Molecular Advances to Combat Different Biotic and Abiotic Stresses in Linseed (Linum usitatissimum L.): A Comprehensive Review
Abstract
:1. Introduction
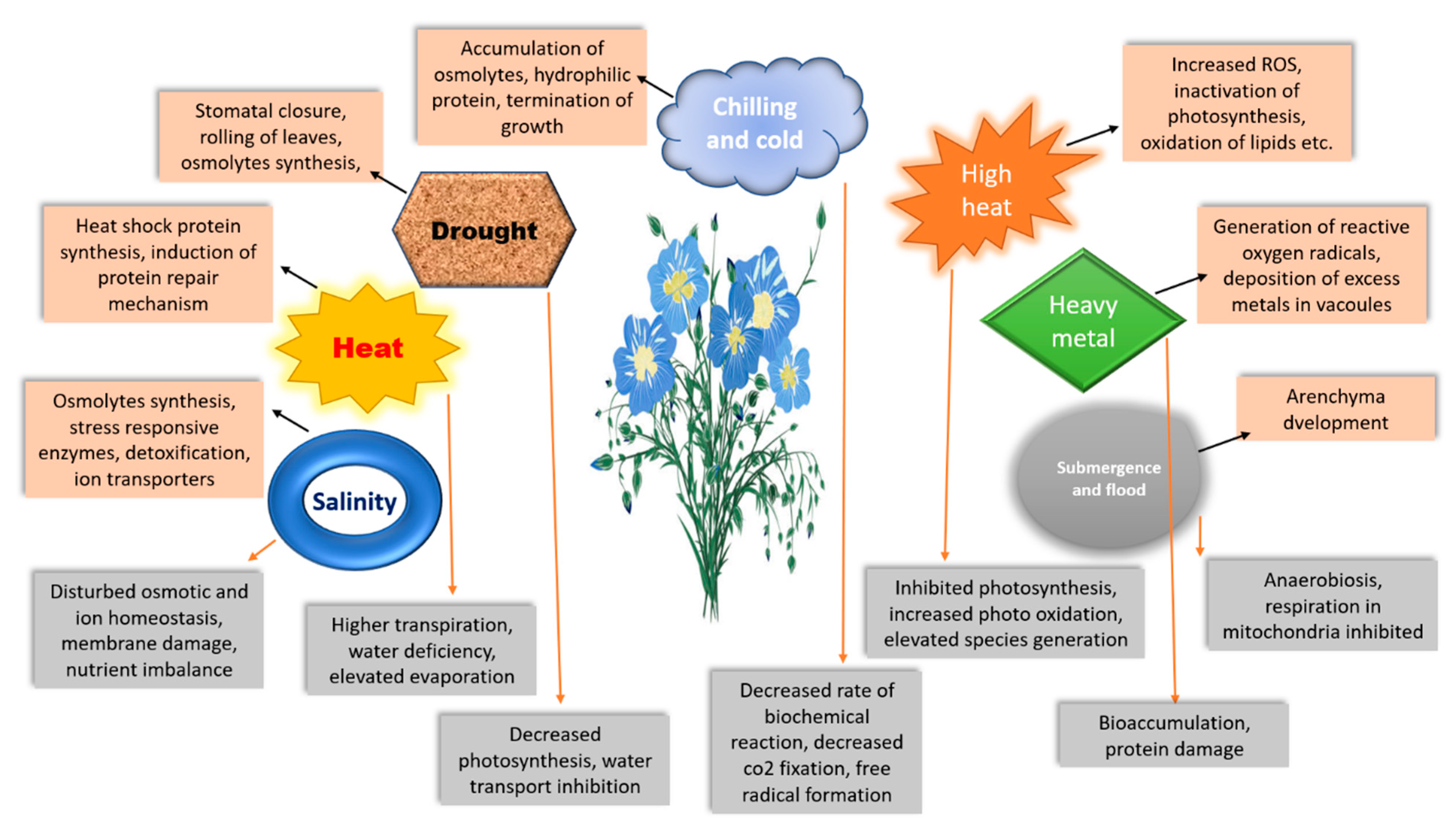
2. Abiotic Stress
2.1. Drought Tolerance
2.2. Salinity Tolerance
2.3. Heavy Metal Stress Tolerance
2.4. Cold Stress Tolerance
2.5. Heat Tolerance
3. Biotic Stresses
3.1. Rust Resistance
3.2. Fusarium Wilt Resistance
3.3. Alternaria Blight Resistance
3.4. Powdery Mildew Resistance
4. Flax Genetic Resources
4.1. International
4.2. National
5. Breeding through Conventional Methods
5.1. Interspecific Hybridisation
5.2. Hybrid Linseed
5.3. Mutation Breeding
6. Advanced Methods in Linseed Breeding
6.1. Molecular Diversity
6.2. Association Mapping
6.3. Extent of Linkage Disequilibrium
6.4. Genetic Loci Identified by GWAS
6.5. Molecular Mapping of QTLs
7. Integrated Omics Approach
7.1. Genomics
7.2. Transcriptomics
7.3. Metabolomics
7.4. Proteomics
7.5. Ionomics
7.6. Phenomics
8. Transgenic Flax/Linseed
9. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kajla, P.; Sharma, A.; Sood, D.R. Flaxseed—A potential functional food source. J. Food Sci. Technol. 2015, 52, 1857–1871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laux, M. 2011. Available online: https://www.agmrc.org/commodities-products/grains-oilseeds/flax-profile (accessed on 13 March 2012).
- Morris, D.H. Linseed in the Ruminant Diet—Adding Linseed to Feed Enhances the Fat Profile of Milk Winnipeg, MB, Flax Council of Canada. 2008. Available online: http://www.flaxcouncil.ca/files/web/Beef_R3_final.pdfm (accessed on 25 May 2012).
- Singh, K.; Mridula, D.; Rehal, J.; Barnwal, P. Flaxseed: A potential source of food, feed and fiber. Crit. Rev. Food Sci. Nutr. 2011, 51, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Friis, I.B. Linaceae: Linum. In Flora of Ethiopia and Eritrea: Mangnoliaceae to Flacourtiaceae; Edwards, S., Tadesse, M., Demissew, S., Hedberg, I., Eds.; The National Herbarium, Addis Ababa and the Department of Systematic Botany: Uppsala, Sweden, 2000; Volume 2, pp. 352–357. [Google Scholar]
- Jhala, A.J.; Hall, L.M.; Hall, J.C. Potential hybridization of flax with weedy and wild relatives: An avenue for movement to engineered genes? Crop Sci. 2008, 48, 825–840. [Google Scholar] [CrossRef] [Green Version]
- Mansby, E.; Diaz, O.; von Bothmer, R. Preliminary study of genetic diversity in Swedish flax (Linum usitatissimum L.). Genet. Resour. Crop Evol. 2000, 47, 417–424. [Google Scholar] [CrossRef]
- Hoque, A.; Fiedler, J.D.; Rahman, M. Genetic diversity analysis of a flax (Linum usitatissimum L.) global collection. BMC Genom. 2020, 21, 557. [Google Scholar] [CrossRef]
- Kurt, O.; Evans, G.M. Genetic Basis of Variation in Linseed (Linum usitatissimum L.) Cultivars. Turk. J. Agric. For. 1998, 22, 373–379. [Google Scholar]
- Mohammed, A.S.; Atnaw, S.M.; Ramaya, A.V.; Alemayehu, G. A comprehensive review on the effect of ethers, antioxidants, and cetane improver additives on biodiesel-diesel blend in CI engine performance and emission characteristics. J. Energy Inst. 2023, 108, 101227. [Google Scholar] [CrossRef]
- Liu, N.; Wan, B.; Zhang, Z.; Fang, X.; Lin, X.; Wang, Y.; Tang, J.; Bai, X.; Li, Y.; Yao, Y.; et al. Self-healing waterborne polyurethane coatings with high transparence and haze via cellulose nanocrystal stabilized linseed oil Pickering emulsion. Int. J. Biol. Macromol. 2023, 235, 123830. [Google Scholar] [CrossRef]
- Goyal, A.; Sharma, V.; Upadhyay, N.; Gill, S.; Sihag, M. Flax and flaxseed oil: An ancient medicine & modern functional food. J. Food Sci. Technol. 2014, 51, 1633–1653. [Google Scholar]
- Parikh, M.; Maddaford, T.G.; Austria, J.A.; Aliani, M.; Netticadan, T.; Pierce, G.N. Dietary flaxseed as a strategy for improving human health. Nutrients 2019, 11, 1171. [Google Scholar] [CrossRef] [Green Version]
- Sethi, S.; Tyagi, S.K.; Anurag, R.K. Plant-based milk alternatives an emerging segment of functional beverages: A review. J. Food Sci. Technol. 2016, 53, 3408–3423. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Leyva, D.; Weighell, W.; Edel, A.L.; LaVallee, R.; Dibrov, E.; Pinneker, R.; Maddaford, T.G.; Ramjiawan, B.; Aliani, M.; Guzman, R.; et al. Potent antihypertensive action of dietary flaxseed in hypertensive patients. Hypertension 2013, 62, 1081–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pohjanheimo, T.A.; Hakala, M.A.; Tahvonen, R.L.; Salminen, S.J.; Kallio, H.P. Flaxseed in bread making: Effects on sensory quality, aging, and composition of bakery products. J. Food Sci. 2006, 71, S343–S348. [Google Scholar] [CrossRef]
- Alam, S.; Sarker, M.; Rahman, M.; Afrin, S.; Richi, F.T.; Zhao, C.; Zhou, J.R.; Mohamed, I.N. Traditional Herbal Medicines, Bioactive Metabolites, and Plant Products Against COVID-19: Update on Clinical Trials and Mechanism of Actions. Front. Pharmacol. 2021, 12, 1248. [Google Scholar] [CrossRef]
- Alagirusamy, R.; Das, A. Yarns: Production, processability and properties. In Fibrous and Composite Materials for Civil Engineering Applications; Woodhead Publishing: Sawston, UK, 2011; Volume 1, pp. 29–61. [Google Scholar]
- Zaidi, Z.; Do Ain UzunAnaf, W.H. Katan (Linum usitatissimum Linn.) a potent unani therapeutic agent for respiratory tract diseases. J. Pharmacognocy 2021, 8, 112–118. [Google Scholar]
- Pramanik, A.; Tiwari, S.; Tripathi, M.K.; Tomar, R.S.; Singh, A.K. Molecular characterization of groundnut (Arachis hypogea L.) germplasm lines for yield attributed traits. Indian J. Genet. 2019, 79, 56–65. [Google Scholar] [CrossRef]
- Mandloi, S.; Tripathi, M.K.; Tiwari, S.; Tripathi, N. Genetic diversity analysis among late leaf spot and rust resistant and susceptible germplasm in groundnut (Arachis hypogea L.). Isr. J. Plant Sci. 2022, 69, 163–171. [Google Scholar] [CrossRef]
- Rajpoot, N.S.; Tripathi, M.K.; Tiwari, S.; Tomar, R.S.; Kandalkar, V.S. Characterization of Indian mustard germplasm on the basis of morphological traits and SSR markers. Curr. J. Appl. Sci. Technol. 2020, 39, 300–311. [Google Scholar] [CrossRef]
- Diederichsen, A.; Raney, J.P. Seed colour, seed weight and seed oil content in Linum usitaitissimum accessions held by plant gene resources of Canada. Plant Breed. 2006, 125, 372–373. [Google Scholar] [CrossRef]
- Saeidi, G. Genetic variation and heritability for germination, seed vigour and field emergence in brown and yellow- seeded genotypes of flax. Int. J. Plant Prod. 2012, 2, 15–22. [Google Scholar]
- Sharma, A.; Tripathi, M.K.; Tiwari, S.; Gupta, N.; Tripathi, N.; Mishra, N. Evaluation of soybean (Glycine max L.) genotypes on the basis of biochemical contents and anti-oxidant enzyme activities. Legume Res. 2021, 44, 1419–1429. [Google Scholar] [CrossRef]
- Baghel, R.; Sharma, A.K.; Tiwari, S.; Tripathi, M.K.; Tripathi, N. Genetic Diversity Analysis of Indian Mustard (Brassica spp.) Germplasm Lines using SSR Molecular Markers. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 137–143. [Google Scholar] [CrossRef]
- Patzk, J. Comparison of RAPD, STS, ISSR and AFLP molecular methods used for assessment of genetic diversity in hop (Humulus lupulus L.). Euphytica 2001, 121, 9–18. [Google Scholar] [CrossRef]
- Krulíckova, K.; Posvec, Z.; Griga, M. Identification of flax and linseed cultivars by isozyme markers. Biol. Plant. 2002, 45, 327–336. [Google Scholar] [CrossRef]
- Fu, Y.B. Geographic patterns of RAPD variation in cultivated flax. Crop Sci. 2005, 45, 1084–1091. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.B. Redundancy and distinctness in flax germplasm as revealed by RAPD dissimilarity. Plant Genet. Resour. 2006, 4, 117–124. [Google Scholar] [CrossRef]
- Singh, P.K.; Akram, M.; Srivastava, R.L. Genetic diversity in linseed (Linum usitatissimum) cultivars based on RAPD-analysis. Indian J. Agric. Sci. 2009, 79, 1046–1049. [Google Scholar]
- Bibi, T.; Mustafa, H.S.B.; Hasan, E.U.; Rauf, S.; Mahmood, T.; Ali, Q. Analysis of genetic diversity in linseed using molecular markers. Life Sci. J. 2015, 12, 28–37. [Google Scholar]
- Nagabhushanam, B.; Mir, M.I.; Nagaraju, M.; Sujatha, E.; Devi, B.R.; Kumar, B.K. Genetic Diversity Analysis of Linseed (Linum usitatissimum L.) Accessions Using RAPD Markers. Emirates J. Food Agric. 2021, 33, 589. [Google Scholar] [CrossRef]
- Everaert, I.; De Riek, J.; De Loose, M.; Van Waes, J.; Van Bockstaele, E. Most similar variety grouping for distinctness evaluation of flax and linseed (Linum usitatissimum L.) varieties by means of AFLP and morphological data. Plant Var. Seeds 2001, 14, 69–87. [Google Scholar]
- Van Treuren, R.; van Soest, L.J.M.; van Hintum, T.J.L. Markerassisted rationalization of genetic resource collections: A case study in flax using AFLPs. Theor. Appl. Genet. 2001, 103, 144–152. [Google Scholar] [CrossRef]
- Wakjira, A.; Viljoen, C.D.; Labuschagne, M.T. Analysis of genetic diversity in linseed using AFLP markers. Ethiop. J. Sci. 2005, 28, 41–50. [Google Scholar] [CrossRef]
- Chandrawati Maurya, R.; Singh, P.K.; Ranade, S.A.; Yadav, H.K. Diversity analysis in Indian genotypes of linseed (Linum usitatissimum L.) using AFLP markers. Gene 2014, 549, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Wiesnerová, D.; Wiesner, I. ISSR-based clustering of cultivated flax germplasm is statistically correlated to thousand seed mass. Mol. Biotechnol. 2004, 3, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Rajwade, A.V.; Arora, R.S.; Kadoo, N.Y.; Harsulkar, A.M.; Ghorpade, P.B.; Gupta, V.S. Relatedness of Indian flax genotypes (Linum usitatissimum L.): An inter-simple sequence repeat (ISSR) primer assay. Mol. Biotechnol. 2010, 45, 161–170. [Google Scholar] [CrossRef]
- El Sayed, A.A.; Ezzat, S.M.; Mostafa, S.H.; Zedan, S.Z.; Abdel-Sattar, E.; El Tanbouly, N. Inter simple sequence repeat analysis of genetic diversity and relationship in four Egyptian flaxseed genotypes. Phcog. Res. 2018, 10, 166–172. [Google Scholar]
- Choudhary, S.B.; Sharma, H.K.; Kumar, A.A.; Maruthi, R.T.; Mitra, J.; Chowdhury, I.; Singh, B.K.; Karmakar, P.G. SSR and morphological trait based population structure analysis of 130 diverse flax (Linum usitatissimum L.) accessions. Comptes Rendus Biol. 2017, 340, 65–75. [Google Scholar] [CrossRef]
- Wu, J.; Zhao, Q.; Wu, G.; Zhang, S.; Jiang, T. Development of Novel SSR Markers for Flax (Linum usitatissimum L.) Using Reduced-Representation Genome Sequencing. Front. Plant Sci. 2017, 7, 2018. [Google Scholar] [CrossRef] [Green Version]
- Green, A.G.; Chen, Y.; Singh, S.P.; Dribnenki, J.C.P. Flax. In Compendium of Transgenic Crop Plants; Kole, C., Hall, T.C., Eds.; Blackwell Publishing Ltd.: Oxford, UK, 2008; pp. 199–226. [Google Scholar]
- Cloutier, S.; Ragupathy, R.; Niu, Z.; Duguid, S. SSR-based link-age map of flax (Linum usitatissimum L.) and mapping of QTLs underlying fatty acid composition traits. Mol. Breed. 2011, 28, 437–451. [Google Scholar] [CrossRef]
- Cloutier, S.; Miranda, E.; Ward, K.; Radovanovic, N.; Reimer, E.; Walichnowski, A.; Datla, R.; Rowland, G.; Duguid, S.; Ragupathy, R. Simple sequence repeat marker development from bacterial artificial chromosome end sequences and expressed sequence tags of flax (Linum usitatissimum L.). Theor. Appl. Genet. 2012, 125, 685–694. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.B.; Diederichsen, A.; Richards, K.W.; Peterson, G. Genetic diversity within a range of cultivars and landraces of flax (Linum usitatissimum L.) as revealed by RAPDs. Genet. Resour. Crop Evol. 2002, 49, 167–174. [Google Scholar] [CrossRef]
- Fu, Y.-B.; Rowland, G.G.; Duguid, S.D.; Richards, K.W. RAPD analysis of 54 north American flax cultivars. Crop Sci. 2003, 43, 1510–1515. [Google Scholar] [CrossRef]
- Cloutier, S.; Niu, Z.; Datla, R.; Duguid, S. Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor. Appl. Genet. 2009, 119, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Asati, R.; Tripathi, M.K.; Tiwari, S.; Yadav, R.K.; Tripathi, N. Molecular approaches in the development of drought tolerance in chickpea. Life 2022, 12, 1846. [Google Scholar] [CrossRef]
- Yadav, P.K.; Singh, A.K.; Tripathi, M.K.; Tiwari, S.; Yadav, S.K.; Tripathi, N. Morpho-Physiological and Molecular Characterization of Maize (Zea mays L.) Genotypes for Drought Tolerance. Eur. J. Appl. Sci. 2022, 10, 65–87. [Google Scholar]
- Shyam, C.; Tripathi, M.K.; Tiwari, S.; Tripathi Niraj Ahuja, A. Identification of Low and High Erucic Acid Containing Genotype(S) in Indian Mustard Employing Molecular Markers. In Recent Progress in Plant and Soil Research; BP International: West Bengal, India, 2022; Volume 5, pp. 18–36. [Google Scholar] [CrossRef]
- Tripathi, N. Biodiversity and conservation of genetic resources: Key factor in crop improvement. In Biodiversity and Crop Improvement; Wani, S.H., Ed.; Scientific Research Publishing: Wuhan, China, 2015; pp. 104–129. ISBN 978-1-61896-068-9. [Google Scholar]
- Diederichsen, A. Ex-situ collections of cultivated flax (Linum usitatissimum L.) and other species of the genus Linum L. Genet. Resour. Crop Evol. 2007, 54, 661–678. [Google Scholar] [CrossRef]
- Diederichsen, A.; Fu, Y.B. Flax genetic diversity as the raw material for future success. In Proceedings of the International Conference on Flax and Other Bast Plants, Saskatoon, SK, Canada, 21–23 July 2008; pp. 270–280. [Google Scholar]
- Heslop-Harrison, J.S.; Schwarzacher, T. Genetics and genomics of crop domestication. In Plant Biotechnology and Agriculture: Prospects for the 21st Century; Altman, A., Hasegawa, P.M., Eds.; Elsevier Academic: Alpharetta, GA, USA, 2012; pp. 3–18. [Google Scholar]
- Brutch, N.B. The flax genetic resources collection held at the N.I. Vavilov Institute, Russian Federation. In Flax Genetic Resources in Europe; Maggioni, L.M., Pavelek, M., van Soest, L.J.M., Lipman, E., Eds.; IPGRI: Maccarese, Italy; Rome, Italy, 2002; pp. 61–65. [Google Scholar]
- Zhuchenko, A.A.; Rozhmina, T.A. Mobilizacija genetičeskich resursov l“na [Mobilization of flax genetic resources]. In VILAR and VNIIL; Starica: Staritsa, Russia, 2000; p. 224. [Google Scholar]
- Rashid, K.Y. Principal diseases of flax. In Flax the Genus Linum; Muir, A.D., Westcott, N.D., Eds.; Taylor and Francis Ltd.: London, UK, 2003; pp. 92–123. [Google Scholar]
- Shivaraj, S.M.; Dhakate, P.; Sonah, H.; Vuong, T.; Nguyen, H.T.; Deshmukh, R. Progress toward development of climate-smart flax: A perspective on omics-assisted breeding. In Genomic Designing Climate-Smart Oilseed Crops; Springer: Cham, Switzerland, 2019; pp. 239–274. [Google Scholar]
- Mishra, N.; Tripathi, M.K.; Tripathi, N.; Tiwari, S.; Gupta, N.; Sharma, A. Screening of soybean genotypes against drought on the basis of gene-linked microsatellite markers. In Book Innovations in Science and Technology; BP International: West Bengal, India, 2022; Volume 3, pp. 49–61. [Google Scholar]
- Tripathi, M.K.; Tripathi, N.; Tiwari, S.; Mishra, N.; Sharma, A.; Tiwari, S.; Singh, S. Identification of Indian soybean (Glycine max [L.] Merr.) genotypes for drought tolerance and genetic diversity analysis using SSR markers. Scientists 2023, 3, 31–46. [Google Scholar]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [Green Version]
- Nasir, M.W.; Toth, Z. Effect of Drought Stress on Potato Production: A Review. Agronomy 2022, 12, 635. [Google Scholar] [CrossRef]
- Molotoks, A.; Stehfest, E.; Doelman, J.; Albanito, F.; Fitton, N.; Dawson, T.P.; Smith, P. Global projections of future cropland expansion to 2050 and direct impacts on biodiversity and carbon storage. Glob. Chang. Biol. 2018, 24, 5895–5908. [Google Scholar] [CrossRef] [Green Version]
- Cui, Z.; Yan, B.; Gao, Y.; Wu, B.; Wang, Y.; Wang, H.; Xu, P.; Zhao, B.; Cao, Z.; Zhang, Y.; et al. Agronomic cultivation measures on productivity of oilseed flax: A review. Oil Crop Sci. 2022, 7, 53–62. [Google Scholar] [CrossRef]
- Zare, S.; Mirlohi, A.; Saeidi, G.; Sabzalian, M.R.; Ataii, E. Water stress intensified the relation of seed color with lignan content and seed yield components in flax (Linum usitatissimum L.). Sci. Rep. 2021, 11, 23958. [Google Scholar] [CrossRef] [PubMed]
- Fila, G.; Bagatta, M.; Maestrini, C.; Potenza, E.; Matteo, R. Linseed as a dual-purpose crop: Evaluation of cultivar suitability and analysis of yield determinants. J. Agric. Sci. 2018, 156, 162–176. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulos, P.; Mavroeidis, A.; Papadopoulos, G.; Roussis, I.; Bilalis, D.; Kakabouki, I. On the Path towards a “Greener” EU: A Mini Review on Flax (Linum usitatissimum L.) as a Case Study. Plants 2023, 12, 1102. [Google Scholar] [CrossRef]
- Heller, K.; Byczyńska, M. The impact of environmental factors and applied agronomy on quantitative and qualitative traits of flax fiber. J. Nat. Fibers 2015, 12, 26–38. [Google Scholar] [CrossRef]
- Kaur, V.; Yadav, R.; Wankhede, D.P. Linseed (Linum usitatissimum L.) genetic resources for climate change intervention and its future breeding. J. Appl. Nat. Sci. 2017, 9, 1112–1118. [Google Scholar] [CrossRef] [Green Version]
- Dash, P.K.; Cao, Y.; Jailani, A.K.; Gupta, P.; Venglat, P.; Xiang, D.; Rai, R.; Sharma, R.; Thirunavukkarasu, N.; Abdin, M.Z.; et al. Genome-wide analysis of drought induced gene expression changes in flax (Linum usitatissimum). GM Crops Food 2014, 5, 106–119. [Google Scholar] [CrossRef] [Green Version]
- Tawfik, R.S.; Badr, A.; Sammour, R.; Ibrahim, U.; Matter, M.; Sakr, M. Improvement of flax drought tolerance using gene transfer. Plant Tissue Cult. Biotechnol. 2016, 26, 197–207. [Google Scholar] [CrossRef] [Green Version]
- Asgarinia, P.; Mirlohi, A.; Saeidi, G.; MohamadiMirik, A.A.; Gheysari, M.; Razavi, V.S. Selection criteria for assessing drought tolerance in a segregating population of flax (Linum usitatissimum L.). Can. J. Plant Sci. 2016, 97, 424–437. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.S.; Wang, X.R.; Xu, J.; Zhang, J.P.; Mi, J. Drought-resistance evaluation of flax germplasm at adult plant stage. Sci. Agric. Sin. 2010, 43, 3076–3087. [Google Scholar]
- Sharma, J.C.; Tomar, S.S.; Shivran, R.K.; Prakash, C. Water requirement water use efficiency consumptive use yield and quality parameters of linseed (Linum usitatissimum L.) varieties as influenced by fertility levels irrigation scheduling. Adv. Life Sci. 2012, 1, 180–182. [Google Scholar]
- Kaur, V.; Yadav, S.K.; Wankhede, D.P.; Pulivendula, P.; Kumar, A.; Chinnusamy, V. Cloning and characterization of a gene encoding MIZ1, a domain of unknown function protein and its role in salt and drought stress in rice. Protoplasma 2020, 257, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Gowda, V.R.P.; Henry, A.; Yamauchi, A.; Shashidhar, H.E.; Serraj, R. Root biology and genetic improvement for drought avoidance in rice. Field Crop Res. 2011, 122, 1–13. [Google Scholar] [CrossRef]
- Tuberosa, R.; Sanguineti, M.C.; Landi, P.; Giuliani, M.M.; Salvi, S.; Conti, S. Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the field at two water regimes. Plant Mol. Biol. 2002, 48, 697–712. [Google Scholar] [CrossRef]
- Manschadi, A.M.; Christopher, J.; deVoil, P.; Hammer, G.L. The role of root architectural traits in adaptation of wheat to water-limited environments. Funct. Plant Biol. 2006, 33, 823–837. [Google Scholar] [CrossRef]
- Soto-Cerda, B.J.; Cloutier, S.; Gajardo, H.A.; Aravena, G.; Quian, R. Identifying drought-resilient flax genotypes and related candidate genes based on stress indices, root traits and selective sweep. Euphytica 2019, 215, 41. [Google Scholar] [CrossRef]
- Soto-Cerda, B.J.; Cloutier, S.; Gajardo, H.A.; Aravena, G.; Quian, R.; You, F.M. Drought response of flax accessions and identification of quantitative trait nucleotides (QTNs) governing agronomic and root traits by genome-wide association analysis. Mol. Breed. 2020, 40, 15. [Google Scholar] [CrossRef]
- Mishra, N.; Tripathi, M.K.; Tiwari, S.; Tripathi, N.; Gupta, N.; Sharma, A. Morphological and physiological performance of Indian soybean [Glycine max (L.) Merrill.] genotypes in respect to drought. Legume Res. 2021, 1–9. [Google Scholar] [CrossRef]
- Choudhary, M.L.; Tripathi, M.K.; Tiwari, S.; Pandya, R.K.; Gupta, N.; Tripathi, N.; Parihar, P. Screening of Pearl Millet [Pennisetum glaucum (L.) R. Br.] Germplam Lines for Drought Tolerance Based on Morpho-physiological Traits and SSR Markers. Curr. J. Appl. Sci. Technol. 2021, 40, 46–63. [Google Scholar] [CrossRef]
- Kachare, S.; Tiwari, S.; Tripathi, N. Expression of DREB1, RBCL, PIP, SGR genes and morpho-physiological changes under water stress in soybean. J. Plant Biochem. Biotechnol. 2022, 32, 338–355. [Google Scholar] [CrossRef]
- Kariuki, L.W.; Masinde, P.; Githiri, S.; Onyango, A.N. Effect of water stress on growth of three linseed (Linum usitatissimum L.) varieties. SpringerPlus 2016, 5, 759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansari, A.; Razmjoo, J.; Karimmojeni, H. Mycorrhizal colonization and seed treatment with salicylic acid to improve physiological traits and tolerance of flaxseed (Linum usitatissimum L.) plants grown under drought stress. Acta Physiol. Plant 2016, 38, 34. [Google Scholar] [CrossRef]
- Dubey, S.; Bhargava, A.; Fuentes, F.; Shukla, S.; Srivastava, S. Effect of salinity stress on yield and quality parameters in flax (Linum usitatissimum L.). Not. Bot. Horti Agrobot. 2020, 48, 954–966. [Google Scholar] [CrossRef]
- Singh, M.; Nara, U.; Kumar, A.; Choudhary, A.; Singh, H.; Thapa, S. Salinity tolerance mechanisms and their breeding implications. J. Genet. Eng. Biotechnol. 2021, 19, 173. [Google Scholar] [CrossRef]
- Tao, R.; Ding, J.; Li, C.; Zhu, X.; Guo, W.; Zhu, M. Evaluating and Screening of Agro-Physiological Indices for Salinity Stress Tolerance in Wheat at the Seedling Stage. Front. Plant Sci. 2021, 12, 646175. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Kocak, M.Z.; Göre, M.; Kurt, O. The effect of different salinity levels on germination development of some flax (Linum usitatissimum L.) varieties. J. Food Sci. Technol. 2022, 10, 657–662. [Google Scholar] [CrossRef]
- El-Afry, M.M.; El-Okkiah, S.A.; El-Kady, E.S.A.; El-Yamanee, G.S.A. Exogenous application of ascorbic acid for alleviation the adverse effects of salinity stress in flax (Linum usitatissimum L.). Middle East J. 2018, 7, 716–739. [Google Scholar]
- Nasri, N.; Maatallah, S.; Saidi, I.; Lachaal, M. Influence of salinity on germination, seedling growth, ion content and acid phosphatase activities of Linum usitatissimum L. J. Anim. Plant Sci. 2017, 27, 517–521. [Google Scholar]
- Patil, N.M.; Datir, S.S.; Shah, P.V. Salt-induced physiological and biochemical changes in two varieties of Linum usitatissimum L. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 296–304. [Google Scholar]
- Kaya, M.D.; Day, S.; Cikili, Y.; Arslan, N. Classification of some linseed (Linum usitatissimum L.) genotypes for salinity tolerance using germination, seedling growth, and ion content. Chil. J. Agric. Res. 2012, 72, 27–32. [Google Scholar] [CrossRef] [Green Version]
- Khan, N. Designing Genomic Solutions for Abiotic Traits in Flax (Linum usitatissimum L.). Ph.D. Thesis, University of Ottawa, Ottawa, ON, Canada, 2022. Submitted. [Google Scholar]
- Guo, R.; Zhou, J.; Ren, G.; Hao, W. Physiological responses of linseed seedlings to iso osmotic polyethylene glycol, salt, and alkali stresses. Agron. J. 2013, 105, 764–772. [Google Scholar] [CrossRef]
- Miart, F.; Fontaine, J.X.; Mongelard, G.; Wattier, C.; Lequart, M.; Bouton, S.; Molinie, R.; Dubrulle, N.; Fournet, F.; Demailly, H. Integument-Specific Transcriptional Regulation in the Mid-Stage of Flax Seed Development Influences the Release of Mucilage and the Seed Oil Content. Cells 2021, 10, 2677. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, H.; Yang, Y.; Lou, D.; Liang, C.; Yuan, H.; Wu, G.; Xu, C. Identification and Characterization of Differentially Expressed microRNAs and Target Gene Related to Flax Stem Development. J. Nat. Fibers 2022, 19, 5974–5990. [Google Scholar] [CrossRef]
- Wei, W.; Qian, D.; Qiaoling, J. Effects of NaCl stress on the biochemical characteristics in six species fiber flax seeding. Chin. Agric. Sci. Bull. 2013, 18, 017. [Google Scholar]
- Belkadhi, A.; De Haro, A.; Soengas, P.; Obregon, S.; Cartea, M.E.; Djebali, W.; Chaïbi, W. Salicylic acid improves root antioxidant defense system and total antioxidant capacities of flax subjected to cadmium. OMICS 2013, 17, 398–406. [Google Scholar] [CrossRef] [Green Version]
- Belkadhi, A.; De Haro, A.; Obregon, S.; Chaïbi, W.; Djebali, W. Positive effects of salicylic acid pretreatment on the composition of flax plastidial membrane lipids under cadmium stress. Environ. Sci. Pollut. Res. 2015, 22, 1457–1467. [Google Scholar] [CrossRef]
- Kaplan, M.E.; Simmons, E.R.; Hawkins, J.C.; Ruane, L.G.; Carney, J.M. Influence of cadmium and mycorrhizal fungi on the fatty acid profile of flax (Linum usitatissimum) seeds. J. Sci. Food Agric. 2015, 95, 2528–2532. [Google Scholar] [CrossRef]
- Grant, C.A.; Dribnenki, J.C.P.; Bailey, L.D. Cadmium and zinc concentrations and ratios in seed and tissue of solin (cv LinolaTM 947) and flax (cvs McGregor and Vimy) as affected by nitrogen and phosphorus fertiliser and Provide (Penicillium bilaji). J. Sci. Food Agric. 2000, 80, 1735–1743. [Google Scholar] [CrossRef]
- Smykalova, I.; Vrbova, M.; Tejklova, E.; Vetrovcova, M.; Griga, M. Large scale screening of heavy metal tolerance in flax/linseed (Linum usitatissimum L.) tested in vitro. Ind. Crops Prod. 2010, 32, 527–533. [Google Scholar] [CrossRef]
- Soudek, P.; Katrušáková, A.; Sedláček, L.; Petrová, Š.; Kočí, V.; Maršík, P.; Griga, M.; Vaněk, T. Effect of heavy metals on inhibition of root elongation in 23 cultivars of flax (Linum usitatissimum L.). Arch. Environ. Contamin. Toxicol. 2010, 59, 194–203. [Google Scholar] [CrossRef]
- Darapuneni, M.K.; Morgan, G.D.; Ibrahim, A.M.; Duncan, R.W. Effect of vernalization and photoperiod on flax flowering time. Euphytica 2014, 195, 279–285. [Google Scholar] [CrossRef]
- Ramirez-Villegas, J.; Molero Milan, A.; Alexandrov, N.; Asseng, S.; Challinor, A.J.; Crossa, J.; Eeuwijk, F.V.; Ghanem, M.E.; Grenier, C.; Heinemann, A.B.; et al. CGIAR modeling approaches for resource-constrained scenarios: I. Accelerating crop breeding for a changing climate. Crop Sci. 2020, 60, 547–567. [Google Scholar] [CrossRef] [Green Version]
- Cross, R.H. Heat Stress Effects on Flowering and Reproduction in Linum usitatissimum (Flax). Master’s Thesis, University of Saskatchewan, Saskatoon, SK, Canada, 2002. [Google Scholar]
- Saha, D.; Mukherjee, P.; Dutta, S.; Meena, K.; Sarkar, S.K.; Mandal, A.B.; Dasgupta, T.; Mitra, J. Genomic insights into HSFs as candidate genes for high-temperature stress adaptation and gene editing with minimal off-target effects in flax. Sci. Rep. 2019, 9, 5581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, D.; Shaw, A.K.; Datta, S.; Mitra, J. Evolution and functional diversity of abiotic stress-responsive NAC transcription factor genes in Linum usitatissimum L. Environ. Exp. Bot. 2021, 188, 104512. [Google Scholar] [CrossRef]
- Cross, R.H.; McKay, S.A.B.; McHughen, A.G.; Bonham-Smith, P.C. Heat stress effects on reproduction and seed set in Linum usitatissimum L. (flax). Plant Cell Environ. 2003, 26, 1013–1020. [Google Scholar] [CrossRef]
- Pokhrel, S.; Meyers, B.C. Heat-responsive microRNAs and phased small interfering RNAs in reproductive development of flax. Plant Direct. 2022, 6, 385. [Google Scholar] [CrossRef]
- Tripathi, N.; Tripathi, M.K.; Tiwari, S.; Payasi, D.K. Molecular Breeding to overcome biotic stresses in Soybean: Update. Plants 2022, 11, 1967. [Google Scholar] [CrossRef]
- Kenaschuk, E.O.; Rowland, G.G. Flax. In Harvest of Gold: The History of Field Crop Breeding in Canada; Slinkard, A.E., Knott, D.R., Eds.; University of Saskatchewan: Saskatoon, SK, Canada, 1993; pp. 173–176. [Google Scholar]
- Russell, G.E. Plant Breeding for Pest and Disease Resistance. Studies in Agriculture Sciences; Butterworth-Heinemann eBook: London, UK, 2013; p. 496. [Google Scholar]
- Lawrence, G.J.; Dodds, P.N.; Ellis, J.G. Transformation of the flax rust fungus, Melampsoralini: Selection via silencing of an avirulence gene. Plant J. 2010, 61, 364–369. [Google Scholar] [CrossRef] [Green Version]
- Flor, H. Tests for allelism of rust-resistance genes in flax. Crop Sci. 1965, 5, 415–418. [Google Scholar] [CrossRef] [Green Version]
- Khan, I.A. Flax, the Genus Linum Edited by Alostar D. Muir and Neil D. Westcott (Agriculture and Agri-Food Canada, Saskatoon); Taylor & Francis: London, UK, 2003; ISBN 0-415-30807-0. [Google Scholar]
- Kanapin, A.; Samsonova, A.; Rozhmina, T.; Bankin, M.; Logachev, A.; Samsonova, M. The Genome Sequence of Five Highly Pathogenic Isolates of Fusarium oxysporum f. sp. lini. Mol. Plant Microbe Interact. 2020, 33, 1112–1115. [Google Scholar] [CrossRef]
- Agrawal, S.; Narware, H.; Sahu, R. Inheritance of Fusarium wilt resistance in linseed. J. Oilseeds Res. 1991, 8, 231. [Google Scholar]
- Spielmeyer, W.; Green, A.; Bittisnich, D.; Mendham, N.; Lagudah, E. Identification of quantitative trait loci contributing to Fusarium wilt resistance on an AFLP linkage map of flax (Linum usitatissimum). Theor. Appl. Genet. 1998, 97, 633–641. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Lagudah, E.; Mendham, N.; Green, A. Inheritance of resistance to flax wilt (Fusarium oxysporum f. sp. lini Schlecht) in a doubled haploid population of Linum usitatissimum L. Euphytica 1998, 101, 287–291. [Google Scholar] [CrossRef]
- Portyankin, D.; Karachan, V. Resistance to Fusarium wilt of perspective fiber flax varieties. Zashchita Rastenij 1999, 79, 308–314. [Google Scholar]
- Diederichsen, A.; Rozhmina, T.A.; Kudrjavceva, L.P. Variation patterns within 153 flax (Linum usitatissimum L.) genebank accessions based on evaluation for resistance to fusarium wilt, anthracnose and pasmo. Plant Genet. Resour. 2008, 6, 22–32. [Google Scholar] [CrossRef]
- Elwakil, M.A.; Arafat, N.S.; Shabana, Y.M. Identification, Variability and Phylogeny of Alternaria linicola Associated with Linseed in Egypt. Plant Pathol. J. 2022, 21, 33–40. [Google Scholar] [CrossRef]
- Singh, R.B.; Singh, R.N. Occurrence Status and Management of Alternaria Blight (Alternaria species) of Linseed (Linum usitatissimum). Indian J. Agric. Sci. 2005, 75, 277–280. [Google Scholar]
- Holi, S.K.; Meena, S. Management of Alternaria Blight of Linseed (Linum usitatissimum) Caused by Alternaria lini. J. Plant Sci. Res. 2015, 31, 47–50. [Google Scholar]
- Fitt, B.D.L.; Coskun, H. Occurrence and pathogenicity of Alternaria spp. on linseed in the UK. In Flax as a Fibre and Oil Bearing Crop, Proceedings of the FAO European Regional Workshop on Flax, Brno, Czech Republic, 18–20 June 1991; University of Hertfordshire Research Archieves; University of Hertfordshire: Hatfield, UK, 1991; pp. 338–346. [Google Scholar]
- Vloutoglou, I.; Fitt, B.; Lucas, J. Infection of Linseed by Alternaria linicola; Effects of Inoculum Density, Temperature, Leaf Wetness and Light Regime. Eur. J. Plant Pathol. 1999, 105, 585–595. [Google Scholar] [CrossRef]
- Singh, R.B.; Singh, H.K.; Parmar, A. Yield loss assessment due to Alternaria blight and its management in linseed. Pak. J. Biol. Sci. 2014, 17, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Speck, A.; Trouvé, J.P.; Enjalbert, J.; Geffroy, V.; Joets, J.; Moreau, L. Genetic Architecture of Powdery Mildew Resistance Revealed by a Genome-Wide Association Study of a Worldwide Collection of Flax (Linum usitatissimum L.). Front. Plant Sci. 2022, 13, 871633. [Google Scholar] [CrossRef] [PubMed]
- Ashry, N.; Mansour, M.; Aly, A.; Zayed, S. Genetic studies on powdery mildew resistance of flax, yield and some yield components. Egypt J. Agric. Res. 2002, 80, 1525–1537. [Google Scholar]
- Rashid, K.; Duguid, S. Inheritance of resistance to powdery mildew in flax. Can. J. Plant Pathol. 2005, 27, 404–409. [Google Scholar] [CrossRef]
- Rashid, K.Y.; Desjardins, M.L.; Duguid, S. Diseases of flax in Manitoba and Saskatchewan in 2009. Can. Plant Dis. Surv. 2010, 90, 136–137. [Google Scholar]
- Cullis, C.A. Mechanisms and control of rapid genomic changes in flax. Ann. Bot. 2005, 95, 201–206. [Google Scholar] [CrossRef]
- Maggioni, L.; Pavelek, M.; van Soest, L.J.M.; Lipman, E. Flax genetic resources in Europe. In Proceedings of the Ad Hoc Meeting of European Cooperative Programme for Crop Genetic Resources Network ECP/GR of the International Plant Genetic Resources Institute, Prague, Czech Republic, 7–8 December 2001. [Google Scholar]
- Shamov, D. Status of the Bulgarian national flax collection. In Proceedings of the Ad Hoc Meeting, Prague, Czech Republic, 7–8 December 2001. [Google Scholar]
- UPOV. Working paper on the revision of test guidelines for flax, linseed (Linum usitatissimum L.). In Proceedings of the International Union for the Protection of New Varieties of Plants, Geneva, Technical Working Party for Agricultural Crops, Twentieth Session, Beltsville, MD, USA, 13–17 May 1991. [Google Scholar]
- Diederichsen, A.; Hammer, K. Variation of cultivated flax (Linum usitatissimum L. subsp. usitatissimum) and its wild progenitor pale flax (subsp. angustifolium (Huds.) Thell.). Genet. Resour. Crop Evol. 1995, 42, 263–272. [Google Scholar] [CrossRef]
- Pavelek, M. Recent state of international flax database and future development’, European cooperative network on flax, breeding research group. In Proceedings of the Report of Flax Genetic Resources Workshop, Second Meeting, Brno, Czech Republic, 8–10 November 1994; pp. 57–63. [Google Scholar]
- Pavelek, M. Further development of International Flax Data Base and special descriptors for more detail evaluation of agronomic and processing characters. In Breeding for Fibre and Oil Quality in Flax’, Proceedings of the Third Meeting of the International Flax Breeding Group, St Valery enCaux, France, 7–8 November 1995; Centre Technique Pour l’étude et l’amélioration du lin (CETEAL): Paris, France, 1995; pp. 1–13. [Google Scholar]
- Rakouský, S.; Tejklova, E.; Wiesner, I.; Wiesnerova, D.; Kocabek, T.; Ondrej, M. TDNA induced mutations and somaclonal variants of flax. Nat. Fibres 1998, 2, 244–246. [Google Scholar]
- Rakouský, S.; Tejklov_a, E.; Wiesner, I. Recent advances in flax biotechnology in the Czech Republic. Nat. Fibres 2001, 1, 151–155. [Google Scholar]
- Jia, G.; You, F.M.; Cloutier, S.; Deyholos, M.K.; Booker, H. The flax breeding database (FlaxDB). In Proceedings of the XXVII Plant and Animal Genome Conference, San Diego, CA, USA, 12–16 January 2019; p. PE1082. [Google Scholar]
- Biradar, S.A.; Ajithkumar, K.; Rajanna, B.; Savitha, A.S.; Shubha, G.V.; Shankergoud, I.; Singh, P.K. Prospects and challenges in linseed (Linum usitatissimum L.) production: A review. J. Oilseeds Res. 2016, 33, 1–13. [Google Scholar]
- Tadesse, N.; Lay, C.; Dybing, C.D. Comparative seed yield performance of high-by-high and low-by-high crosses in flax. Plant Breed. 1997, 116, 561–566. [Google Scholar] [CrossRef]
- Pavelek, M. International flax database. In Proceedings of the Flax Genetic Resources in Europe, Ad Hoc Meeting, Prague, Czech Republic, 7–8 December 2001; Maggioni, L., Pavelek, M., van Soest, L.J.M., Lipman, E., Eds.; International Plant Genetic Resources Institute: Rome, Italy, 2001. [Google Scholar]
- Tejklova, E. Some factors affecting anther cultures in Linum usitatissimum L. Rostl. Vyrob. 1996, 42, 249–260. [Google Scholar]
- Tejklova, E. Study of anther culture in flax (Linum usitatissimum L.). Nat. Fibres 1998, 2, 202–209. [Google Scholar]
- Yurenkova, S.I.; Kubrak, S.V.; Titok, V.V.; Khotyljova, L.V. Flax species polymorphism for isozyme and metabolic markers. Genetika 2005, 41, 334–340. [Google Scholar] [CrossRef]
- Ijaz, A.; Shahbaz, A.; Ullah, I.; Ali, S. Molecualar Characterization of Linseed Germplasm Using RAPD DNA Fingerprinting Markers. Am.-Eurasian J. Agric. Environ. Sci. 2013, 13, 1266–1274. [Google Scholar]
- Rowland, G.G. Growing Flax: Production, Management and Diagnostic Guide; Flax Council of Canada and Saskatchewan Flax Development Commission: Saskatoon, SK, Canada, 1998. [Google Scholar]
- Gill, K.S. Evolutionary Relationships Among Linum Species. Ph.D. Thesis, University of California, Riverside, CA, USA, 1966. [Google Scholar]
- Carnahan, H.L. Combining Ability in Flax (Linum usitatissimum). Master’s Thesis, University of Minnesota, Minneapolis, MN, USA, 1947. [Google Scholar]
- Dubey, D.K.; Singh, S.P. Extent of heterosis and problems of hybrid seed production in linseed (Linum usitatissimum L.). Balwant Vidyapeeth J. Agric. Sci. Res. 1969, 6, 83–87. [Google Scholar]
- Kumar, S.; Singh, S.P. Inheritance of male sterility in some introduced varieties of linseed (Linum usitatissimum L.). Indian J. Agric. Sci. 1970, 40, 184–191. [Google Scholar]
- Kumar, S.; Singh, S.P. Inheritance of partial male fertility in linseed (Linum usitatissimum L.). Indian J. Agric. Sci. 1972, 42, 34–38. [Google Scholar]
- Dubey, D.K.; Singh, S.P. Mechanism of pollen abortion in three male sterile lines of flax (Linum usitatissimum L.). Crop Sci. 1965, 5, 121–124. [Google Scholar] [CrossRef] [Green Version]
- Dubey, D.K.; Singh, S.P. Use of cytoplasmatic male sterility for the production of hybrid seeds in flax Linum usitatissimum L. Crop Sci. 1966, 6, 125–126. [Google Scholar] [CrossRef]
- Kumar, S. Haploid linseed. Agra Univ. J. Res. 1971, 40, 23–25. [Google Scholar]
- Pant, S.C.; Mishra, V.K. Heterosis over superior parents under diallel cross in linseed X flax (Linum usitatissimum L.). Indian J. Plant Genet. Resour. 2008, 21, 93–97. [Google Scholar]
- Sood, S.; Kalia, N.R.; Bhateria, S.; Kumar, S. Detection of genetic components of variation for some biometrical traits in Linum usitatissimum L. in sub-mountain Himalayan region. Euphytica 2007, 155, 107–115. [Google Scholar] [CrossRef]
- Bhateria, S.; Sood, S.P.; Pathania, A. Genetic analysis of quantitative traits across environments in linseed (Linum usitatissimum L.). Euphytica 2006, 150, 185–194. [Google Scholar] [CrossRef]
- George, K.P.; Nayar, K.K. Early dwarf mutants in linseed induced by gamma rays. Curr. Sci. 1973, 42, 137–138. [Google Scholar]
- Rai, M.; Das, K. Potentiality and genetic variability in irradiated population of linseed. Indian J. Genet. Plant Breed. 1976, 36, 20–25. [Google Scholar]
- Pospisil, B. The significance of chemical muatgens in breeding flax. Genet. Slechteni 1974, 10, 187–196. [Google Scholar]
- Bianu, M.; Marki, A.; Ochesanu, C. The flexuous stem-a new gene in Linum usitatissimum induced by the alkylating agents. Botanique 1972, 25, 171–175. [Google Scholar]
- Deshpande, R.B. A note on the occurrence of chlorophyll deficiency in linseed. Curr. Sci. 1939, 8, 168–169. [Google Scholar]
- Levan, A. Experimentally induced chlorophyll mutants in flax. Hereditas 1944, 30, 225–230. [Google Scholar] [CrossRef]
- Sharov, I. The mutation spectrum of fibre flax induced by chemical mutagens. Refrat. Zh 1971, 8, 55–119. [Google Scholar]
- Green, A.G. A mutant genotype of flax (Linum usitatissimum) containing very low levels of linolenic acid in its seed oil. Can. J. Plant Sci. 1986, 66, 499–503. [Google Scholar] [CrossRef]
- Gill, K.S. Linseed; Indian Council of Agricultural Research: New Delhi, India, 1987. [Google Scholar]
- Cloutier, S.; Miranda, E.; Ragupathy, R.; Radovanovic, N.; Reimer, E.; Walichnowski, A. Integrated consensus genetic and physical maps of flax (Linum usitatissimum L.). Theor. Appl. Genet. 2012, 125, 1783–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Huang, Y.; Huang, R.; Wu, Y.; Wang, W. The genetic architecture of amylose biosynthesis in maize kernel. Plant Biotechnol. J. 2018, 16, 688–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remington, D.L.; Thornsberry, J.M.; Matsuoka, Y.; Wilson, L.M.; Whitt, S.R.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 2001, 98, 11479–11484. [Google Scholar] [CrossRef]
- Sonah, H.; O’Donoughue, L.; Cober, E.; Rajcan, I.; Belzile, F. Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol. J. 2015, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Soto-Cerda, B.J.; Diederichsen, A.; Ragupathy, R.; Cloutier, S. Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types. BMC Plant Biol. 2013, 13, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto-Cerda, B.J.; Diederichsen, A.; Duguid, S.; Booker, H.; Rowland, G.; Cloutier, S. The potential of pale flax as a source of useful genetic variation for cultivated flax revealed through molecular diversity and association analyses. Mol. Breed. 2014, 34, 2091–2107. [Google Scholar] [CrossRef] [Green Version]
- Soto-Cerda, B.J.; Duguid, S.; Booker, H.; Rowland, G.; Diederichsen, A.; Cloutier, S. Association mapping of seed quality traits using the Canadian flax (Linum usitatissimum L.) core collection. Theor. Appl. Genet. 2014, 127, 881–896. [Google Scholar] [CrossRef] [Green Version]
- Cullis, C.A. Genetics and Genomics of Linum; Springer International Publishing: Cham, Switzerland, 2019. [Google Scholar]
- Kenaschuk, E.O. High linolenic acid flax. Google Patents Cullis CA (2007) Flax. In Genome Mapping and Molecular Breeding in Plants; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; Volume 2, pp. 275–295. [Google Scholar]
- Zhang, J.; Xie, Y.; Dang, Z.; Wang, L.; Li, W.; Zhao, W.; Zhao, L.; Dang, Z. Oil content and fatty acid components of oilseed flax under different environments in China. Agron. J. 2016, 108, 365–372. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Amer. J. Hum. Genet. 1980, 32, 314. [Google Scholar]
- Paterson, A.H.; Lander, E.S.; Hewitt, J.D.; Peterson, S.; Lincoln, S.E.; Tanksley, S.D. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 1988, 335, 721–726. [Google Scholar] [CrossRef]
- Oh, T.J.; Gorman, M.; Cullis, C.A. RFLP and RAPD mapping in flax (Linumusitatisimum). Theor. Appl. Genet. 2000, 101, 590–593. [Google Scholar] [CrossRef]
- Kumar, S.; You, F.M.; Cloutier, S. Genome wide SNP discovery in flax through next generation sequencing of reduced representation libraries. BMC Genom. 2012, 13, 684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukushima, A.; Kusano, M.; Redestig, H.; Arita, M.; Saito, K. Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol. 2009, 13, 532–538. [Google Scholar] [CrossRef]
- Weckwerth, W.; Ghatak, A.; Bellaire, A.; Chaturvedi, P.; Varshney, R.K. PANOMICS meets germplasm. Plant Biotechnol. J. 2020, 18, 1507–1525. [Google Scholar] [CrossRef] [Green Version]
- Pazhamala, L.T.; Kudapa, H.; Weckwerth, W.; Millar, A.H.; Varshney, R.K. Systems biology for crop improvement. Plant Genome 2021, 14, e20098. [Google Scholar] [CrossRef]
- Kumari, J.; Mahatman, K.K.; Sharma, S.; Singh, A.K.; Adhikari, S.; Bansal, R.; Kaur, V.; Kumar, S.; Yadav, M.C. Recent advances in different omics mechanism for drought stress tolerance in rice. Russ. J. Plant Physiol. 2022, 69, 18. [Google Scholar] [CrossRef]
- Chaudhary, J.; Patil, G.B.; Sonah, H.; Deshmukh, R.K.; Vuong, T.D.; Valliyodan, B.; Nguyen, H.T. Expanding omics resources for improvement of soybean seed composition traits. Front. Plant Sci. 2015, 6, 1021. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, J.; Khatri, P.; Singla, P.; Kumawat, S.; Kumari, A.; Vikram, A.; Jindal, S.K.; Kardile, H.; Kumar, R.; Sonah, H.; et al. Advances in omics approaches for abiotic stress tolerance in tomato. Biology 2019, 8, 90. [Google Scholar] [CrossRef] [Green Version]
- Akhmetshina, A.O.; Strygina, K.V.; Khlestkina, E.K.; Porokhovinova, E.A.; Brutch, N.B. High-throughput sequencing techniques to flax genetics and breeding. Ecol. Genet. 2020, 18, 103–124. [Google Scholar] [CrossRef] [Green Version]
- Yadav, B.; Kaur, V.; Narayan, O.P.; Yadav, S.K.; Kumar, A.; Wankhede, D.P. Integrated omics approaches for flax improvement under abiotic and biotic stress: Current status and future prospects. Front. Plant Sci. 2022, 13, 931275. [Google Scholar] [CrossRef]
- Saroha, A.; Pal, D.; Kaur, V.; Kumar, S.; Bartwal, A.; Aravind, J.; Radhamani, J.; Rajkumar, S.; Kumar, R.; Gomashe, S.S.; et al. Agro-morphological variability and genetic diversity in linseed (Linum usitatissimum L.) germplasm accessions with emphasis on flowering and maturity time. Genet. Resour. Crop Evol. 2022, 69, 315–333. [Google Scholar] [CrossRef]
- Kaur, V.; Kumar, S.; Yadav, R.; Wankhede, D.P.; Aravind, J.; Radhamani, J.; Rana, J.C.; Kumar, A. Analysis of genetic diversity in Indian and exotic linseed germplasm and identification of trait specific superior accessions. J. Environ. Biol. 2018, 39, 702–709. [Google Scholar] [CrossRef]
- Uysal, H.; Fu, Y.-B.; Kurt, O.; Peterson, G.W.; Diederichsen, A.; Kusters, P. Genetic diversity of cultivated flax (Linum usitatissimum L.) and its wild progenitor pale flax (Linum bienne mill.) as revealed by ISSR markers. Genet. Resour. Crop Evol. 2010, 57, 1109–1119. [Google Scholar] [CrossRef]
- Fu, Y.-B.; Peterson, G.; Diederichsen, A.; Richards, K. RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genet. Resour. Crop Evol. 2002, 49, 253–259. [Google Scholar] [CrossRef]
- Wang, Z.; Hobson, N.; Galindo, L.; Zhu, S.; Shi, D.; McDill, J.; Yang, L.; Hawkins, S.; Neutelings, G.; Datla, R.; et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 2012, 72, 461–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spindel, J.E.; Dahlberg, J.; Colgan, M.; Hollingsworth, J.; Sievert, J.; Staggenborg, S.H.; Hutmacher, R.; Jansson, C.; Vogel, J.P. Association mapping by aerial drone reveals 213 genetic associations for Sorghum bicolor biomass traits under drought. BMC Genom. 2018, 19, 679. [Google Scholar] [CrossRef] [Green Version]
- Lasky, J.R.; Upadhyaya, H.D.; Ramu, P.; Deshpande, S.; Hash, C.T.; Bonnette, J.; Juenger, T.E.; Hyma, K.; Acharya, C.; Mitchell, S.E.; et al. Genome-environment associations in sorghum landraces predict adaptive traits. Sci. Adv. 2015, 1, e1400218. [Google Scholar] [CrossRef] [Green Version]
- Badigannavar, A.; Teme, N.; de Oliveira, A.C.; Li, G.; Vaksmann, M.; Viana, V.E.; Ganapathi, T.R.; Sarsu, F. Physiological, genetic and molecular basis of drought resilience in sorghum (Sorghum bicolor (L.)). Indian J. Plant Physiol. 2018, 23, 670–688. [Google Scholar] [CrossRef]
- Guo, Z.; Yang, W.; Chang, Y.; Ma, X.; Tu, H.; Xiong, F.; Jiang, N.; Feng, H.; Huang, C.; Yang, P.; et al. Genome-wide association studies of image traits reveal genetic architecture of drought resistance in rice. Mol. Plant 2018, 11, 789–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangin, B.; Bonnafous, F.; Blanchet, N.; Boniface, M.C.; Bret-Mestries, E.; Carrère, S.; Cottret, L.; Legrand, L.; Marage, G.; Pegot-Espagnet, P.; et al. Genomic prediction of sunflower hybrids oil content. Front. Plant Sci. 2017, 8, 1633. [Google Scholar] [CrossRef] [Green Version]
- Dossa, K.; Li, D.; Zhou, R.; Yu, J.; Wang, L.; Zhang, Y.; You, J.; Liu, A.; Mmadi, M.A.; Fonceka, D.; et al. The genetic basis of drought tolerance in the high oil crop Sesamum indicum. J. Plant Biotechnol. 2019, 17, 1788–1803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Wang, L.; Zhang, Y.; Zhu, G.; Zhu, X.; Xia, Y.; Li, J.; Gao, X.; Wang, S.; Zhang, J.; et al. Genome-Wide Identification and Function of Aquaporin Genes During Dormancy and Sprouting Periods of Kernel-Using Apricot (Prunus armeniaca L.). Front. Plant Sci. 2021, 12, 690040. [Google Scholar] [CrossRef] [PubMed]
- Shivaraj, S.M.; Deshmukh, R.; Bhat, J.A.; Sonah, H.; Bélanger, R.R. Understanding aquaporin transport system in eelgrass (Zostera marina L.), an aquatic plant species. Front. Plant Sci. 2017, 8, 1334. [Google Scholar] [CrossRef] [Green Version]
- Thambugala, D.P. Analysis of Genetic Diversity and Expression of Genes Involved in Fatty Acid Composition in Flax (Linum usitatissimum L.) and Comparative Genomic Analysis of their Loci. Ph.D. Thesis, Department of Plant Science, University of Manitoba, Winnipeg, MB, Canada, 2015. [Google Scholar]
- He, L.; Xiao, J.; Rashid, K.Y.; Yao, Z.; Li, P.; Jia, G.; Wang, X.; Cloutier, S.; You, F.M. Genome-wide association studies for pasmo resistance in flax (Linum usitatissimum L.). Front. Plant Sci. 2019, 9, 1982. [Google Scholar] [CrossRef]
- You, F.M.; Rashid, K.Y.; Zheng, C.; Khan, N.; Li, P.; Xiao, J.; He, L.; Yao, Z.; Cloutier, S. Insights into the Genetic Architecture and Genomic Prediction of Powdery Mildew Resistance in Flax (Linum usitatissimum L.). Int. J. Mol. Sci. 2022, 23, 4960. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; Campos, G.D.L.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Abed, A.; Pérez-Rodríguez, P.; Crossa, J.; Belzile, F. When less can be better: How can we make genomic selection more cost-effective and accurate in barley? Theor. Appl. Genet. 2018, 131, 1873–1890. [Google Scholar] [CrossRef]
- Deshmukh, R.; Sonah, H.; Patil, G.; Chen, W.; Prince, S.; Mutava, R.; Vooung, T.; Valliyodan, D.; Nguyen, H.T. Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 2014, 5, 244. [Google Scholar] [CrossRef]
- Budhlakoti, N.; Kushwaha, A.K.; Rai, A.; Chaturvedi, K.K.; Kumar, A.; Pradhan, A.K.; Kumar, U.; Kumar, R.R.; Juliana, P.; Mishra, D.C.; et al. Genomic selection: A tool for accelerating the efficiency of molecular breeding for development of climate-resilient crops. Front. Genet. 2022, 13, 832153. [Google Scholar] [CrossRef] [PubMed]
- Sertse, D.; You, F.M.; Ravichandran, S.; Soto-Cerda, B.J.; Duguid, S.; Cloutier, S. Loci harboring genes with important role in drought and related abiotic stress responses in flax revealed by multiple GWAS models. Theoret. Appl. Genet. 2021, 134, 191–212. [Google Scholar] [CrossRef] [PubMed]
- You, F.M.; Cloutier, S. Mapping quantitative trait loci onto chromosome-scale pseudomolecules in flax. Methods Protoc. 2020, 3, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asgarinia, P.; Cloutier, S.; Duguid, S.R.; Rashid, K.; Mirlohi, A.; Banik, M.; Saeidi, G. Mapping quantitative trait loci for powdery mildew resistance in flax. Crop. Sci. 2013, 53, 2462–2472. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Zhao, Q.; Sun, D.; Wu, G.; Zhang, L.; Yuan, H.; Yu, Y.; Zhang, S.; Yang, X.; Li, Z.; et al. Transcriptome analysis of flax (Linum usitatissimum L.) undergoing osmotic stress. Ind. Crop Prod. 2018, 116, 215–223. [Google Scholar] [CrossRef]
- Cartagena, J.A.; Marquez, G.P.B. Transcriptome analysis in Jatropha during abiotic stress response. In Oil Crop Genomics; Tombuloglu, H., Unver, T., Tombuloglu, G., Hakeem, K.R., Eds.; Springer: Cham, Switzerland, 2021; pp. 317–337. [Google Scholar]
- Moschen, S.; Di Rienzo, J.A.; Higgins, J.; Tohge, T.; Watanabe, M.; González, S.; Rivarola, M.; García-García, F.; Dopazo, J.; Hopp, H.E.; et al. Integration of transcriptomic and metabolic data reveals hub transcription factors involved in drought stress response in sunflower (Helianthus annuus L.). Plant Mol. Biol. 2017, 94, 549–564. [Google Scholar] [CrossRef]
- Leisner, C.P.; Yendrek, C.R.; Ainsworth, E.A. Physiological and transcriptomic responses in the seed coat of field-grown soybean (Glycine max L. Merr.) to abiotic stress. BMC Plant Biol. 2017, 17, 242. [Google Scholar] [CrossRef] [Green Version]
- Dmitriev, A.A.; Krasnov, G.S.; Rozhmina, T.A. Flax (Linum usitatissimum L.) response to non-optimal soil acidity and zinc deficiency. BMC Plant Biol. 2019, 19, 54. [Google Scholar] [CrossRef] [Green Version]
- Aghaee, P.; Rahmani, F. Biochemical and molecular responses of flax to 24-epibrassinosteroide seed priming under drought stress. J. Plant Interact. 2019, 14, 242–253. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Wu, G.; Yuan, H.; Cheng, L.; Zhao, D.; Huang, W.; Zhang, S.; Zhang, L.; Chen, H.; Zhang, J.; et al. Identification and characterization of miRNAs and targets in flax (Linum usitatissimum) under saline, alkaline, and saline-alkaline stresses. BMC Plant Biol. 2016, 16, 124. [Google Scholar] [CrossRef] [Green Version]
- Dash, P.K.; Rai, R.; Mahato, A.K.; Gaikwad, K.; Singh, N.K. Transcriptome landscape at different developmental stages of a drought tolerant cultivar of flax (Linum usitatissimum). Front. Chem. 2017, 5, 82. [Google Scholar] [CrossRef] [Green Version]
- Melnikova, N.V.; Dmitriev, A.A.; Belenikin, M.S.; Koroban, N.V.; Speranskaya, A.S.; Krinitsina, A.A.; Krasnov, G.S.; Lakunina, V.A.; Snezhkina, A.V.; Sadritdinova, A.F.; et al. Identification, expression analysis, and target prediction of flax genotroph MicroRNAs under normal and nutrient stress conditions. Front. Plant Sci. 2016, 7, 399. [Google Scholar] [CrossRef] [Green Version]
- Melnikova, N.V.; Dmitriev, A.A.; Belenikin, M.S.; Speranskaya, A.S.; Krinitsina, A.A.; Rachinskaia, O.A. Excess fertilizer responsive miRNAs revealed in Linum usitatissimum L. Biochimie 2015, 109, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Zyablitsin, A.V.; Dmitriev, A.A.; Krasnov, G.S.; Bolsheva, N.L.; Rozhmina, T.A.; Muravenko, O.V.; Fedorova, M.S.; Snezhkina, A.V.; Kudryavtseva, A.V.; Melnikova, N.V. CAX3 gene is involved in flax response to high soil acidity and aluminium exposure. Mol. Biol. 2018, 52, 514–519. [Google Scholar] [CrossRef]
- Shivaraj, S.M.; Deshmukh, R.K.; Rai, R.; Bélanger, R.; Agrawal, P.K.; Dash, P.K. Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum). Sci. Rep. 2017, 7, 46137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Zhao, Q.; Wu, G.; Yuan, H.; Ma, Y.; Lin, H.; Pan, L.; Li, S.; Sun, D. Comprehensive analysis of differentially expressed unigenes under NaCl stress in flax (Linum usitatissimum L.) using RNA-Seq. Int. J. Mol. Sci. 2019, 20, 369. [Google Scholar] [CrossRef] [Green Version]
- Kostyn, K.; Czemplik, M.; Kulma, A.; Bortniczuk, M.; Skała, J.; Szopa, J. Genes of phenylpropanoid pathway are activated in early response to fusarium attack in flax plants. Plant Sci. 2012, 190, 103–115. [Google Scholar] [CrossRef]
- Dmitriev, A.A.; Krasnov, G.S.; Rozhmina, T.A.; Novakovskiy, R.O.; Snezhkina, A.V.; Fedorova, M.S.; Yurkevich, O.Y.; Muravenko, O.V.; Bolsheva, N.L.; Kudryavtseva, A.V.; et al. Differential gene expression in response to Fusarium oxysporum infection in resistant and susceptible genotypes of flax (Linum usitatissimum L.). BMC Plant Biol. 2017, 17, 253. [Google Scholar] [CrossRef] [Green Version]
- Boba, A.; Kostyn, K.; Kozak, B.; Wojtasik, W.; Preisner, M.; Prescha, A.; Gola, E.M.; Lysh, D.; Dudek, B.; Szopa, J.; et al. Fusarium oxysporum infection activates the plastidial branch of the terpenoid biosynthesis pathway in flax, leading to increased ABA synthesis. Planta 2020, 251, 50. [Google Scholar] [CrossRef] [Green Version]
- Wróbel-Kwiatkowska, M.; Lorenc-Kukula, K.; Starzycki, M.; Oszmiański, J.; Kepczyńska, E.; Szopa, J. Expression of β-1, 3-glucanase in flax causes increased resistance to fungi. Physiol. Mol. Plant Pathol. 2004, 65, 245–256. [Google Scholar] [CrossRef]
- Boba, A.; Kostyn, K.; Kozak, B.; Zalewski, I.; Szopa, J.; Kulma, A. Transcriptomic profiling of susceptible and resistant flax seedlings after Fusarium oxysporum lini infection. PLoS ONE 2021, 16, e0246052. [Google Scholar] [CrossRef]
- Yu, Y.; Huang, W.; Chen, H.; Wu, G.; Yuan, H.; Song, X.; Kang, Q.; Zhao, D.; Jiang, W.; Liu, Y.; et al. Identification of differentially expressed genes in flax (Linum usitatissimum L.) under saline–alkaline stress by digital gene expression. Gene 2014, 549, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, L.; Wang, L.; Tan, M.; Ogutu, C.O.; Yin, Z.; Zhou, J.; Wang, J.; Wang, L.; Yan, X. Transcriptome analysis and molecular mechanism of linseed (Linum usitatissimum L.) drought tolerance under repeated drought using single-molecule long-read sequencing. BMC Genom. 2021, 22, 109. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Nemri, A.; Blackman, L.M.; Catanzariti, A.-M.; Sperschneider, J.; Lawrence, G.J.; Dodds, P.N.; Jones, D.A.; Hardham, A.R. Flax rust infection transcriptomics reveals a transcriptional profile that may be indicative for rust Avr genes. PLoS ONE 2019, 14, e0226106. [Google Scholar] [CrossRef] [PubMed]
- Nemri, A.; Saunders, D.G.; Anderson, C.; Upadhyaya, N.M.; Win, J.; Lawrence, G.J.; Jones, D.A.; Kamoun, S.; Ellis, J.G.; Dodds, P.N. The genome sequence and effector complement of the flax rust pathogen Melampsoralini. Front. Plant Sci. 2014, 5, 98. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Gonzalez, L.; Deyholos, M.K. RNA-seq transcriptome response of flax (Linum usitatissimum L.) to the pathogenic fungus Fusarium oxysporum f. sp. lini. Front. Plant Sci. 2016, 7, 1766. [Google Scholar] [CrossRef] [Green Version]
- Pontarin, N.; Molinié, R.; Mathiron, D.; Tchoumtchoua, J.; Bassard, S.; Gagneul, D.; Thiombiano, B.; Demailly, H.; Fontaine, J.X.; Guillot, X.; et al. Age-dependent metabolic profiles unravel the metabolic relationships within and between flax leaves (Linum usitatissimum). Metabolites 2020, 10, 218. [Google Scholar] [CrossRef]
- Weckwerth, W. Metabolomics in systems biology. Annu. Rev. Plant Biol. 2003, 54, 669–689. [Google Scholar] [CrossRef]
- Ghatak, A.; Chaturvedi, P.; Weckwerth, W. Metabolomics in plant stress physiology. Adv. Biochem. Eng./Biotechnol. 2018, 164, 187–236. [Google Scholar] [CrossRef]
- Hall, R.D. Plant metabolomics: From holistic hope, to hype, to hot topic. New Phytol. 2006, 169, 453–468. [Google Scholar] [CrossRef]
- Ibáñez, C.; García-Cañas, V.; Valdés, A.; Simó, C. Novel MS-based approaches and applications in food metabolomics. Trends Anal. Chem. 2013, 52, 100–111. [Google Scholar] [CrossRef]
- Kouba, M.; Mourot, J. A review of nutritional effects on fat composition of animal products with special emphasis on n-3 polyunsaturated fatty acids. Biochimie 2011, 93, 13–17. [Google Scholar] [CrossRef]
- McCann, M.J.; Gill, C.I.R.; McGlynn, H.; Rowland, I.R. Role of mammalian lignans in the prevention and treatment of prostate cancer. Nutr. Cancer 2005, 52, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, A.; Fliniaux, O.; Fang, J.; Molinie, R.; Roscher, A.; Grand, E.; Guillot, X.; Kovensky, J.; Fliniaux, M.A.; Schneider, S.; et al. Development of an NMR metabolomics-based tool for selection of flaxseed varieties. Metabolomics 2014, 10, 1258–1267. [Google Scholar] [CrossRef]
- El Sabagh, A.; Hossain, A.; Barutcular, C.; Gormus, O.; Ahmad, Z.; Hussain, S.; Islam, M.S.; Alharby, H.; Bamagoos, A.; Kumar, N.; et al. Effects of drought stress on the quality of major oilseed crops: Implications and possible mitigation strategies—A review. Appl. Ecol. Environ. Res. 2019, 17, 4019–4043. [Google Scholar] [CrossRef]
- Jakab, G.; Ton, J.; Flors, V.; Zimmerli, L.; Metraux, J.P.; Mauch-Mani, B. Enhancing Arabidopsis salt and drought stress synthase gene in transgenic tobacco plants: Pleiotropic phenotypes include drought tolerance. Planta 2005, 201, 293–297. [Google Scholar] [CrossRef] [Green Version]
- Macarisin, D.; Wisniewski, M.E.; Bassett, C.; Thannhauser, T.W. Proteomic analysis of β-aminobutyric acid priming and abscisic acid—Induction of drought resistance in crabapple (Malus pumila): Effect on general metabolism, the phenyl-propanoid pathway and cell wall enzymes. Plant Cell Environ. 2009, 32, 1612–1631. [Google Scholar] [CrossRef]
- Garg, A.K.; Kim, J.K.; Owens, T.G.; Ranwala, A.P.; Do Choi, Y.; Kochian, L.V.; Wu, R.J. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. USA 2002, 99, 15898–15903. [Google Scholar] [CrossRef]
- Du, Y.L.; Wang, Z.Y.; Fan, J.W.; Turner, C.; Wang, T.; Li, F.M. B-aminobutyric acid increases abscisic acid accumulation and desiccation tolerance and decreases water use but fails to improve grain yield in two spring wheat cultivars under soil drying. J. Exp. Bot. 2012, 63, 4849–4860. [Google Scholar] [CrossRef]
- Bengtsson, T.; Weighill, D.; Proux-Wéra, E.; Levander, F.; Resjö, S.; Burra, D.D.; Moushib, L.I.; Hedley, P.E.; Liljeroth, E.; Jacobson, D.; et al. Proteomics and transcriptomics of the BABA-induced resistance response in potato using a novel functional annotation approach. BMC Genom. 2014, 15, 315. [Google Scholar] [CrossRef] [Green Version]
- Cortina, C.; Culianez-Macia, F.A. Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Sci. 2005, 169, 75–82. [Google Scholar] [CrossRef]
- Jakab, G.; Cottier, V.; Toquin, V.; Rigoli, G.; Zimmerli, L.; Metraux, J.P.; Mauch-Mani, B. β-Aminobutyric acid-induced resistance in plants. Eur. J. Plant Pathol. 2001, 107, 29–37. [Google Scholar] [CrossRef]
- Singh, P.; Wu, C.C.; Zimmerli, L. β-Aminobutyric acid priming by stress imprinting. Plant Signal. Behav. 2010, 5, 878–880. [Google Scholar] [CrossRef] [Green Version]
- Quéro, A.; Fliniaux, O.; Elboutachfaiti, R.; Petit, E.; Guillot, X.; Hawkins, S.; Courtois, J.; Mesnard, F. Β-aminobutyric acid increases drought tolerance and reorganizes solute content and water homeostasis in flax (Linum usitatissimum). Metabolomics 2015, 11, 1363–1375. [Google Scholar] [CrossRef]
- Qayyum, M.A.; Bashir, F.; Maqbool, M.M.; Ali, A.; Bashir, S.; Abbas, Q. Implications of saline water irrigation for linseed on seed germination, seedling survival and growth potential. Sarhad J. Agric. 2019, 35, 1289–1297. [Google Scholar] [CrossRef]
- Cha-um, S.; Supaibulwatana, K.; Kirdmanee, C. Water relation, photosynthetic ability and growth of Thai jasmine rice (Oryza sativa L. ssp. indica cv. KDML105) to salt stress by application of exogenous glycine betaine and choline. J. Agron. Crop Sci. 2006, 192, 25–36. [Google Scholar] [CrossRef]
- Naz, T.; Akhtar, J.; Iqbal, M.M.; Haq, M.A.U.; Saqib, M. Boron toxicity in salt-affected soils and effects on plants. In Soil Science: Agricultural and Environmental Prospectives; Hakeem, K.R., Akhtar, J., Sabir, M., Eds.; Springer: Cham, Switzerland, 2016; pp. 259–286. [Google Scholar]
- Hamade, K.; Fliniaux, O.; Fontaine, J.X.; Molinié, R.; OtogoNnang, E.; Bassard, S.; Guénin, S.; Gutierrez, L.; Lainé, E.; Hano, C.; et al. NMR and LC-MS-based metabolomics to study osmotic stress in lignan-deficient flax. Molecules 2021, 26, 767. [Google Scholar] [CrossRef]
- Vo KT, X.; Rahman, M.M.; Rahman, M.M.; Trinh KT, T.; Kim, S.T.; Jeon, J.S. Proteomics and metabolomics studies on the biotic stress responses of rice: An update. Rice 2021, 14, 30. [Google Scholar] [CrossRef]
- Gevaert, K.; Vandekerckhove, J. Gel-Free Proteomics: Methods and Protocols; Springer: Berlin, Germany, 2011. [Google Scholar]
- Baggerman, G.; Vierstraete, E.; De Loof, A.; Schoofs, L. Gel-based versus gel-free proteomics: A review. Comb. Chem. High Throughput Screen 2005, 8, 669–677. [Google Scholar] [CrossRef]
- Klubicová, K.; Danchenko, M.; Skultéty, L.; Berezhna, V.V.; Hricová, A.; Rashydov, N.M.; Hajduch, M. Agricultural recovery of a formerly radioactive area: II. Systematic proteomic characterization of flax seed development in the remediated Chernobyl area. J. Proteom. 2011, 74, 1378–1384. [Google Scholar] [CrossRef]
- Hradilová, J.; Řehulka, P.; Řehulková, H.; Vrbová, M.; Griga, M.; Brzobohatý, B. Comparative analysis of proteomic changes in contrasting flax cultivars upon cadmium exposure. Electrophoresis 2010, 31, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Balbuena, T.S.; Salas, J.J.; Martinez-Force, E.; Garces, R.; Thelen, J.J. Proteome analysis of cold acclimation in sunflower. J. Proteome Res. 2011, 10, 2330–2346. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.; Berla, B.M.; Sheffield, J.; Cahoon, R.E.; Jez, J.M.; Hicks, L.M. Comprehensive analysis of the Brassica juncea root proteome in response to cadmium exposure by complementary proteomic approaches. Proteomics 2009, 9, 2419–2431. [Google Scholar] [CrossRef] [PubMed]
- Wendelboe-Nelson, C.; Morris, P.C. Proteins linked to drought tolerance revealed by DIGE analysis of drought resistant and susceptible barley varieties. Proteomics 2012, 12, 3374–3385. [Google Scholar] [CrossRef]
- Pang, Q.; Chen, S.; Dai, S.; Chen, Y.; Wang, Y.; Yan, X. Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J. Proteome Res. 2010, 9, 2584–2599. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Prášil, I.T. Proteomics of stress responses in wheat and barley—Search for potential protein markers of stress tolerance. Front. Plant Sci. 2014, 5, 711. [Google Scholar] [CrossRef] [Green Version]
- Caruso, G.; Cavaliere, C.; Foglia, P.; Gubbiotti, R.; Samperi, R.; Laganà, A. Analysis of drought responsive proteins in wheat (Triticum durum) by 2D-PAGE and MALDI-TOF mass spectrometry. Plant Sci. 2009, 177, 570–576. [Google Scholar] [CrossRef]
- Caruso, G.; Cavaliere, C.; Guarino, C.; Gubbiotti, R.; Foglia, P.; Laganà, A. Identification of changes in Triticum durum L. leaf proteome in response to salt stress by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Anal. Bioanal. Chem. 2008, 391, 381–390. [Google Scholar] [CrossRef]
- Rinalducci, S.; Egidi, M.G.; Karimzadeh, G.; Jazii, F.R.; Zolla, L. Proteomic analysis of a spring wheat cultivar in response to prolonged cold stress. Electrophoresis 2011, 32, 1807–1818. [Google Scholar] [CrossRef]
- Majoul, T.; Bancel, E.; Triboi, E.; Ben Hamida, J.; Branlard, G. Proteomic analysis of the effect of heat stress on hexaploid wheat grain: Characterization of heat-responsive proteins from non-prolamins fraction. Proteomics 2004, 4, 505–513. [Google Scholar] [CrossRef]
- Li, H.; Yang, M.; Zhao, C.; Wang, Y.; Zhang, R. Physiological and proteomic analyses revealed the response mechanisms of two different drought-resistant maize varieties. BMC Plant Biol. 2021, 21, 513. [Google Scholar] [CrossRef]
- Halder, T.; Choudhary, M.; Liu, H.; Chen, Y.; Yan, G.; Siddique, K. Wheat proteomics for abiotic stress tolerance and root system architecture: Current status and future prospects. Proteomes 2022, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Melnikova, N.V.; Belenikin, M.S.; Bolsheva, N.L.; Dmitriev, A.A.; Speranskaya, A.S.; Krinitsina, A.A.; Nadezda, V.K.; Samatadze, T.E.; Amosova, A.V.; Muravenko, O.V.; et al. Flax inorganic phosphate deficiency responsive miRNAs. J. Agric. Sci. 2014, 6, 156–160. [Google Scholar] [CrossRef] [Green Version]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Qiu, S.Q.; Rollins, M.; Datla, R.; Gupta, V.S.; Kadoo, N.Y. Genome-wide identification and characterization of microRNA genes and their targets in flax (Linum usitatissimum). Planta 2013, 237, 1149–1161. [Google Scholar] [CrossRef]
- Young, L.W. High Temperature Stress and Flowering in Brassica napus L. Ph.D. Thesis, University of Saskatchewan, Saskatoon, SK, Canada, 2003. [Google Scholar]
- Dmitriev, A.A.; Kudryavtseva, A.V.; Krasnov, G.S.; Koroban, N.V.; Speranskaya, A.S.; Krinitsina, A.A.; Belenikin, M.S.; Snezhkina, A.V.; Sadritdinova, A.F.; Kishlyan, N.V.; et al. Gene expression profiling of flax (Linum usitatissimum L.) under edaphic stress. BMC Plant Biol. 2016, 16, 237. [Google Scholar] [CrossRef] [Green Version]
- Hano, C.; Addi, M.; Fliniaux, O.; Bensaddek, L.; Duverger, E.; Mesnard, F.; Lamblin, F.; Laine, E. Molecular characterization of cell death induced by a compatible interaction between Fusarium oxysporum f. sp. linii and flax (Linum usitatissimum) cells. Plant Physiol. Biochem. 2008, 46, 590–600. [Google Scholar] [CrossRef]
- Fujita, Y.; Venterink, H.O.; Van Bodegom, P.M.; Douma, J.C.; Heil, G.W.; Hölzel, N.; Jablonska, E.; Kotowski, W.; Okruszko, T.; Pawlikowski, P.; et al. Low investment in sexual reproduction threatens plants adapted to phosphorus limitation. Nature 2013, 505, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.W.; Shi, Y.; Zhu, Y.X.; Wang, Y.C.; Gong, H.J. Mechanisms of enhanced heavy metal tolerance in plants by silicon, a review. Pedosphere 2013, 23, 815–825. [Google Scholar] [CrossRef]
- Guo, P.; Baum, M.; Grando, S.; Ceccarelli, S.; Bai, G.; Li, R.; von Korff, M.; Varshney, R.K.; Graner, A.; Valkoun, J. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J. Exp. Bot. 2009, 60, 3531–3544. [Google Scholar] [CrossRef]
- Shi, D.C.; Wang, D. Effects of various salt-alkaline mixed stresses on Aneurolepidium chinense (Trin.) Kitag. Plant Soil 2005, 271, 15–26. [Google Scholar] [CrossRef]
- Guo, B.; Fedorova, N.D.; Chen, X.; Wan, C.-H.; Wang, W.; Nierman, W.C.; Bhatnagar, D.; Yu, J. Gene Expression Profiling and Identification of Resistance Genes to Aspergillus flavus Infection in Peanut through EST and Microarray Strategies. Toxins 2011, 3, 737–753. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Hao, W.; Gong, D.Z. Effects of water stress on germination and growth of linseed seedlings (Linum usitatissimum L.), photosynthetic efficiency and accumulation of metabolites. J. Agric. Sci. 2012, 4, 253–265. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Zhou, J.; Hao, W.; Gu, F.; Liu, Q.; Li, H.; Xia, X.; Mao, L. Germination, growth, chlorophyll fluorescence and ionic balance in linseed seedlings subjected to saline and alkaline stresses. Plant Prod. Sci. 2014, 17, 20–31. [Google Scholar] [CrossRef] [Green Version]
- White, J.W.; Andrade-Sanchez, P.; Gore, M.A.; Bronson, K.F.; Coffelt, T.A.; Conley, M.M.; Feldmann, K.A.; French, A.N.; Heun, J.T.; Hunsaker, D.J.; et al. Field-based phenomics for plant genetics research. Field Crops Res. 2012, 133, 101–112. [Google Scholar] [CrossRef]
- Pratap, A.; Gupta, S.; Nair, R.M.; Gupta, S.K.; Schafleitner, R.; Basu, P.S.; Singh, C.M.; Prajapati, U.; Gupta, A.K.; Nayyar, H.; et al. Using Plant Phenomics to Exploit the Gains of Genomics. Agronomy 2019, 9, 126. [Google Scholar] [CrossRef] [Green Version]
- Walter, A.; Liebisch, F.; Hund, A. Plant phenotyping: From bean weighing to image analysis. Plant Methods 2015, 11, 14. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Doonan, J.H.; Hawkesford, M.J.; Pridmore, T.; Zhou, J. Editorial: State-of-the-Art Technology and Applications in Crop Phenomics. Front. Plant Sci. 2021, 12, 767324. [Google Scholar] [CrossRef]
- Tao, H.; Xu, S.; Tian, Y.; Li, Z.; Ge, Y.; Zhang, J.; Wang, Y.; Zhou, G.; Deng, X.; Zhang, Z.; et al. Proximal and remote sensing in plant phenomics: 20 years of progress, challenges, and perspectives. Plant Commun. 2022, 3, 100344. [Google Scholar] [CrossRef]
- Bekkering, C.S.; Huang, J.; Tian, L. Image-Based, Organ-Level Plant Phenotyping for Wheat Improvement. Agronomy 2020, 10, 1287. [Google Scholar] [CrossRef]
- Sirault, X.R.R.; James, R.A.; Furbank, R.T. A new screening method for osmotic component of salinity tolerance in cereals using infrared thermography. Funct. Plant Biol. 2009, 36, 970–977. [Google Scholar] [CrossRef]
- Jordan, M.C.; McHughen, A. Glyphosate tolerant flax plants from mediated gene transfer. Plant Cell Rep. 1988, 7, 281–284. [Google Scholar] [CrossRef] [PubMed]
- McHughen, A.; Holm, F. Development and preliminary field testing of a glufosinate-ammonium tolerant transgenic flax. Can. J. Plant Sci. 1995, 75, 117–120. [Google Scholar] [CrossRef]
- Haughn, G.W.; Smith, J.; Mazur, B.; Somerville, C. Transformation with a mutant Arabidopsis acetolactate synthase gene renders tobacco resistant to sulfonylurea herbicides. Mol. Gen. Genet. 1988, 211, 266–271. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshmukh, R.K.; Sonah, H.; Bélanger, R.R. Plant Aquaporins: Genome-Wide Identification, Transcriptomics, Proteomics, and Advanced Analytical Tools. Front. Plant Sci. 2016, 7, 1896. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Bhatt, V.; Kumar, V.; Kumawat, S.; Khatri, P.; Singla, P.; Shivaraj, S.M.; Nadaf, A.; Deshmukh, R.; Sharma, T.R.; et al. Evolutionary Understanding of Aquaporin Transport System in the Basal Eudicot Model Species Aquilegia coerulea. Plants 2020, 9, 799. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Mishra, A. Plant aquaporins alleviate drought tolerance in plants by modulating cellular biochemistry, root-architecture, and photosynthesis. Physiol. Plant. 2021, 172, 1030–1044. [Google Scholar] [CrossRef]




| Species | No. of Accessions | Life Form | 2n | Origin |
|---|---|---|---|---|
| L. campanulatum L. | 1 | Perennial | 28 | West Mediterranean |
| L. austriacum L. | 3 | Perennial | 18 | Asia, Siberia, Central and East Europe |
| L. bienne Mill. | 11 | Winter-annual | 30 | Mediterranean, West Europe, West Asia |
| L. decumbens Desf. | 1 | Annual | 30 | South Europe |
| L. capitatum | 1 | Perennial | 34 | Balkan |
| L. altaicum Ledeb. | 2 | Perennial | 18 | West Siberia |
| L. leonii | 1 | Perennial | 18 | France, Germany |
| L. flavum L. | 4 | Perennial | 28 (30) | Caucasus, South and Central Europe |
| L. trigynum L. | 2 | Annual | 20 | Mediterranean, South and Central Europe, West Asia |
| L. hirsutum L. | 1 | Perennial | 16 | Central and East Europe, West Asia |
| L. narbonense L. | 3 | Perennial | 18 (20) | Mediterranean |
| L. rigidum Pursh | 1 | Perennial | 20 | North America |
| L. lewisii Pursh | 11 | Perennial | 18 | North America |
| L. pallescens Bunge | 1 | Perennial | 30 | West Siberia |
| L. tenuifolium L. | 1 | Perennial | 16 | Europe, West Asia, Mediterranean, |
| L. perenne L. | 10 | Perennial | 18 | Siberia, West Asia, Central and East Europe |
| L. strictum L. | 3 | Annual | 18 (30, 22) | Mediterranean, West Asia, South Europe, |
| L. grandiflorum Desf. | 7 | Annual | 16 | Algeria |
| Varietal Characters | Name of Variety |
|---|---|
| Seed type | JLS95, JLS66, LSL93, RLC 148, Pratap 2, T 397, JLS27, PKV-NL 260, Kota Barani Alsi 4, JL41, JLS67, JLS73, Arpita, Padmini, Divya, Priyam, Indu (LCK 1108), Rajan, Surya, TL 99, Kota Barani Alsi 5, Kota Barani Alsi 6, Jawahar Linseed 165, RLC 164, RLC 167, RLC 171, Sabour Tisi-2, Sabour Tisi-3, Birsa Tisi-1, LCK 1611, JLS122, SHA5 |
| Irrigated conditions | Pratap 2, JLS27, Kota Alsi 6, Priyam, Indu (LCK 1108), Rajan, Surya, TL 99, SHA5, LCK 1611, JLS95, JLS66 |
| Rainfed condition | JLS95, JLS66, LSL93, RLC 148, T 397, Sharda, Padmini, KV-NL 260, Kota Barani Alsi 4, JL 41, JLS67, JLS73, Arpita, Padmini, Divya, JLS122 |
| Utera situation | RLC 143, RLC 153, Sabour Tisi-2, Sabour Tisi-3, R 552 |
| Cultivars with dual purpose (seed + fibre) | Him Alsi-2, Nagarkot, Jeevan, RLU-6, Rashmi, Parvati, Meera, Shikha, Tiara (JRF 2) |
| S. No. | Name of Variety | Year of Release and Notification No. | Pedigree | Originating Centre | Duration | Avg. Yield kg/ha | Oil Content (%) | Recommended States | Special Features |
|---|---|---|---|---|---|---|---|---|---|
| 1 | K-2 | 1975 44 E, 21 August 1975 | Rust-resistant strain from UP x Kangra local | Ludhiana (Punjab) | 170–175 | 1110(I) | 40.04 | Punjab, Haryana, and Uttar Pradesh | White flower, dark brown, resistant to wilt and rust |
| 2 | LC-185 | 1975 44 E, 21 August 1975 | NP(RR)-37x Kangra local | Ludhiana (Punjab) | 150–160 | 500(U) | 38.89 | Punjab | Blue flower, resistant to powdery mildew and rust |
| 3 | HIRA | 1978 429 E, 19 December 1978 | H 342 x NP(RR)-9 | Kanpur (U.P.) | 130–135 | 1200(R) | 36.36 | Bundelkhand area and U.P. | White flower, resistant to wilt and rust |
| 4 | MUKTA | 1978 429 E, 19 December 1978 | H 626 x NP(RR)-9 | Kanpur (U.P.) | 127–132 | 1200(I) | 41.40 | Eastern U.P. | Large white flower, medium- and brown-seeded, resistant to wilt and rust |
| 5 | CHAMBAL | 1978 13 E, 19 December 1978 | Local x RR45 | Kota Rajasthan | 125–130 | 900(I) | 40.11 | Rajasthan | Blue flower, brown- and bold-seeded, moderately resistant to rust, wilt, and powdery mildew |
| 6 | NEELUM | 1978 13 E, 19 December 1978 | T-1 x NP(RR)-9 | Kanpur (U.P.) | 140–145 | 1500(I) | 43.00 | Mid Central and Western Uttar Pradesh | Blue flower, brown- and bold-seeded, tolerant to wilt and rust |
| 7 | JAWAHAR-1 | 1982 19 E, 14 January 1982 | Selection of KP 29 | Jabalpur (M.P.) | 116–120 | 900(R) | 38.34 | Madhya Pradesh | Red-violet flower, dark-brown- and bold-seeded, resistant to rust |
| 8 | JAWAHAR-7 (JLS-1) | 1982 19 E, 14 January 1982 | Selection of No. 55 | Raipur (C.G.) | 116–120 | 700(R), 300(U) | 37.79 | Madhya Pradesh | Red-violet flower, dark-brown and bold-seeded, resistant to rust |
| 9 | JAWAHAR-17 (JLS-7) | 1982 19 E, 14 January 1982 | Selection of No. 55 | Raipur (C.G.) | 117–120 | 1300(I), 800(R) | 37.61 | Madhya Pradesh | Red-violet flower, dark-brown- and bold-seeded, resistant to rust |
| 10 | NEELA | 1982 19 E, 14 January 1982 | SPS from indigenous collection | Behrampore (W.B.) | 127–135 | 850(R) | 34.93 | West Bengal | Blue flower, brown- and medium-seeded, resistant to bud fly |
| 11 | LC-54 | 1982 19 E, 14 January 1982 | K2 x Kangra local | Ludhiana (Punjab) | 155–165 | 1330(I) | 42.00 | Punjab, H.P. and Haryana | White flower, brown- and medium-seeded resistant to wilt and rust |
| 12 | C-429 | 1983 429 E, 3 January 1983 | No.3 x IP-135 | Nagpur (M.S.) | 125–130 | 1000(R) | 39.07 | Maharashtra | Red-violet flower, dark-brown- andsmall-seeded, resistant to rust |
| 13 | T-397 | 1984 596 E, 13 August 1984 | T-491 x T-1103-1 | Kanpur (U.P.) | 120–125 | 1100(I) | 44.00 | Bundelkhand of U.P., Bihar, Assam, M.P. and Rajasthan | Violet flower, dark-brown-seeded, tolerant to rust, wilt, and drought |
| 14 | JAWAHAR-552 (R-552) | 1984 596 E, 13 August 1984 | Sel. Number 55x B-67 | Raipur (C.G.) | 118–125 | 900(R) | 44.00 | Madhya Pradesh | Violet flower, light-brown-seeded, medium toleranceto rust, wilt, and powdery mildew |
| 15 | PUSA-2 | 1985 295 E, 9 April 1985 | Selection of BS 12 | New Delhi | 125–150 | 730(R) | 38.31 | Punjab, H.P., Haryana and Rajasthan | White flower, brown-seeded, medium height, resistant to rust |
| 16 | PUSA-3 | 1985 295 E, 9 April 1985 | K2 x T-603 | New Delhi | 125–150 | 800(I) | 37.65 | Punjab, H.P., Haryana and Rajasthan | White flower, brown-seeded, medium height, resistant to rust |
| 17 | S-36 | - | Selection of Local Solapur | Pune (MH) | 130–135 | 400(R) | 34.80 | Karnataka | Blue flower, light-brown- and small-seeded |
| 18 | HIMALINI | 1985 295 E, 9 April 1985 | K2 x Kangra local | Palampur (H.P.) | 150–175 | 1310(I) | 42.00 | Punjab, H.P., Haryana and adjoining of Rajasthan | White flower, brown-seeded, medium height, resistant to rust and wilt |
| 19 | JAWAHAR-23 | 1985 540 E, 24 July 1985 | EC 9832 x Hira | Jabalpur (MP) | 120–130 | 1000(I) | 43.00 | M.P., Odisha, Raj. Bundelkhand of U.P., M.S., Karnataka | White flower, brown- andmedium-seeded, resistant to wilt and rust and powdery mildew |
| 20 | GARIMA | 1985 540 E, 24 July 1985 | T-126 x Neelum | Kanpur (UP) | 120–130 | 1490(I) | 42.00 | U.P. (Excl. Bundelkhand), Bihar, W.B. and Assam | Blue flower, medium height, brown-seeded, tolerant to PM, altenararia blight, and wilt, resistant to rust |
| 21 | SWETA | 1985 540 E, 24 July 1985 | Mukta x T-1206 | Kanpur (UP) | 130–135 | 880(R) | 44.00 | U.P. (Excl. Bundelkhand), Bihar, W.B. and Assam | Blue flower, mediumheight, brown-seeded, tolerant to PM, altenararia blight, and wilt, resistant to rust |
| 22 | SHUBHRA | 1985 540 E, 24 July 1985 | Mukta x K 2 | Kanpur (UP) | 130–135 | 1390(I), 870 (R) | 45.00 | U.P. (Excl. Bundelkhand), Bihar, W.B and Assam | Dark-brown-seeded, medium, white flower, resistant to rust |
| 23 | LAXMI-27 | 1987 165 E, 6 March 1987 | Neelum/R1//Neelum x NPRR-9// Neelum x R1// Neelum x Afg-8 | Mauranipur (UP) | 110–120 | 1260(I),1020(R) | 45.00 | Bundelkhand of U.P. | Red-violet flower, dark-brown-seeded medium height, resistant to rust, wilt, and PM |
| 24 | GAURAV | 1987 834 E, 18 September 1987 | Sel.3 x EC-1552 | Kanpur (UP) | 137–140 | 1050 (S) 950(F) | 43.00 | Assam, U.P. (Excl. Bundelkhand), Bihar, West Bengal | Violet-red flower, brown- and medium-seeded, resistant to wilt and rust |
| 25 | KIRAN | 1988 10 E, 1 January 1988 | (R1x Afg-8) x R1 | Raipur (C.G.) | 120–126 | 750(R) | 43.00 | M.P., Odisha, Bundelkhand of U.P., M.S., Karnataka, and Rajasthan | Violet-red flower, brown- and medium-seeded, resistant to wilt, powdery mildew, and rust |
| 26 | JANKI | 1988 | New River x LC 216 | Palampur (H.P.) | 165–170 | 1200(I) | 42.00 | Himachal Pradesh | Violet-red flower, brown- and medium-seeded, resistant to wilt, powdery mildew, and rust |
| 27 | JEEVAN | 1988 10 E, 1 January 1988 | Summit x LC-216 | Palampur (H.P.) | 175–180 | 1090(S 1100(F) | 45.00 | Punjab and Himachal Pradesh | Violet-red flower, brown- and medium-seeded, resistant to wilt, powdery mildew, and rust |
| 28 | SURABHI | 1995 408 E, 4 May 1995 | LC 216 x LC 185 | Palampur (H.P.) | 165–170 | 1000(U) | 44.00 | Himachal Pradesh | White flower, yellow- and small-seeded, resistant to wilt, powdery mildew, and rust |
| 29 | NAGARKOT | 1995 400 E, 4 May 1995 | New river x LC 216 | Palampur (H.P.) | 165–170 | 1150 (S) 950 (F) | 43.00 | Panjab, U.P., H.P., W.B., Assam, Haryana and Raj. | Blue flower, brown- and medium-seeded, resistant to wilt, powdery mildew, and rust |
| 30 | SHIKHA | 1997 401 E, 15 May 1998 | Hira xCrista | Kanpur (U.P.) | 135–140 | 1233 (S) 1033 (F) | 42.00 | U.P. (Excl. Bundelkhand), Bihar, W.B. and Assam | Blue flower, brown- and medium-seeded, resistant to wilt and rust |
| 31 | LC 2023 | 1998 | Flake x LC 54 | Ludhiana (Punjab) | 158–163 | 1294 (I) | 38.8 | Punjab | - |
| 32 | PADMINI | 1998 1050 E, 26 October 1999 | (EC 41,628 x EC 77959) x (DPL 20 x Neelum) | Manranipur (U.P.) | 120–125 | 943 (R) | 43.00 | M.P., Rajasthan, Bundelkhand of U.P., Maharashtra, Odissa and Karnataka | Blue flower, brown-seeded, resistant to powdery mildew |
| 33 | JAWAHAR LINSEED SAGAR-9 (JLS 9) | 1998 425 E, 8 June 1999 | (RL-102 x R7) x J 23 | Sagar (M.P.) | 115–125 | 1250 (I), 1000 (R) | 42.00 | Madhya Pradesh | White flower, mediumheight, brown- and bold-seeded, resistant to wilt, powdery mildew, and rust |
| 34 | RASHMI | 1999 1050 E, 26 October 1999 | Gaurav x Janki | Kanpur (U.P.) | 135–140 | 1003 (S), 719 (F) | 41.00 | U.P., Bihar, W.B. and Rajasthan | Very light blue flower, brown-seeded, resistant to wilt, powdery mildew, and rust |
| 35 | MEERA | 2000 340 E, 3 April 2000 | (RL75-6-2 x RL 29-8) x LCK 8528 | Kota (Raj.) | 135–140 | 1439 (S), 1011 (F) | 42.00 | U.P., Bihar, W.B. and Rajasthan | Violet-blue flower, brown-seeded, resistant to wilt, powdery mildew, and rust |
| 36 | RL-914 | 2000 973 E, 4 September 2002 | (RR 9 x R-93) x (Flake 1 x LC 54) | Kota (Raj.) | 135–140 | 1617(I) | 41.10 | Kota command area of Rajasthan | Violet-blue flower, medium height, brown-seeded, resistant to wilt, powdery mildew, and rust |
| 37 | PARVATI | 2001 92 E, 2 February 2001 | EC 41628/ EC77959//DPL20 x Neelam///EC216 x Hira//BR 1x NP 440 | Manranipur (U.P.) | 140–146 | 1600 (S), 1020 (F) | 42.00 | U.P., Bihar, W.B. and Rajasthan | Blue flower, medium height, brown-seeded, resistant to powdery mildew and rust |
| 38 | SHEELA | 2001 92 E, 2 February 2001 | Gaurav x Janki | Kanpur (U.P.) | 155–160 | 1379 (R) | 41.00 | Himachal Pradesh, Panjab, Haryana, and Jammu and Kashmir | Blue flower, shining-brown-seeded, resistant to wilt, powdery mildew, and rust |
| 39 | SHEKHAR | 2001 92 E, 2 February 2001 | Laxmi 27 x EC 1387 | Kanpur (U.P.) | 135–140 | 1555(I), 920 (R) | 43.00 | U.P. (Excl. Bundelkhand), Bihar, W.B. and Assam | Violet-blue flower, shining-brown-seeded, moderately resistant to bud fly, alternaria blight, powdery mildew, and rust |
| 40 | NL-97 | 2001 92 E, 2 February 2001 | R-7 x RLC 4 | Nagpur (M.H.) | 115–120 | 641 (R) | 42.00 | Bidarbha Region | Blue flower, brown- and medium-seeded |
| 41 | JAWAHAR LINSEED SAGAR-27 (SUYOG) | 2002 161 E, 4 February 2004 | (Kran x KL-168) x Kiran | Sagar (M.P.) | 118–125 | 1509 (I) | 41.43 | Raj. Bundelkhand of U.P., M.P., C.G., M.H., Odissa, A.P., and Karnataka | White flower, light-brown-seeded, resistant to rust, powdery mildew, and bud fly |
| 42 | BINWA (KL-210) | 2004 122 E, 2 February 2005 | Flake 1 x SPS-47/7-10-3 | Palampur (H.P.) | 179–186 | 858 (I) | 40.00 | Haryana, Panjab, Himachal Pradesh, and J&K | Blue flower, yellow-seeded, resistant to wilt and rust |
| 43 | BANER (KL-224) | 2005 1177 E, 25 August 2005 | EC 21741 x LC 216 | Palampur (H.P.) | 171–203 | 511 (U) | 39.70 | Haryana, Panjab, Himachal Pradesh, and J&K | Purple flower, brown-coloured seeds, resistant to rust |
| 44 | INDIRA ALSI-32 (RLC-76) | 2005 1566 E, 5 November 2005 | Kiran x RLC 29 | Raipur (C.G.) | 110–115 | 780(R) | 39.18 | CG, Maharashtra, Karnataka, and Odisha | Blue flower, dark-brown-seeded, powdery mildew |
| 45 | KARTIKA (RLC-76) | 2005 1566 E, 5 November 2005 | Kiran x LCK-88062 | Raipur (C.G.) | 103–108 | 1078(R) | 42.93 | Rainfed areas of Chhattisgarh | Blue flower, light-brown and medium-seeded, resistant to bud fly and powdery mildew |
| 46 | DEEPIKA (RLC 78) | 2006 1178 E, 20 July 2007 | Kiran x Ayogi | Raipur (C.G.) | 110–115 | 1272 (SI&U) | 41.39 | Partially irrigated as well as Utera Situation of CG | Blue flower, brown-seeded, resistant to powdery mildew |
| 47 | SHARDA (LMS-4-27) | 2006 1572 E, 20 September 2006 | (Shubhra x J1) x (J1 x Kiran) | Manranipur (C.G.) | 100–105 | 762 (R) | 41.32 | CG, Maharashtra, Karnataka, AP and Odissa | White flower, brown-seeded, MR to wilt, powdery mildew, and bud fly |
| 48 | PRATAP ALSI-1 (RLU-6) | 2007 1703 E, 5October 2007 | Acc. 750 x RL 29-8 | Kota (Raj.) | 129–135 | 1997 (S) 834 (F) | 41.08 | Rajasthan Kota command area | White flower, brown-seeded, MR to rust, wilt, and bud fly |
| 49 | LC-2063 | 2007 1108 E, 8 May 2008 | 1509 x LC 54 | Ludhiana (Punjab) | 115–125 | 1200 (I) | 37.40 | Irrigated area of Punjab State | Blue flower, dark-brown-seeded, MR to bud fly |
| 50 | HIMANI KL-214 | 2008 2458 E, 16 October 2008 | DPL 20 x KLS-1 | Palampur (H.P.) | 177–200 | 583 (U) | 36.40 | HP, PB, Haryana and J&K | Blue flower, small- andbrown-seeded, MR to powdery mildew and rust |
| 51 | AZAD ALSI-1 (LMS 9-2K) | 2008 2458 E, 16 October 2008 | RL 904 x Kiran | Manranipur (U.P.) | 125–130 | 1610 (I) | 39.92 | BKD area of UP, MP and Rajasthan | Red violet, MR to wilt, PM blue flower, dark-brown-seeded, resistant to bud fly and rust |
| 52 | RLC 92 | 2008 2458 E, 16 October 2008 | Jeevan x LCK 9209 | Raipur (C.G.) | 111 | 1196 (I) | 37.70 | CG, Maharashtra, Karnataka, AP, and Odissa | Tinyblue flower, light-violet flower, brown- and medium-seeded, moderately resistant to powdery mildew, rust, and bud fly |
| 53 | SHIVAL (SLS-67) | 2010 733 E, 1 April 2010 | LCK 9610 x LMS 127 | Sagar (M.P.) | 108–110 | 1252 (R) | 40.16 | BKD area of UP, MP, and Rajasthan | White flower, light-brown-seeded, MR to powdery mildew and rust |
| 54 | JAWAHAR LINSEED-41 | 2011 2326 E, 10 October2011 | Kiran x Acc. No.443 | Hoshangabad (M.P.) | 115–120 | 1600 (I) | 40.00 | Area of MP state with limited facility | White flower, brown- and bold-seeded, resistant to rust, powdery mildew, and rust |
| 55 | BHAGSU (KL-215) | 2010 2137 E, 31 August 2010 | RL-50-3 x Surbhi | Palampur (H.P.) | 175–201 | 428(U) | 36.38 | Himachal Pradesh, J&K, Uttaranchal, Punjab, and Haryana | Blue flower, brown- and small-seeded, MR to rust |
| 56 | RUCHI (LCK 5021) | 2011 283 E, 7 February 2011 | Garima x LCK 88062 | Kanpur (U.P.) | 134 | 1366 (S), 1055 (F) | 39.84 | UP. (Excl. Bundelkhand), Bihar, WB and Assam | Blue flower, shining-brown-seeded, MR to rust, powdery mildew, and bud fly |
| 57 | JAWAHAR LINSEED SAGAR-73 (JLS 73) | 2011 632 E, 25 March 2011 | Padmini x Laxmi 27 | Sagar (M.P.) | 111–114 | 1090 (R) | 38.82 | Bundelkh and region of UP, MP, Rajasthan | Large blue flower, bold-seeded, high omega 3(58.20), high oil content (42%), light-brown-seeded, resistant to rust, powdery mildew, and bud fly |
| 58 | MAU AZAD ALSI-2 (LMS 149-4) | 2011 2326 E, 10 October 2011 | KL 178 x Hira | Manranipur (U.P.) | 105–110 | 815 (R) | 40.17 | CG, Maharashtra, Karnataka, AP, Odisha | Medium white flower, medium- and brown-seeded, resistant to rust |
| 59 | NDL 2004-05 | 2011 | Garima x RL 993 | Faizabaad (U.P.) | 125–130 | 1800(I) | 40.0 | Eastern Uttar Pradesh | Light flower, medium- and dark-brown-seeded, resistant to Alternaria blight, rust, and powdery mildew |
| 60 | NDL 2002 | 2011 | Garima x EC 44 | Faizabaad (U.P.) | 130–135 | 1600 (I & R) | 39.3 | Eastern Uttar Pradesh | Light-blue flower, medium- and dark-brown-seeded, resistant to rust and powdery mildew and tolerant to bud fly |
| 61 | Pratap Alsi 2 (RL 26016) | 2012 268 E, 28 January 2015 | RL 914 x NL 93 | Kota (Raj.) | 129–135 | 1957 (I) | 41.81 | Rajasthan | Blue flower, shining-brown- and bold-seeded, powdery mildew, wilt, and bud fly. Moderately resistant to Alternaria blight, powdery mildew, wilt, and bud fly |
| 62 | PKVNL-260 | 2015 112 E, 12 January 2015 | R 552 x RLC6 | Nagpur (M.S.) | 102–106 | 963 (R) | 37.67 | Maharashtra | Light flower, brown-seeded, moderately resistant to powdery mildew and bud fly |
| 63 | Kota Barani Alsi-3 (RL-292002) | 2015 | RL 903 x Ayogi | Kota (Raj.) | 119–124 | 1370 (R) | 38.73 | Rajasthan | Blue flower, shining dark-brown-seeded, resistant to rust and moderately resistant to altarnaria blight, powdery mildew, and bud fly |
| 64 | Kota Barani Alsi-4 (RL-10193) | 2015 | Triveni x RL-1011 | Kota (Raj.) | 120–126 | 1100 (R) | 40.37 | UP, MP, Rajasthan | White flower, shining dark-brown-seeded, moderately resistant to altarnaria blight and powdery mildew |
| 65 | ChattisgadhAlsi-1 (RLC-133) | 2015 | NL-14 x ACC-26 | Raipur (C.G.) | 110–113 | 844 (R) | 36.4 | CG | White flower, early, moderately resistant to bud fly, altarnaria blight, powdery mildew, and rust |
| 66 | Tiara (JRF-2) | 2015 1228 E | FT-889 x FT-895 | CRIJEF (W.B.) | 135–150 | 1290 | H.P., Uttarakhand. Sikkim, A.P., U.P, Kashmir, and W.B. | White flower, tall, suitable for fibre yield | |
| 67 | Arpita (OL 98-13-1) | 2016 3540 E, 22 November 2016 | RLC 29 x R 1871 | Keonjhar (Odisha) | 102–106 | 849 (R) | 35.67 | Odisha | Blue flower, light-brown-seeded, resistant to wilt and powdery mildew |
| 68 | UteraAlsi (RLC-143) | 2016 | LCK-88062 x T-397 | Raipur (C.G.) | 115–118 | 570 (U) | 34.1 | C.G., M.P., U.P., Jharkhand, Maharashtra, and Odisha | Suitable for Utera/para, blue flower, resistant to powdery mildew and rust, moderately resistant to budfly and altarnaria blight |
| 69 | Jawahar Linseed Sagar-79 (JLS-79) | 2016 3540 E, 22 November 2016 | Padmini x Laxmi-27 | Sagar (M.P.) | 1762(I) | 40.35 | Madhya Pradesh | Blue flower, suitable for irrigated conditions, moderately tolerant to powdery mildew, wilt, Alternaria blight, and bud fly | |
| 70 | PKV-NL-260 (NL-260) | 2016 3540 E, 2 November 2016 | R552 x RLC-6 | Nagpur (M.S.) | 102–106 | 940(R) | 35.56 | Maharashtra | Blue flower, suitable for rainfed conditions, moderately tolerant to powdery mildew, wilt, Alternaria blight, and bud fly |
| 71 | DIVYA(BAU-06-03) | 2016 3540 E, 22 November 2016 | BAU-1008 x Kiran | Kanke (J.K.) | 180 | 1538(I) | 39.80 | Punjab, Haryana, Himachal Pradesh, and Uttarakhand | White flower, resistant to rust and wilt |
| 72 | INDU (LCK 1108) | 2016 1007 E, 30 March 2017 | LMS1-2Kx LMS4-27 | Kanpur (U.P.) | 133–137 | 955 (I) | 40.00 | Whole of U.P. | Blue flower, bold- and brown-seeded, highly resistant to rust, resistant to alternaria blight and powdery mildew. |
| 73 | UMA (LCK-1101) | 2016 1007 E, 30 March 2017 | Polf 34 x Padmini | Kanpur (U.P.) | 120–123 | 868 (R) | 37.60 | Whole of U.P. | Blue flower, medium- and light-brown-seeded, moderately resistant to rust, wilt, and Alternaria blight |
| 74 | VARSHA ALSI (RLC-148) | 2017 | SIKO 10 x Kiran | Raipur (C.G.) | 110–114 | 1033 (R) | 35.70 | CG, MP, UP, JH, M.S., and Odisha | Blue flower, suitable for rainfed situation, moderately resistant to budfly, moderately susceptible to wilt and powdery mildew |
| 75. | PRIYAM (BAU-2012-1) | 2017 2805 E, 25 August 2017 | BAU-2k-14 x Garima | Ranchi (Jharkhand) | 173 | 1151(R) | 37.15 | Punjab, Haryana, H.P., and Uttarakhand. | White flower, dark brown seed |
| 76. | JAWAHAR LINSEED SAGAR-95 (JLS-95) | 2018 1379 E, 27 March 2018 | JLS-27x GS-281 | Sagar (M.P.) | 133 | 1240(R) | 39.40 | Bundelkhand part of Uttar Pradesh, Raj. Madhya Pradesh, Karnataka, Odisha, Maharashtra, Chhattisgarh | White flower, bold-seeded, high omega 3 (53.50), high oil content (40%), suitable for rainfed and mechanical harvesting, resistant to rust, moderately resistant to wilt and powdery mildew, alternaria blight, and budfly |
| 77 | JAWAHAR LINSEED SAGAR-66 (JLS 66) | 2018 399 E, 24 January 2018 | Selection from Accn. 2511 | Sagar (MP) | 114 | 1200 (R) | 40.50 | Madhya Pradesh | Blue flower, suitable for rainfed conditions, bold-seeded, high omega 3 (55.96), high oil content (42%), resistant to rust, moderately resistant to wilt, powdery mildew, Alternaria blight, and budfly |
| 78 | SABOUR TISI-1 (BAUP-101) | 2018 1379 E, 27 March 2018 | Selection from Kishanganj local | Patna (Bihar) | 124 | 453(U) | 32.68 | U.P., excluding Bundelkhand, Bihar, Jharkhand, W.B, and Assam | Blue flower, suitable for Utera, moderately resistant to rust and powdery mildew |
| 80 | HIM PALAM ALSI-2 (KL-263) | 2018 1379 E, 27 March 2018 | KL-223 x KL-224 | Palampur | 130 | 1633(I) | 35.58 | H.P., Haryana, and Jammu and Kashmir | Blue flower, suitable for rainfed conditions, moderately resistant to rust and powdery mildew |
| 81 | RAJAN | 2018 1498 E, 1 April 2019 | Gaurav x GS 234 | Kanpur | 130–135 | 1528(S) 1093(F) | 38.01 | Whole of U.P. | Blue flower, brown- and medium-seeded, highly resistant to rust, moderately resistant to alternaria blight |
| 82 | SURYA (LCK 1404) | 2018 3220 E, 5 September 2019 | Shubhra x H-25 | Kanpur | 155–160 | 1431(I) | 35.49 | H.P., Punjab, Haryana, and J&K | Blue flower, brown- and medium-seeded, highly resistant to rust, moderately resistant to wilt |
| 83 | UTERA ALSI-2 (RLC-153) | 2019 1326 E, 2 April 2019 | LCK88062x EC-1424 | Raipur | 118–120 | 519(U) | 34.8 | Assam, Bihar, Chhattisgarh, Jharkhand, Orrisa, M.P., M.H. and U.P. | Suitable for Utera conditions, moderately resistant to budfly |
| 84 | LSL-93 | 2019 3220 E, 5 September 2019 | Selection from SLS-93 | Latur | 106 | 846(R) | 37.07 | Maharashtra | White flower, suitable for rainfed conditions, resistant to rust, moderately resistant to wilt, powdery mildew, alternaria blight, and bud fly |
| 85 | TL 99 | 2019 3220 E, 5 September 2019 | RLC 6 X Solin | BARC Trombey | 120 | 1274(I) | 36.56 | U.P., Bihar, Jharkhand, W.B., Assam, Nagaland | Low linolenic acid content (2–3%), edible oil purpose, resistant to wilt and rust, moderately resistant to budfly |
| 86 | JAWAHAR LINSEED 165 | 2019 3220 E, 5 September 2019 | Meera x Polf 17 | Hoshangabad (M.P.) | 125 | 1392(I) | 33.84 | H.P, Punjab, and Jammu and Kashmir | Blue flower, suitable for irrigated conditions, resistant to rust and moderately resistant to powdery mildew, wilt, and bud fly |
| 87 | KOTA BARANI ALSI 5 (RL 29005) | 2020 99 E, 9 November 2020 | PKDL 41 x Meera | Kota (Raj.) | 115–120 | 1150(R) | 36.00 | Rajasthan | Moderately resistant to wilt, alternaria blight, and bud fly |
| 88 | KOTABARANI ALSI 6 (RL 15584) | 2020 3482 E, 7 October 2020 | RL 101 x RLU 6 | Kota (Raj.) | 115–120 | 1200(R) | 32.00 | H.P, Punjab, and Jammu and Kashmir | Moderately resistant to wilt, powdery mildew, rust, alternaria blight, and bud fly |
| 89 | KOTA ALSI 6 (RL 13165) | 2020 3482 E, 7 October 2020 | RLC 25127 x RL 26016 | Kota (Raj.) | 1200(R) | 32.00 | UP, Jharkhand, Bihar, W.B., and Assam | Moderately resistant to wilt and powdery mildew, rust, alternaria blight, and bud fly | |
| 90 | RLC 161 (SUVEE) | 2020 3482 E, 7 October 2020 | Ayogi x GS 234 | Raipur (C.G.) | 1262(R) | 32.10 | H.P, Punjab, and Jammu | Moderately resistant to alternaria blight and budfly | |
| 92 | RLC 164 | 2020 500 E, 29 January 2021 | Polf 22 x JRF 5 | Raipur (C.G.) | 1161(R) | 32.60 | H.P, Punjab and Jammu | Resistant to rust, moderately resistant to alternaria blight and bud fly | |
| 93 | RLC 167 | 2020 500 E, 29 January 2021 | T 397 x Polf 22 | Raipur (C.G.) | 1297(R) | 34.20 | H.P, Punjab and Jammu | Moderately resistant to wilt, powdery mildew, and budfly | |
| 93 | APARNA (LCK1611) | 2020 500 E, 29 January 2021 | BAU610 x Parvati | Kanpur (U.P.) | 1342(I) | 33.00 | H.P, Punjab, and Jammu | Resistant to Alternaria blight, moderately resistant to wilt and powdery mildew | |
| 95 | BUAT ALSI-4 (LMS 15-31) | 2020 500 E, 29 January 2021 | P 18 x Accn. No. 2299 | Mauranipur (U.P.) | 1271(I) | 31.80 | Bundelkhand part of U.P., M.P., MH, K.N., Orisha, C.G. | Moderately resistant to powdery mildew and budfly | |
| 96 | BIRSA TISI 1 (BAU 15-03) | 2021 8 E, 24 December 2021 | (Shekhar x RLC 80) x Garima | Ranchi, Jharkhand | 128–130 | 1141(R) | 41.80 | Jharkhand | Resistant to wilt and powdery mildew, moderately resistant to alternaria blight and bud fly |
| 97 | PRIYAM (BAU 12-1) | 2021 8 E, 24 December 2021 | BAU 2K-1 x Garima | Ranchi, Jharkhand | 128–130 | 1253(R) | 40.70 | H.P., Punjab, and Jammu and Jharkhand | Resistant to rust, wilt, alternaria blight, and powdery mildew, moderately resistant to bud fly |
| 98 | DIVYA (BAU 06-3) | 2021 8 E, 24 December 2021 | BAU 1008 x Kiran | Ranchi, Jharkhand | 128–130 | 1538(I) | 39.80 | H.P., Punjab, Jammu, and Jharkhand | Resistant to powdery mildew and rust, moderately resistant to alternaria blight and wilt |
| 99 | SHUATS ALSI 2 (SHA-2) | 2021 8 E, 24 December 2021 | LCK 8879 x CI 1399 | Naini (U.P.) | 123–125 | 1110(R) | 37.40 | Uttar Pradesh | Resistant to powdery mildew and wilt, moderately resistant to rust |
| 100 | SABOUR TISI 2 (BRLS-101) | 2021 8 E, 24 December 2021 | SLS 72 x Shekhar | Sabour, Bihar | 118 | 547(U) | 38.20 | U.P., Bihar, Jharkhand, W.B., Assam, M.P., M.H., C.G., Nagaland, Raj., Odisha, Karnataka | Resistant to rust and wilt, moderately resistant to Alternaria blight |
| 101 | SABOUR TISI 3 (BRLS-107-1) | 2021 8 E, 24 December 2021 | LCK 7035 x Shekhar | Sabour, Bihar | 122 | 1883(I) | 37.80 | U.P., Bihar, Jharkhand, W.B., Assam, M.P., Nagaland, Raj., M.H., C.G., Odisha, Karnataka | Resistant to powdery mildew and wilt, moderately resistant to rust |
| 102 | (RLC 171) | 2022 3254 E, 27 July 2022 | Polf 22 x JRF 22 | Raipur (C.G.) | 132 | 1073(R) | 34.53 | Assam, Bihar, C.G., H.P., Jammu, JK, K.N., M.P., M.H., Nagaland, Orrisa, Punjab, Raj., and U.P. | Blue flower, moderately resistant to rust and bud fly |
| 103 | SABOUR TISI 4 (BRLS 121) | 2023 1056 E, 6 March 2023 | (NL 260 x Shekhar) x PKDL 71 | Sabour, Bihar | 120 | 1000 | 34.00 | UP., Bihar, Jharkhand, W.B., Assam and Nagaland | Blue flower, moderately resistant to rust and bud fly |
| 104 | JAWAHAR LINSEED SAGAR 122 (JLS 122) | 2023 1056 E, 6 March 2023 | JLS 73 x JLS 66 | Sagar M.P. | 116 | 964(R) | 41.00 | Madhya Pradesh | Blue flower, semi-dehiscent capsule suitable for mechanical harvesting, bold-seeded (8 g), high omega 3 (54.20), resistant to rust, moderately resistant to powdery mildew, Alternaria blight, and bud fly |
| 105 | AZAD PRAGYA (LCK 1516) | 1056 E, 6 March 2023 | Shubhra x Shikha | Kanpur (U.P.) | 128 | 1345(I) | 35.00 | Uttar Pradesh | Blue flower, resistant to rust and powdery mildew, moderately resistant to wilt and bud fly |
| 106 | SHUATS ALSI 5 (SHA-5) | 2023 1056 E, 6 March 2023 | RL 10205 x IC 564620 | Naini (U.P.) | 125–128 | 1250(I) | 35.80 | Uttar Pradesh | Blue flower, resistant to rust and powdery mildew, moderately resistant to wilt and bud fly |
| Trait | Candidate Gene | Type | QTN/QTL | Function | Reference |
|---|---|---|---|---|---|
| Root surface area stability | Lus10034840 | Calcium transporting ATPase9, Plasma membrane type | Lu5-4,774,423 | Pollen development | [82] |
| Lus10016017 | Catalase isozyme 3 | Lu6-15,939,492 | Response to water deprivation, promotion of drought stress tolerance | [81] | |
| Total root length stability | Lus10039723 | IAA amidosynthetaseGH3.6 | Lus-20,209,630 | Response to stress and root development | [82] |
| Lus10039747 | Diacylglycerol kinase 5 | Cold and drought stress tolerance | [81] | ||
| Lus10021019 | Allene oxide synthase | Lu6-19,733,117 | Stomatal closure and drought tolerance | [180] | |
| Lus10020997 | S/Tproteinkinase SRK2E | Response to water deprivation and regulation of stomatal closure | [81] | ||
| Stress tolerance index | Lus10019811 | Probable cinnamyl alcohol dehydrogenase1 | Lu6-17,376,408 | Drought tolerance | [82] |
| Lus10014978 | aquaporinPIP2-2 | Lu14-23,517,150 | Drought tolerance | [180] | |
| Lus10019781 | L-Ascorbate peroxidase | Enhanced salt tolerance, drought tolerance, and cold tolerance | [181] | ||
| Total root volume stability | Lus10016017 | Catalase Isozyme C | Lu6-15,961,789 | Promotion of drought stress tolerance and response to water deprivation | [81] |
| Bundle weight under drought stress | Lus10040333 | 3-Ketoacyl-CoA synthase 19 | Chr9:4203006 | Drought tolerance and biomass-related traits | [217] |
| Canopy temperature under drought stress | Lus10013240 | Xyloglucan endotransglucosylase/hydrolase protein 27 | Chr2:23123754 | Leaf size and veins and drought susceptibility index | [82] |
| Lus10019365 | Stromal cell-derived factor 2 | Chr3:9279281 | Heat stress and better stress tolerance indices | [217] | |
| Lus10024816 | Cytochrome | Chr9:18937269 | Moisture stress tolerance | [81] | |
| Seeds per boll | Lus10021766 | Mitogen-activated protein kinase kinasekinase 5 | Chr9:15446958 | Drought susceptibility index | [81] |
| Grain yield | Lus10042229 | CBL-interacting protein kinase | Chr11:3972867 | Drought | [180] |
| Lus10042231 | Translocon chloroplast 110 | Heat shock and drought susceptibility index | [181] | ||
| Thousand-seed weight under drought stress | Lus10029127 | Kelchrepeat F-box | Chr1:7029139 | Ovule development and stress tolerance index | [82] |
| Lus10029115 | Ribosomal pentatricopeptide repeat protein 4 | Seed development and stress tolerance | [82] | ||
| Lus10030137 | Nuclear factor Y subunit A1 | Chr12:10910146 | Seed development and drought stress tolerance | [81] | |
| Lus10030142 | Translocated promoter region, nuclear pore anchor | Flowering, auxin signalling | [81] | ||
| Plant height under drought stress | Lus10029690/1 | Cellulose synthase interactive | Chr5:1375386 | Flax fibre and stress tolerance index | [82] |
| Yield | Lus10031398 | Inositol Monophosphatase 1 | Chr12:20557728 | Drought tolerance | [81] |
| Trait | QTN/QTL | Candidate Gene | Type | Function | References |
|---|---|---|---|---|---|
| Pasmo resistance | QTL 45 | Lus10031043 | LRR receptor kinase | Bacterial pathogen-associated molecular pattern | [211] |
| (PAMP) receptor | |||||
| Lus10031058 | Elongation Factor | Effector-trigger immunity | [135] | ||
| Fusarium wilt resistance | afB13 | -- | -- | [218] | |
| Powdery mildew resistance | QPM-crc-LG1 | -- | -- | [219] | |
| - | Pm1 | [135] | |||
| Lu4-12,432,479 | Lus10036891 | RGA (WRKY transcription factor) | [212] |
| Trait | Tissue | DEGs/DEUs | Key Points | Platform/Tool | References |
|---|---|---|---|---|---|
| Response to salt stress | Flaxseeds | 77,361,566 in neutral-salt stress, alkaline-salt stress, and alkaline stress | Photosynthesis-, pathogen-, and wax-related genes | Illumina HiSeq 2000 | [238] |
| Flaxseeds | 3374 upregulated and 18,040 downregulated | Provide high-impact gene expression profile | Illumina high-throughput sequencing | [232] | |
| Response to drought stress | Seeds, roots, and shoots | 183:72 upregulated and 111 downregulated | Maintain homeostasis and growth | Combi matrix 90 K array | [72] |
| Response to drought-sensitive and tolerant varieties | Flax leaves | In cv.Z1413,245 upregulated and 4167 downregulated | DNA repair from ROS damage and proline biosynthesis | PacBIo ISO-seq | [239] |
| Response to osmotic stress under PEG-induced condition | Flaxseeds | 3922:1487 upregulated and 2432 downregulated | Signal transduction and biochemical pathway | Illumina platform | [240] |
| Response to aluminium stress and high soil acidity | Flax seedlings | - | Compartmentalisation of Ca2+ in vacuoles | Illumina Platform | [230] |
| Response against flax rust (Melanosporalini) | Leaf tissues | - | Gene encoding PR protein and hydrolysis and uptake of nutrients | Illumina genome analyzer II | [241] |
| Response to Fusarium wilt | Roots, stem | Transduction and reception of pathogen signals | Illumina HiSeq 2000 | [242] |
| Character | Upregulated/Downregulated Genes | References |
|---|---|---|
| Salinity and alkalinity | HSP70 NAC family members, ABA, WRKY, PrxR | [238] |
| LUS-miRNAs and miRNA-targeted genes | [281] | |
| UBE2 gene, mitochondrial termination factor family protein and auxin signalling F-box, Myb domain protein | [282] | |
| Drought | Lipid transferase protein, cell wall synthesis genes, RUBISCO, PS1, r2r3-MYB transcription factor, EF-tu, LEA5, cytochrome P450 family proteins, brassinosteroid insensitive I-associated receptor kinase1, and AP2/ERF domain-containing transcription factor | [72] |
| NAC domain protein | [112] | |
| Heat | HSF | [283] |
| Heat shock factors (HSFs) | [111] | |
| miRNAs and phasiRNAs | [114] | |
| GUS activity in sepals, petals, and pistils | [110] | |
| Nutrient stress | WRKY, ING1 family JAZ and HARB11 | [284] |
| Aluminium stress | miR319, UDP-glycosyl-transferasemiR390, glutathione-S-transferase and miR393 | [234] |
| Rust | Avrs and CWDEs | [240] |
| Fusarium wilt | PAL, PCBER, SRG1, UGT73C3, AAA-ATPase ASD, mitochondrial (AATPA), glucan endo-1,3-β-glucosidase, MYB transcription factors, ERD dehydrins, and auxin-responsive protein SAUR, WKY3, WRKY70, WRKY75, MYB113, and MYB108 | [234,285] |
| Fusarium culmorum | PAL, CCR, CAD, UGT, and TD | [233] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paliwal, S.; Tripathi, M.K.; Tiwari, S.; Tripathi, N.; Payasi, D.K.; Tiwari, P.N.; Singh, K.; Yadav, R.K.; Asati, R.; Chauhan, S. Molecular Advances to Combat Different Biotic and Abiotic Stresses in Linseed (Linum usitatissimum L.): A Comprehensive Review. Genes 2023, 14, 1461. https://doi.org/10.3390/genes14071461
Paliwal S, Tripathi MK, Tiwari S, Tripathi N, Payasi DK, Tiwari PN, Singh K, Yadav RK, Asati R, Chauhan S. Molecular Advances to Combat Different Biotic and Abiotic Stresses in Linseed (Linum usitatissimum L.): A Comprehensive Review. Genes. 2023; 14(7):1461. https://doi.org/10.3390/genes14071461
Chicago/Turabian StylePaliwal, Shruti, Manoj Kumar Tripathi, Sushma Tiwari, Niraj Tripathi, Devendra K. Payasi, Prakash N. Tiwari, Kirti Singh, Rakesh Kumar Yadav, Ruchi Asati, and Shailja Chauhan. 2023. "Molecular Advances to Combat Different Biotic and Abiotic Stresses in Linseed (Linum usitatissimum L.): A Comprehensive Review" Genes 14, no. 7: 1461. https://doi.org/10.3390/genes14071461
APA StylePaliwal, S., Tripathi, M. K., Tiwari, S., Tripathi, N., Payasi, D. K., Tiwari, P. N., Singh, K., Yadav, R. K., Asati, R., & Chauhan, S. (2023). Molecular Advances to Combat Different Biotic and Abiotic Stresses in Linseed (Linum usitatissimum L.): A Comprehensive Review. Genes, 14(7), 1461. https://doi.org/10.3390/genes14071461






