardi, R.; Lorenzi, C.; Calesella, F.; Cazzetta, S.; Branchi, I.; Colombo, C.; Furlan, R.; et al. A Peripheral Inflammatory Signature Discriminates Bipolar from Unipolar Depression: A Machine Learning Approach. Prog. Neuropsychopharmacol. Biol. Psychiatry 2021, 105, 110136. [Google Scholar] [CrossRef] [PubMed]
- Frydecka, D.; Krzystek-Korpacka, M.; Lubeiro, A.; Stramecki, F.; Stańczykiewicz, B.; Beszłej, J.A.; Piotrowski, P.; Kotowicz, K.; Szewczuk-Bogusławska, M.; Pawlak-Adamska, E.; et al. Profiling Inflammatory Signatures of Schizophrenia: A Cross-Sectional and Meta-Analysis Study. Brain Behav. Immun. 2018, 71, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Kalmady, S.V.; Shivakumar, V.; Jose, D.; Ravi, V.; Keshavan, M.S.; Gangadhar, B.N.; Venkatasubramanian, G. Plasma Cytokines in Minimally Treated Schizophrenia. Schizophr. Res. 2018, 199, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Tansey, K.E.; Schalkwyk, L.C.; Powell, T.R. The Inflammatory Cytokines: Molecular Biomarkers for Major Depressive Disorder? Biomark. Med. 2015, 9, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Rizavi, H.S.; Ren, X.; Zhang, H.; Bhaumik, R.; Pandey, G.N. Abnormal Gene Expression of Proinflammatory Cytokines and Their Membrane-Bound Receptors in the Lymphocytes of Depressed Patients. Psychiatry Res. 2016, 240, 314–320. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.-P.; Liang, S.; Pan, M.; Sun, R.-L.; Chen, C.; Luan, H.; Jiang, M.-H. Interleukin-6 Genotypes and Serum Levels in Chinese Hui Population. Int. J. Clin. Exp. Med. 2014, 7, 2851–2857. [Google Scholar]
- Köhler, C.A.; Freitas, T.H.; Maes, M.; de Andrade, N.Q.; Liu, C.S.; Fernandes, B.S.; Stubbs, B.; Solmi, M.; Veronese, N.; Herrmann, N.; et al. Peripheral Cytokine and Chemokine Alterations in Depression: A Meta-Analysis of 82 Studies. Acta Psychiatr. Scand. 2017, 135, 373–387. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-M.; Kang, H.-J.; Kim, J.-W.; Bae, K.-Y.; Kim, S.-W.; Kim, J.-T.; Park, M.-S.; Cho, K.-H. Associations of Tumor Necrosis Factor-α and Interleukin-1β Levels and Polymorphisms with Post-Stroke Depression. Am. J. Geriatr. Psychiatry 2017, 25, 1300–1308. [Google Scholar] [CrossRef]
- Tartter, M.; Hammen, C.; Bower, J.E.; Brennan, P.A.; Cole, S. Effects of Chronic Interpersonal Stress Exposure on Depressive Symptoms Are Moderated by Genetic Variation at IL6 and IL1β in Youth. Brain Behav. Immun. 2015, 46, 104–111. [Google Scholar] [CrossRef] [Green Version]
- Lezheiko, T.V.; Andryushchenko, A.V.; Korovaitseva, G.I.; Kondratiev, N.V.; Gabaeva, M.V.; Krikova, E.V.; Golimbet, V.E. A Study on the Association of Genes for pro-Inflammatory Cytokines and Depression. Zhurnal Nevrol. Psikhiatrii Im. S.S. Korsakova 2018, 118, 89–93. [Google Scholar] [CrossRef] [PubMed]
- McQuaid, R.J.; Gabrys, R.L.; McInnis, O.A.; Anisman, H.; Matheson, K. Understanding the Relation Between Early-Life Adversity and Depression Symptoms: The Moderating Role of Sex and an Interleukin-1β Gene Variant. Front. Psychiatry 2019, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, D.; Eszlari, N.; Petschner, P.; Pap, D.; Vas, S.; Kovacs, P.; Gonda, X.; Juhasz, G.; Bagdy, G. Effects of IL1B Single Nucleotide Polymorphisms on Depressive and Anxiety Symptoms Are Determined by Severity and Type of Life Stress. Brain Behav. Immun. 2016, 56, 96–104. [Google Scholar] [CrossRef]
- Kang, H.-J.; Bae, K.-Y.; Kim, S.-W.; Shin, I.-S.; Hong, Y.J.; Ahn, Y.; Jeong, M.H.; Yoon, J.-S.; Kim, J.-M. Relationship between Interleukin-1β and Depressive Disorder after Acute Coronary Syndrome. Prog. Neuropsychopharmacol. Biol. Psychiatry 2017, 72, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Draganov, M.; Arranz, M.J.; Salazar, J.; de Diego-Adeliño, J.; Gallego-Fabrega, C.; Jubero, M.; Carceller-Sindreu, M.; Portella, M.J. Association Study of Polymorphisms within Inflammatory Genes and Methylation Status in Treatment Response in Major Depression. Eur. Psychiatry 2019, 60, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-K.; Hong, J.-P.; Hwang, J.-A.; Lee, H.-J.; Yoon, H.-K.; Lee, B.-H.; Jung, H.-Y.; Hahn, S.-W.; Na, K.-S. TNF-Alpha -308G>A Polymorphism Is Associated with Suicide Attempts in Major Depressive Disorder. J. Affect. Disord. 2013, 150, 668–672. [Google Scholar] [CrossRef]
- Tsai, S.-J.; Kao, C.-F.; Su, T.-P.; Li, C.-T.; Lin, W.-C.; Hong, C.-J.; Bai, Y.-M.; Tu, P.-C.; Chen, M.-H. Cytokine- and Vascular Endothelial Growth Factor-Related Gene-Based Genome-Wide Association Study of Low-Dose Ketamine Infusion in Patients with Treatment-Resistant Depression. CNS Drugs 2023, 37, 243–253. [Google Scholar] [CrossRef]
- Kim, J.-M.; Stewart, R.; Kim, S.-Y.; Kang, H.-J.; Jang, J.-E.; Kim, S.-W.; Shin, I.-S.; Park, M.-H.; Yoon, J.-H.; Park, S.-W.; et al. A One Year Longitudinal Study of Cytokine Genes and Depression in Breast Cancer. J. Affect. Disord. 2013, 148, 57–65. [Google Scholar] [CrossRef]
- Lückhoff, H.K.; Kidd, M.; van Rensburg, S.J.; van Velden, D.P.; Kotze, M.J. Apolipoprotein E Genotyping and Questionnaire-Based Assessment of Lifestyle Risk Factors in Dyslipidemic Patients with a Family History of Alzheimer’s Disease: Test Development for Clinical Application. Metab. Brain Dis. 2016, 31, 213–224. [Google Scholar] [CrossRef]
- Lu, D.; Wang, M.; Yang, T.; Wang, J.; Lin, B.; Liu, G.; Liang, Q. Association of Interleukin-6 Polymorphisms with Schizophrenia and Depression: A Case-Control Study. Lab. Med. 2023, 54, 250–255. [Google Scholar] [CrossRef]
- Golimbet, V.E.; Volel, B.A.; Korovaitseva, G.I.; Kasparov, S.V.; Kondratiev, N.V.; Kopylov, F.Y. Association of Inflammatory Genes with Neuroticism, Anxiety and Depression in Male Patients with Coronary Heart Disease. Zhurnal Nevrol. Psikhiatrii Im. S.S. Korsakova 2017, 117, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, D.; Eszlari, N.; Petschner, P.; Pap, D.; Vas, S.; Kovacs, P.; Gonda, X.; Bagdy, G.; Juhasz, G. Interleukin-6 Promoter Polymorphism Interacts with Pain and Life Stress Influencing Depression Phenotypes. J. Neural Transm. 2016, 123, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Gal, Z.; Torok, D.; Gonda, X.; Eszlari, N.; Anderson, I.M.; Deakin, B.; Juhasz, G.; Bagdy, G.; Petschner, P. Inflammation and Blood-Brain Barrier in Depression: Interaction of CLDN5 and IL6 Gene Variants in Stress-Induced Depression. Int. J. Neuropsychopharmacol. 2023, 26, 189–197. [Google Scholar] [CrossRef]
- Udina, M.; Moreno-España, J.; Navinés, R.; Giménez, D.; Langohr, K.; Gratacòs, M.; Capuron, L.; de la Torre, R.; Solà, R.; Martín-Santos, R. Serotonin and Interleukin-6: The Role of Genetic Polymorphisms in IFN-Induced Neuropsychiatric Symptoms. Psychoneuroendocrinology 2013, 38, 1803–1813. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, Z.; Zhao, G.; Wang, F.; Fang, Y. Identification of IL6 as a Susceptibility Gene for Major Depressive Disorder. Sci. Rep. 2016, 6, 31264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciukiewicz, M.; Marshe, V.S.; Tiwari, A.K.; Fonseka, T.M.; Freeman, N.; Rotzinger, S.; Foster, J.A.; Kennedy, J.L.; Kennedy, S.H.; Müller, D.J. Genetic Variation in IL-1β, IL-2, IL-6, TSPO and BDNF and Response to Duloxetine or Placebo Treatment in Major Depressive Disorder. Pharmacogenomics 2015, 16, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Khandaker, G.M.; Zammit, S.; Burgess, S.; Lewis, G.; Jones, P.B. Association between a Functional Interleukin 6 Receptor Genetic Variant and Risk of Depression and Psychosis in a Population-Based Birth Cohort. Brain Behav. Immun. 2018, 69, 264–272. [Google Scholar] [CrossRef]
- Dunn, L.B.; Aouizerat, B.E.; Langford, D.J.; Cooper, B.A.; Dhruva, A.; Cataldo, J.K.; Baggott, C.R.; Merriman, J.D.; Dodd, M.; West, C.; et al. Cytokine Gene Variation Is Associated with Depressive Symptom Trajectories in Oncology Patients and Family Caregivers. Eur. J. Oncol. Nurs. 2013, 17, 346–353. [Google Scholar] [CrossRef] [Green Version]
- Doong, S.-H.; Dhruva, A.; Dunn, L.B.; West, C.; Paul, S.M.; Cooper, B.A.; Elboim, C.; Abrams, G.; Merriman, J.D.; Langford, D.J.; et al. Associations between Cytokine Genes and a Symptom Cluster of Pain, Fatigue, Sleep Disturbance, and Depression in Patients Prior to Breast Cancer Surgery. Biol. Res. Nurs. 2015, 17, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Santos, M.; Carvalho, S.; Lima, L.; Mota-Pereira, J.; Pimentel, P.; Maia, D.; Correia, D.; Gomes, S.; Cruz, A.; Medeiros, R. The Role of IL18-607C>A and IL18-137G>C Promoter Polymorphisms in Antidepressant Treatment Phenotypes: A Preliminary Report. Neurosci. Lett. 2016, 622, 107–112. [Google Scholar] [CrossRef] [Green Version]
- Sandoval-Carrillo, A.; Alvarado-Esquivel, C.; Salas-Martinez, C.; Mendez-Hernandez, E.M.; Sifuentes-Alvarez, A.; Martínez-Martinez, A.L.; Castillo-Orona, J.M.; Hernandez-Tinoco, J.; Antuna-Salcido, E.I.; Sanchez-Anguiano, L.F.; et al. TNF-α Polymorphisms and Maternal Depression in a Mexican Mestizo Population. CNS Neurol. Disord. Drug Targets 2018, 17, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Saad, S.; Dunn, L.B.; Koetters, T.; Dhruva, A.; Langford, D.J.; Merriman, J.D.; West, C.; Paul, S.M.; Cooper, B.; Cataldo, J.; et al. Cytokine Gene Variations Associated with Subsyndromal Depressive Symptoms in Patients with Breast Cancer. Eur. J. Oncol. Nurs. 2014, 18, 397–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bialek, K.; Czarny, P.; Watala, C.; Wigner, P.; Talarowska, M.; Galecki, P.; Szemraj, J.; Sliwinski, T. Novel Association between TGFA, TGFB1, IRF1, PTGS2 and IKBKB Single-Nucleotide Polymorphisms and Occurrence, Severity and Treatment Response of Major Depressive Disorder. PeerJ 2020, 8, e8676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mihailova, S.; Ivanova-Genova, E.; Lukanov, T.; Stoyanova, V.; Milanova, V.; Naumova, E. A Study of TNF-α, TGF-β, IL-10, IL-6, and IFN-γ Gene Polymorphisms in Patients with Depression. J. Neuroimmunol. 2016, 293, 123–128. [Google Scholar] [CrossRef]
- Talaei, A.; Tavakkol Afshari, J.; Fayyazi Bordbar, M.R.; Pouryousof, H.; Faridhosseini, F.; Saghebi, A.; Rezaei Ardani, A.; Talaei, A.; Tehrani, M. A Study on the Association of Interleukin-1 Cluster with Genetic Risk in Bipolar I Disorder in Iranian Patients: A Case-Control Study. Iran. J. Allergy Asthma Immunol. 2016, 15, 466–475. [Google Scholar]
- Perry, B.I.; Upthegrove, R.; Kappelmann, N.; Jones, P.B.; Burgess, S.; Khandaker, G.M. Associations of Immunological Proteins/Traits with Schizophrenia, Major Depression and Bipolar Disorder: A Bi-Directional Two-Sample Mendelian Randomization Study. Brain Behav. Immun. 2021, 97, 176–185. [Google Scholar] [CrossRef]
- Ben Afia, A.; Aflouk, Y.; Saoud, H.; Zaafrane, F.; Gaha, L.; Bel Hadj Jrad, B. Inteurleukin-8 Gene Variations and the Susceptibility to Schizophrenia. Psychiatry Res. 2020, 293, 113421. [Google Scholar] [CrossRef]
- Kang, H.-J.; Kim, J.-M.; Kim, S.-W.; Shin, I.-S.; Park, S.-W.; Kim, Y.-H.; Yoon, J.-S. Associations of Cytokine Genes with Alzheimer’s Disease and Depression in an Elderly Korean Population. J. Neurol. Neurosurg. Psychiatry 2015, 86, 1002–1007. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Gibby, C.C.; Swartz, M.D.; Yu, X.; Wu, X.; Yennurajalingam, S.; Anderson, K.O.; Spitz, M.R.; Shete, S. Symptom Clusters of Pain, Depressed Mood, and Fatigue in Lung Cancer: Assessing the Role of Cytokine Genes. Support. Care Cancer 2013, 21, 3117–3125. [Google Scholar] [CrossRef] [Green Version]
- Lan, B.; Lv, D.; Sun, X.; Yang, M.; Zhang, L.; Ma, F. Genetic Variations in IFNGR1, BDNF and IL-10 May Predict the Susceptibility to Depression and Anxiety in Chinese Women with Breast Cancer. Clin. Breast Cancer 2022, 22, 674–680. [Google Scholar] [CrossRef]
- Liu, F.-R.; Yang, L.-Y.; Zheng, H.-F.; Zhou, Y.; Chen, B.-B.; Xu, H.; Zhang, Y.-W.; Shen, D.-Y. Plasma Levels of Interleukin 18 but Not Amyloid-β or Tau Are Elevated in Female Depressive Patients. Compr. Psychiatry 2020, 97, 152159. [Google Scholar] [CrossRef] [PubMed]
- Swartz, J.R.; Prather, A.A.; Di Iorio, C.R.; Bogdan, R.; Hariri, A.R. A Functional Interleukin-18 Haplotype Predicts Depression and Anxiety through Increased Threat-Related Amygdala Reactivity in Women but Not Men. Neuropsychopharmacology 2017, 42, 419–426. [Google Scholar] [CrossRef] [Green Version]
- Goldsmith, D.R.; Rapaport, M.H.; Miller, B.J. A Meta-Analysis of Blood Cytokine Network Alterations in Psychiatric Patients: Comparisons between Schizophrenia, Bipolar Disorder and Depression. Mol. Psychiatry 2016, 21, 1696–1709. [Google Scholar] [CrossRef]
- Jeleń, A.; Żebrowska-Nawrocka, M.; Szmajda-Krygier, D.; Mazur, K.; Gałecki, P.; Balcerczak, E. Preliminary Investigation of Two Promoter Region Polymorphisms of the TNFA Gene in Patients with Recurrent Depressive Disorder. Biomed. Rep. 2021, 15, 105. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Cao, X.; Shi, W.; Zhou, X.; Chen, Q.; Ma, K. Gene–Disease Association Study of Tumor Necrosis Factor-α G-308A Gene Polymorphism with Risk of Major Depressive Disorder: A Systematic Review and Meta-analysis. Brain Behav. 2020, 10, e01628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halstead, S.; Siskind, D.; Amft, M.; Wagner, E.; Yakimov, V.; Shih-Jung Liu, Z.; Walder, K.; Warren, N. Alteration Patterns of Peripheral Concentrations of Cytokines and Associated Inflammatory Proteins in Acute and Chronic Stages of Schizophrenia: A Systematic Review and Network Meta-Analysis. Lancet Psychiatry 2023, 10, 260–271. [Google Scholar] [CrossRef]
- Srinivas, L.; Vellichirammal, N.N.; Alex, A.M.; Nair, C.; Nair, I.V.; Banerjee, M. Pro-Inflammatory Cytokines and Their Epistatic Interactions in Genetic Susceptibility to Schizophrenia. J. Neuroinflammation 2016, 13, 105. [Google Scholar] [CrossRef] [Green Version]
- Kapelski, P.; Skibinska, M.; Maciukiewicz, M.; Pawlak, J.; Dmitrzak-Weglarz, M.; Szczepankiewicz, A.; Zaremba, D.; Twarowska-Hauser, J. An Association Between Functional Polymorphisms of the Interleukin 1 Gene Complex and Schizophrenia Using Transmission Disequilibrium Test. Arch. Immunol. Ther. Exp. 2016, 64, 161–168. [Google Scholar] [CrossRef]
- Paul-Samojedny, M.; Owczarek, A.; Kowalczyk, M.; Suchanek, R.; Palacz, M.; Kucia, K.; Fila-Daniłow, A.; Borkowska, P.; Kowalski, J. Association of Interleukin 2 (IL-2), Interleukin 6 (IL-6), and TNF-Alpha (TNFα) Gene Polymorphisms with Paranoid Schizophrenia in a Polish Population. J. Neuropsychiatry Clin. Neurosci. 2013, 25, 72–82. [Google Scholar] [CrossRef]
- Kapelski, P.; Skibinska, M.; Maciukiewicz, M.; Wilkosc, M.; Frydecka, D.; Groszewska, A.; Narozna, B.; Dmitrzak-Weglarz, M.; Czerski, P.; Pawlak, J.; et al. Association Study of Functional Polymorphisms in Interleukins and Interleukin Receptors Genes: IL1A, IL1B, IL1RN, IL6, IL6R, IL10, IL10RA and TGFB1 in Schizophrenia in Polish Population. Schizophr. Res. 2015, 169, 1–9. [Google Scholar] [CrossRef]
- Pu, X.; Li, J.; Ma, X.; Yang, S.; Wang, L. The Functional Polymorphisms Linked with Interleukin-1β Gene Expression Are Associated with Bipolar Disorder. Psychiatr. Genet. 2021, 31, 72–78. [Google Scholar] [CrossRef]
- Frydecka, D.; Misiak, B.; Beszlej, J.A.; Karabon, L.; Pawlak-Adamska, E.; Tomkiewicz, A.; Partyka, A.; Jonkisz, A.; Kiejna, A. Genetic Variants in Transforming Growth Factor-β Gene (TGFB1) Affect Susceptibility to Schizophrenia. Mol. Biol. Rep. 2013, 40, 5607–5614. [Google Scholar] [CrossRef] [Green Version]
- Golimbet, V.; Lezheiko, T.; Mikhailova, V.; Korovaitseva, G.; Kolesina, N.; Plakunova, V.; Kostyuk, G. A Study of the Association between Polymorphisms in the Genes for Interleukins IL-6 and IL-10 and Negative Symptoms Subdomains in Schizophrenia. Indian J. Psychiatry 2022, 64, 484–488. [Google Scholar] [CrossRef]
- Al-Asmary, S.M.; Kadasah, S.; Arfin, M.; Tariq, M.; Al-Asmari, A. Genetic Variants of Interleukin-10 Gene Promoter Are Associated with Schizophrenia in Saudi Patients: A Case-Control Study. N. Am. J. Med. Sci. 2014, 6, 558–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiu, M.H.; Tian, L.; Chen, S.; Tan, Y.L.; Chen, D.C.; Chen, J.; Chen, N.; De Yang, F.; Licinio, J.; Kosten, T.R.; et al. Contribution of IL-10 and Its -592 A/C Polymorphism to Cognitive Functions in First-Episode Drug-Naive Schizophrenia. Brain Behav. Immun. 2016, 57, 116–124. [Google Scholar] [CrossRef]
- Subbanna, M.; Shivakumar, V.; Talukdar, P.M.; Narayanaswamy, J.C.; Venugopal, D.; Berk, M.; Varambally, S.; Venkatasubramanian, G.; Debnath, M. Role of IL-6/RORC/IL-22 Axis in Driving Th17 Pathway Mediated Immunopathogenesis of Schizophrenia. Cytokine 2018, 111, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Tan, Y.-L.; Chen, D.-C.; Tan, S.-P.; Malouta, M.Z.; Bernard, J.D.; Combs, J.L.; Bhatti, S.; Davis, M.C.; Kosten, T.R.; et al. Serum IL-18 Level, Clinical Symptoms and IL-18-607A/C Polymorphism among Chronic Patients with Schizophrenia in a Chinese Han Population. Psychoneuroendocrinology 2016, 68, 140–147. [Google Scholar] [CrossRef]
- Kordi-Tamandani, D.M.; Bahrami, A.R.; Sabbaghi-Ghale-No, R.; Soleimani, H.; Baranzehi, T. Analysis of IL-33 Gene Polymorphism (Rs11792633 C/T) and Risk of Schizophrenia. Mol. Biol. Res. Commun. 2016, 5, 45–48. [Google Scholar] [PubMed]
- Strenn, N.; Pålsson, E.; Liberg, B.; Landén, M.; Ekman, A. Influence of Genetic Variations in IL1B on Brain Region Volumes in Bipolar Patients and Controls. Psychiatry Res. 2021, 296, 113606. [Google Scholar] [CrossRef] [PubMed]
- Shonibare, D.O.; Patel, R.; Islam, A.H.; Metcalfe, A.W.S.; Fiksenbaum, L.; Kennedy, J.L.; Freeman, N.; MacIntosh, B.J.; Goldstein, B.I. Preliminary Study of Structural Magnetic Resonance Imaging Phenotypes Related to Genetic Variation in Interleukin-1β Rs16944 in Adolescents with Bipolar Disorder. J. Psychiatr. Res. 2020, 122, 33–41. [Google Scholar] [CrossRef]
- Sundaresh, A.; Oliveira, J.; Chinnadurai, R.K.; Rajkumar, R.P.; Hani, L.; Krishnamoorthy, R.; Leboyer, M.; Negi, V.S.; Tamouza, R. IL6/IL6R Genetic Diversity and Plasma IL6 Levels in Bipolar Disorder: An Indo-French Study. Heliyon 2019, 5, e01124. [Google Scholar] [CrossRef] [Green Version]
- Majd, M.; Saunders, E.F.H.; Engeland, C.G. Inflammation and the Dimensions of Depression: A Review. Front. Neuroendocr. 2020, 56, 100800. [Google Scholar] [CrossRef] [PubMed]
- Perry, B.I.; Burgess, S.; Jones, H.J.; Zammit, S.; Upthegrove, R.; Mason, A.M.; Day, F.R.; Langenberg, C.; Wareham, N.J.; Jones, P.B.; et al. The Potential Shared Role of Inflammation in Insulin Resistance and Schizophrenia: A Bidirectional Two-Sample Mendelian Randomization Study. PLoS Med. 2021, 18, e1003455. [Google Scholar] [CrossRef] [PubMed]
- Asevedo, E.; Rizzo, L.B.; Gadelha, A.; Mansur, R.B.; Ota, V.K.; Berberian, A.A.; Scarpato, B.S.; Teixeira, A.L.; Bressan, R.A.; Brietzke, E. Peripheral Interleukin-2 Level Is Associated with Negative Symptoms and Cognitive Performance in Schizophrenia. Physiol. Behav. 2014, 129, 194–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mørch, R.H.; Dieset, I.; Færden, A.; Reponen, E.J.; Hope, S.; Hoseth, E.Z.; Gardsjord, E.S.; Aas, M.; Iversen, T.; Joa, I.; et al. Inflammatory Markers Are Altered in Severe Mental Disorders Independent of Comorbid Cardiometabolic Disease Risk Factors. Psychol. Med. 2019, 49, 1749–1757. [Google Scholar] [CrossRef] [PubMed]
- Haapakoski, R.; Mathieu, J.; Ebmeier, K.P.; Alenius, H.; Kivimäki, M. Cumulative Meta-Analysis of Interleukins 6 and 1β, Tumour Necrosis Factor α and C-Reactive Protein in Patients with Major Depressive Disorder. Brain Behav. Immun. 2015, 49, 206–215. [Google Scholar] [CrossRef] [Green Version]
- Solmi, M.; Suresh Sharma, M.; Osimo, E.F.; Fornaro, M.; Bortolato, B.; Croatto, G.; Miola, A.; Vieta, E.; Pariante, C.M.; Smith, L.; et al. Peripheral Levels of C-Reactive Protein, Tumor Necrosis Factor-α, Interleukin-6, and Interleukin-1β across the Mood Spectrum in Bipolar Disorder: A Meta-Analysis of Mean Differences and Variability. Brain Behav. Immun. 2021, 97, 193–203. [Google Scholar] [CrossRef]
- Ermakov, E.A.; Melamud, M.M.; Boiko, A.S.; Kamaeva, D.A.; Ivanova, S.A.; Nevinsky, G.A.; Buneva, V.N. Association of Peripheral Inflammatory Biomarkers and Growth Factors Levels with Sex, Therapy and Other Clinical Factors in Schizophrenia and Patient Stratification Based on These Data. Brain Sci. 2023, 13, 836. [Google Scholar] [CrossRef]
- Gallego, J.A.; Blanco, E.A.; Husain-Krautter, S.; Madeline Fagen, E.; Moreno-Merino, P.; Del Ojo-Jiménez, J.A.; Ahmed, A.; Rothstein, T.L.; Lencz, T.; Malhotra, A.K. Cytokines in Cerebrospinal Fluid of Patients with Schizophrenia Spectrum Disorders: New Data and an Updated Meta-Analysis. Schizophr. Res. 2018, 202, 64–71. [Google Scholar] [CrossRef]
- Zazula, R.; Dodd, S.; Dean, O.M.; Berk, M.; Bortolasci, C.C.; Verri, W.A.; Vargas, H.O.; Nunes, S.O. V Cognition-Immune Interactions between Executive Function and Working Memory, Tumour Necrosis Factor-Alpha (TNF-Alpha) and Soluble TNF Receptors (STNFR1 and STNFR2) in Bipolar Disorder. World J. Biol. Psychiatry 2022, 23, 67–77. [Google Scholar] [CrossRef]
- Wray, N.R.; Ripke, S.; Mattheisen, M.; Trzaskowski, M.; Byrne, E.M.; Abdellaoui, A.; Adams, M.J.; Agerbo, E.; Air, T.M.; Andlauer, T.M.F.; et al. Genome-Wide Association Analyses Identify 44 Risk Variants and Refine the Genetic Architecture of Major Depression. Nat. Genet. 2018, 50, 668–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Westrhenen, R.; van Schaik, R.H.N.; van Gelder, T.; Birkenhager, T.K.; Bakker, P.R.; Houwink, E.J.F.; Bet, P.M.; Hoogendijk, W.J.G.; van Weelden-Hulshof, M.J.M. Policy and Practice Review: A First Guideline on the Use of Pharmacogenetics in Clinical Psychiatric Practice. Front Pharmacol 2021, 12, 640032. [Google Scholar] [CrossRef] [PubMed]
- Van Westrhenen, R.; Aitchison, K.J.; Ingelman-Sundberg, M.; Jukić, M.M. Pharmacogenomics of Antidepressant and Antipsychotic Treatment: How Far Have We Got and Where Are We Going? Front. Psychiatry 2020, 11, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Research | Analyzed Polymorphisms | Sample | Results |
|---|---|---|---|
| Kim et al. (2017) [29] | TNFA: rs1799724 (−850C/T; −308G/A); IL1B: rs16944 (−511C/T), +3953C/T | 286 patients with PSD | −511C/T polymorphism was associated with primary depression and PSD at 2 weeks; higher TNF-α levels were associated with PSD at 2 weeks in in patients carried −850T allele. |
| Tartter et al. (2015) [30] | IL6: rs1800795 (−174G/C); IL1B: rs16944 (−511C/T); TNF: rs1800629 (−308G/A) | 444 young adults whose exposure to chronic stress in the past 6 months; Australian cohort | Patients with the −174G allele had fewer depressive symptoms after interpersonal stress compared with CC homozygotes with equal exposure to interpersonal stress. The −511C allele in IL1B was associated with more severe depression after chronic interpersonal stress compared with TT homozygotes. |
| Lezheiko et al. (2018) [31] | IL1B: rs16944 (−511T/C); TNFA: rs1800629 (−308A/G) | 139 patients with depression vs. 530 HS; Russian cohort | The −511T/C and −308A/G polymorphisms were associated with depression; CC genotype and GG genotype are the risk factors of depression. |
| McQuaid et al. (2019) [32] | IL1B: rs16944; IL6: rs1800795; TNFA: rs1800629 | 475 university students | Depressive symptoms were higher among individuals who experienced childhood adversity with the GG genotype of the IL1B rs16944. |
| Kovacs et al. (2016) [33] | IL1B: rs16944, rs1143643 | 1053 persons; Hungarian cohort | The rs16944*A allele was associated with childhood adversity increasing anxiety and depressive symptoms. The A allele of rs1143643 demonstrated protective effect against depressive symptoms after recent life stress. |
| Bialek et al. (2020) [18] | IL1B: rs1143623 (−1560G/C), rs1143627 (−118C/T); IL1A: rs17561 (c.340G/T); TNFA: rs1799964 (−1211T/C), rs1800629 (−488G/A) | 270 patients with depression vs. 231 HS; Polish cohort | It was shown an association between the T allele and the TT genotype of rs1799964 TNFA and low effectiveness of pharmacotherapy; the C allele and CT genotype were associated with good response to therapy. Carryer of GC and CC genotypes of rs1143623 IL1B showed varying levels of disease severity ccording to the HDRS. The combined genotypes of rs1143627–rs17561, rs1143627–rs1799964 and rs1143623–rs1799964, decreased the risk of depression occurrence, rs1143627–rs1800629 increased the risk. |
| Kang et al. (2017) [34] | IL1B: rs16944 (−511C/T), +3953C/T | 969 patients at 2 weeks after ACS, 711—at 1 year later | Depression during the acute ACS was associated with the −511T allele and the IL-1β levels. There was no association with depression in chronic ACS. There was no association with depression in the acute or chronic phase and the +3953C/T genotype. |
| Draganov et al. (2019) [35] | 41 SNPs in IL1B, IL2, IL6, IL6R, IL10, IL18, TNFA, IFNG | 153 patients with MDD | Polymorphic variant rs1143643 of IL1B was associated with MSM scores. Allelic distribution of rs57569414 IL6R demonstrates a trend to significance with MSM scores. Combinations of alleles of IL1B and IL10 were associated with response to treatment. |
| Kim et al. (2013) [36] | TNFA: rs1800629 (−308G/A); IL10: rs1800896 (−1082A/G); IFNG: rs2430561 (+874T/A) | 301 patients with MDD (204 attempted suicide, 97 not attempted suicide); Korean cohort | Among patients with MDD the TNFA −308GG genotype was associated with an increased risk of suicide; IL10-1082A/G were not associated with that risk. |
| Tsai et al. (2023) [37] | GWAS involving 684,616 SNPs | 65 patients with TRD; Chinese cohort | Two SNPs (rs2540315 and rs75746675) in IL1R1 were associated with a rapid (within 240 min) antidepressant effect of ketamine infusion in patients with TRD. |
| Kim et al. (2013) [38] | TNFA: rs1799724 (−850C/T); IL1B: rs16944 (−511C/T), +3953C/T; IL6: rs1800795 (−174G/C); IL8: −251T/A; IL4: +33T/C; IL10: rs1800896 (−1082A/G) | 309 women with breast cancer at one week after surgery, 244 (79%)—at one year later. | IL1B-511TT was associated with depression at one week after surgery with breast cancer and persistent depression at one year follow-up. |
| Luckhoff et al. (2016) [39] | TNFA: rs1800629 (−308G/A) | 94 patients with MDD vs. 97 HS; South African cohort | The rs1800629*A-allele in TNFA was associated with early-onset of MDD. |
| Lu et al. (2023) [40] | IL6: rs1800795; rs1800796 | 114 patients with depression vs. 110 HS; Han Chinese cohort | The CC genotype and the C allele of rs1800796 were associated with depression. |
| Golimbet et al. (2017) [41] | IL4: −589C/T; IL6: rs1800795 (−174G/C); TNFA: rs1800629 (−308G/A); CRP: −717A/G | 78 male CHD patients with depression; 91—without depression; 127 HS; Russian cohort | The IL6-174G/C was associated with depression comorbid to CHD. The IL4-589C/T was associated with CHD. No association between the TNFA-308G/A and the CRP-717A/G with depression in CHD. |
| Kovacs et al. (2016) [42] | IL6: rs1800795 | 1053 volunteers; Hungarian cohort | The IL6 rs1800795 in common with various stressors increases the risk of depression and has a greater impact measured by the ZSDS symptoms. |
| Gal et al. (2023) [43] | IL6: rs1800795 | UK Biobank, n = 277 501 | The rs1800795 was associated with recent stress on current depressive symptoms and lifetime depression. |
| Udina et al. (2013) [44] | IL6: rs1800795 | 385 patients with chronic hepatitis; Caucasian cohort | The rs1800795 IL6 increases the risk of induced by IFN depression and anxiety. It was associated with fatigue rates in patients with chronic hepatitis C before treatment. |
| Zhang et al. (2016) [45] | IL6: 1800797 | 772 patients with MDD vs. 759 HS; Han Chinese cohort | Association between rs1800797 and the risk of MDD. |
| Maciukiewicz et al. (2015) [46] | Twenty SNPs in IL1B, IL2, IL6, TSPO and BDNF | MDD patients treated with duloxetine (n = 215) or placebo (n = 235) | Association IL6 (−63G/A, rs2066992; +1984T/G, rs10242595) with response to duloxetine therapy in MDD patients. IL6 rs2066992 and rs10242595 were associated with duloxetine response. The rs2066992 was associated with placebo response. |
| Khandaker at al. (2018) [47] | IL6R: rs2228145 (Asp358Ala) | 9912 unselected participants from the ALSPAC birth cohort | Asp358Ala was associated with a reduced risk of severe depression and/or psychosis. Asp358Ala was not associated with total depression score and with the risk factors related with inflammation, depression or psychosis. |
| Dunn et al. (2013) [48] | 104 SNPs and haplotypes in 15 cytokine genes | 167 oncology patients with prostate, breast, lung, or brain cancer and 85 of their FCs | Significant associations of cytokine gene variants with trajectories of depressive symptoms in cancer patients and their FC have been identified. Two of these associations were in genes with anti-inflammatory functions (IL1R2, IL10), and one was with a gene with proinflammatory functions (TNFA). |
| Doong et al. (2015) [49] | 82 SNPs in 15 genes of cytokine | 398 breast cancer patients | Significant associations between IL6 rs2069845, IL13 rs1295686, and TNFA rs18800610 with a symptom cluster of pain, sleep disturbance, fatigue and depression. |
| Santos et al. (2016) [50] | IL18: rs1946518 (−607A/C), rs187238 (−137C/G) | 80 MDD patients; Portuguese cohort | IL18-607A/C and IL18-137C/G were associated with the effect of the AD therapy. Patients carrying CA or AA genotypes of -607A/C and patients carrying GC or CC genotypes of -137C/G were significantly more prone to relapse after therapy and present a significantly lower time to relapse. |
| Sandoval-Carrillo et al. (2018) [51] | TNFA: rs1799724 (−857C/T), rs1800629 (−308G/A), rs361525 (−238G/A) | 153 pregnant women with depression vs. 177 HS | The −857CT genotype increased the risk for depression. The −238GA genotype reduced the risk. No association between the −308G/A polymorphism and depression risk. The C857-G308-A238 haplotype was associated with a decrease of depression risk. |
| Saad et al. (2014) [52] | 82 SNPs in 15 cytokine genes | 155 patients with resilient and 180 patients with subsyndromal depressive symptom classes | In patients with breast cancer variation in three cytokine genes IFNGR1 rs937626, IL6 rs2069840, TNFA rs1799964, predicted membership in the Subsyndromal versus the Resilient class as well as age and functional status. |
| Bialek et al. (2020) [53] | TGFB1: rs1800469 (g.41354391A/G); IRF: rs2070729 (g.132484229C/A); PTGS2: rs5275 (186643058A/G); PTGS2: rs4648308 (g.186640617C/T); TGF-α: rs2166975 (g.70677994G/A); IKBKB: rs5029748 (g.42140549G/T). | 80 patients with depression vs. 180 HS | The AG genotype of rs2166975 TGFA was associated with an increased risk of depression, the GG genotype reduced the risk. The AG genotype and G allele of the rs2166975 TGFA was associated with increased risk of depression development in men. Genotype rs1800469*AA of TGFB1 was associated with earlier age of onset of the disease, GG genotype increased severity of the depressive episode. |
| Mihailova et al. (2016) [54] | TNFA, TGFB, IL10, IL6, IFNG | 80 patients with depression vs. 50 HS; Bulgarian cohort | The TGFB + 869TT genotype (rs1800470) prevailed in patients compared with HS. The TT-GC combined genotype (+869T/C, +915G/C) was associated with disease recurrence. |
| Research | Analyzed Polymorphisms | Samples | Results |
|---|---|---|---|
| Srinivas et al. (2016) [67] | IL1A: rs1800587; IL1B: rs1143634, rs1143627, rs16944; IL1RN: rs2234663; IL3: rs31400, rs31480, rs40401; IL4: rs2243250, rs2070874; IL6: rs1800797, rs1800796, rs1800795; IL10: rs1800872, rs1800871, rs1800896; IFNG: rs2069718, rs2430561 TNFA: rs1800629, rs361525; TGFB1: rs1800471, rs1800470, rs1800469 | 248 SCZ vs. 24 HS; South Indian cohort | Only IL1A rs1800587, IL6 rs1800796, TNFA rs361525, and IFNG rs2069718 polymorphisms were associated with SCZ. In silico analysis showed altered transcriptional activity for IL1A (rs1800587), IL6 (rs1800796, rs1800795) and TNFA (rs361525). |
| Kapelski et al. (2016) [68] | IL1A: rs1800587, rs17561, rs11677416; IL1B: rs1143634, rs1143643, rs16944, rs4848306, rs1143623, rs1143633, rs1143627; IL1RN: rs419598, rs315952, rs9005, rs4251961 | 143 SCZ patients and their healthy parents | There is a trend toward an association of rs1143627, rs16944, rs1143623 in IL1B gene with the risk of SCZ. Alleles rs1143627*G, rs16944*A, and rs1143623*G were more frequently transmitted by parents to children with SCZ. |
| Paul-Samojedny et al. (2013) [69] | IL6: rs1800795; TNFA: rs1800629; IL2: rs2069762 | 115 SCZ vs. 135 HS; Polish cohort | Genotype TT and allele T of rs2069762 IL2, and genotype AA and allele A of rs1800629 TNFA were significantly associated with paranoid SCZ. Patients with haplotype CTA (rs1800795–rs1800629–rs2069762) showed higher scores on the Negative and General subscales of PANSS. |
| Kapelski et al. (2015) [70] | IL1N: rs1800587, rs17561; IL1B: rs1143634, rs1143643, rs16944, rs4848306, rs1143623, rs1143633, rs1143627; IL1RN: rs419598, rs315952, rs9005, rs4251961; IL6: rs1800795, rs1800797; IL6R: rs4537545, rs4845617, rs2228145; IL10: rs1800896, rs1800871, rs1800872, rs1800890, rs6676671; IL10RA: rs2229113, rs3135932; TGFB1: rs1800469, rs1800470 | 621 SCZ vs. 531 HS; Polish cohort | An association of genotype rs4848306*AG of IL1B, allele rs4251961*T of IL1RN gene, allele C and genotype AC of rs2228145 IL6R, allele T and genotyper CT of 4537545 IL6R with SCZ have been observed. Allele A of rs6676671 in IL10 was associated with early age of onset. |
| Pu et al. (2021) [71] | IL6: rs1800795, rs1800796 | 113 SCZ vs. 110 HS; Han Chinese cohort | CC and CG genotypes of rs1800796 IL6 was significantly associated with chronic SCZ |
| Frydecka et al. (2013) [72] | IL2: rs2069756 (−330T/G); IL6: rs1800795 (−174G/C); IFNG: rs2430561 (+874T/A); TGF1B: rs1800470 (+869T/C), rs1800471 (+916G/C) | 151 SCZ vs. 279 HS; Caucasian cohort | The T carriers (CT and TT genotypes) of +869T/C were significantly more frequent in SCZ (especially in females) than in HS. Association of polymorphisms in IL2, IL6 and IFNG genes with SCZ was not found. |
| Golimbet et al. (2022) [73] | IL6: rs1800795 (−174G/C); IL10: rs1800872 (−592C/A), rs1800896 (−1082G/A) | 275 SCZ | Mean score on the AA subdomain of the Positive and Negative Syndromes Scale was higher in the AA genotype of IL10-592C/A compared with the CC genotype and in the GG genotype of IL6-174G/C compared with the AA genotype. AA score decreases with the number of the copies of an A allele of −1082G/A. |
| Ben Afia et al. (2020) [57] | IL8: rs4073, rs2227306, rs1126647 | 206 SCZ vs. 195 HS; Tunisian cohort | In the patients group it was observed an increased frequency of the T allele and the TT genotype of rs1126647. The T allele and the TT genotype of rs1126647 was associated with paranoid SCZ, and more specifically in females. The haplotypes TTT, ACT and TCT (rs4073-rs2227306-rs1126647) were associated with increased risk for paranoid SCZ, and only the TCT haplotype howed as a risk factor for SCZ. |
| Al-Asmary et al. (2014) [74] | IL10: rs1800896 (−1082A/G), rs1800871 (−819T/C), rs1800872 (−592A/C) | 181 SCZ vs. 211 HS; Saudi Arabian cohort | Genotypes −1082GA, −819CC and −592CC are susceptible to SCZ, while genotypes −1082GG, −1082AA, −819CT and −592CA are resistant to SCZ. |
| Xiu et al. (2016) [75] | IL10: rs1800872 (−592A/C) | 256 first-episode drug-naive SCZ vs. 540 HS; Han Chinese cohort | The A allele and AC genotype of −592A/C were associated with worse attentional performance in SCZ and reduced serum IL-10 levels. The AC genotype was associated with SCZ. |
| Subbanna et al. (2018) [76] | IL17A: rs2275913 (−197G/A) | 221 SCZ vs. 223 HS | The AA genotype was associated with higher total scores of bizarre behavior and apathy in female SCZ patients. There was no significant difference in distribution of genotypes and alleles between SCZ and HS. |
| Zhang et al. (2016) [77] | IL18: rs1946518 (−607A/C) | 772 SCZ vs. 775 HS; Han Chinese cohort | There were no significant differences in the distribution of the allele and genotype frequencies of −607A/C between SCZ and HS. The −607CC genotype was associated with higher PANSS general psychopathology subscore and the PANSS total score than both AC and AA genotypes. |
| Kordi-Tamandani et al. (2016) [78] | IL33: rs11792633 | 70 SCZ vs. 70 HS; Iranian cohort | CT and TT genotypes of rs11792633 significantly decreased the risk of SCZ. |
| Research | Analyzed Polymorphisms | Samples | Results |
|---|---|---|---|
| Talaei et al. (2016) [55] | IL1A: rs1800587 (−889G/A); IL1B: rs1143634 (+3954C/T), rs16944 (−511C/T); IL1RN | 48 BD vs. 47 HS; Iranian cohort | The frequency CC and CT genotype of IL1B-511C/T were significantly different between BD patients and healthy controls. The T allele of IL1B-511C/T was significantly more frequent in patients with a positive history of MDD. The T allele of IL1B+3954C/T was significantly more frequent in early onset BD patients. |
| Strenn et al. (2021) [79] | IL1B: rs1143623, rs1143627, rs16944, rs1143634 | 188 BD vs. 54 HS | The genotype distribution did not differ between patients with BD and the control group. |
| Pu et al. (2021) [71] | IL1B: rs1143643, rs1143634, rs1143627, rs16944, rs1143623, rs4848306, rs12621220 | 930 BD vs. 912 HS; Han Chinese cohort | The minor alleles of four polymorphisms of IL1B were associated with risk of BD (rs1143627*G, rs16944*A, rs1143623*G, rs12621220*T). |
| Shonibare et al. (2020) [80] | IL1B: rs16944 | 38 adolescents with BD vs. 32 HS | The rs16944 was associated with greater lateral occipital cortex surface area and volume in BD adolescents. |
| Sundaresh et al. (2018) [81] | IL6: rs1800795; IL6R: rs2228145 | 565 BD vs. 201 HS; French cohort | No association of the IL6 rs1800795 and BD was found. The C allele and CC genotype of IL6R rs2228145 were associated with early onset of disease. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikhalitskaya, E.V.; Vyalova, N.M.; Ermakov, E.A.; Levchuk, L.A.; Simutkin, G.G.; Bokhan, N.A.; Ivanova, S.A. Association of Single Nucleotide Polymorphisms of Cytokine Genes with Depression, Schizophrenia and Bipolar Disorder. Genes 2023, 14, 1460. https://doi.org/10.3390/genes14071460
Mikhalitskaya EV, Vyalova NM, Ermakov EA, Levchuk LA, Simutkin GG, Bokhan NA, Ivanova SA. Association of Single Nucleotide Polymorphisms of Cytokine Genes with Depression, Schizophrenia and Bipolar Disorder. Genes. 2023; 14(7):1460. https://doi.org/10.3390/genes14071460
Chicago/Turabian StyleMikhalitskaya, Ekaterina V., Natalya M. Vyalova, Evgeny A. Ermakov, Lyudmila A. Levchuk, German G. Simutkin, Nikolay A. Bokhan, and Svetlana A. Ivanova. 2023. "Association of Single Nucleotide Polymorphisms of Cytokine Genes with Depression, Schizophrenia and Bipolar Disorder" Genes 14, no. 7: 1460. https://doi.org/10.3390/genes14071460
APA StyleMikhalitskaya, E. V., Vyalova, N. M., Ermakov, E. A., Levchuk, L. A., Simutkin, G. G., Bokhan, N. A., & Ivanova, S. A. (2023). Association of Single Nucleotide Polymorphisms of Cytokine Genes with Depression, Schizophrenia and Bipolar Disorder. Genes, 14(7), 1460. https://doi.org/10.3390/genes14071460







