Multiple Chromosome Fissions, Including That of the X Chromosome, in Aulacocyclus tricuspis Kaup (Coleoptera, Passalidae) from New Caledonia: Characterization of a Rare but Recurrent Pathway of Chromosome Evolution in Animals
Abstract
:1. Introduction
2. Material and Methods
3. Results
3.1. Spermatocytes I at Pachynema
3.2. Spermatocytes II
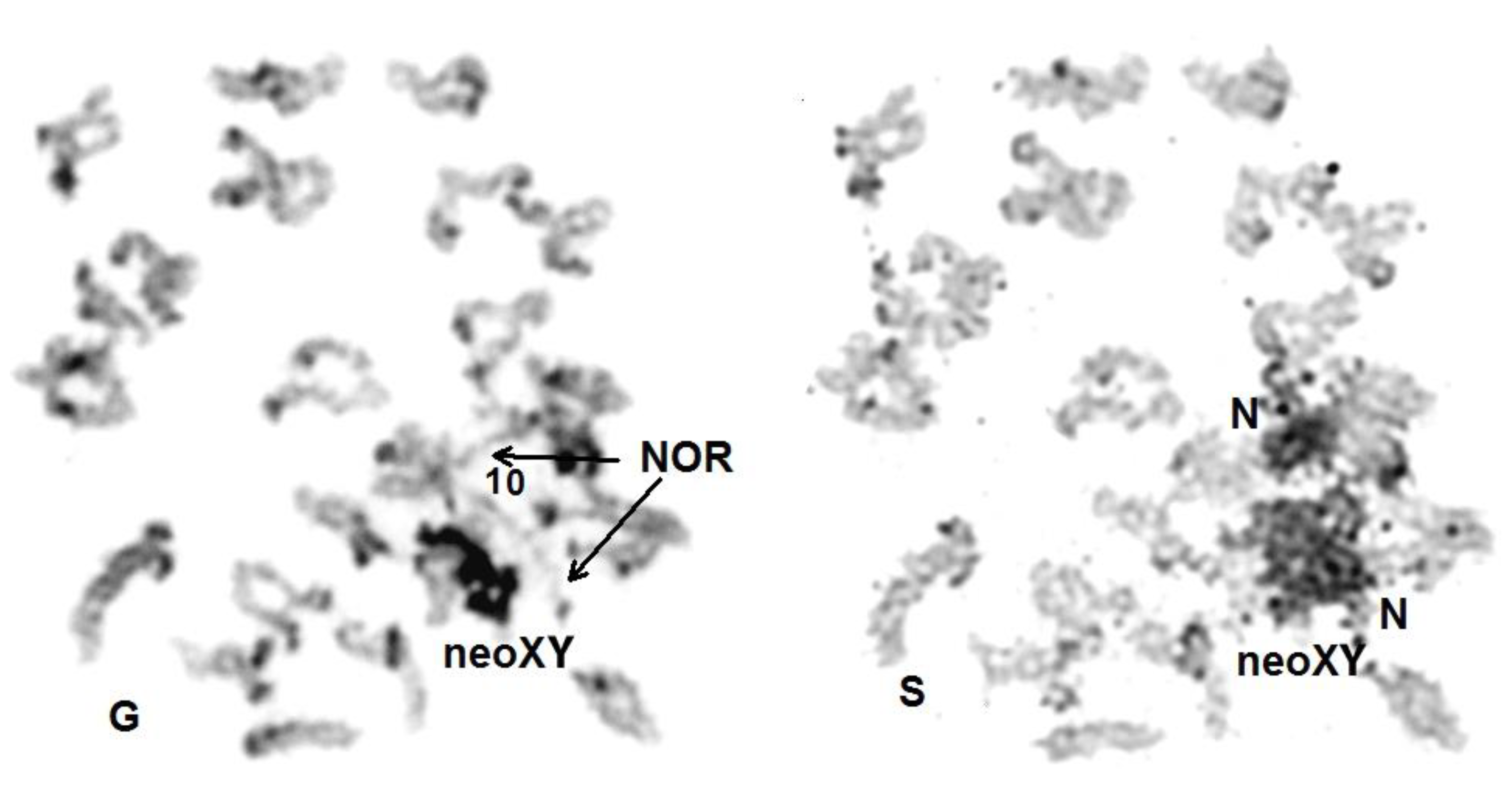
3.3. Diakinesis
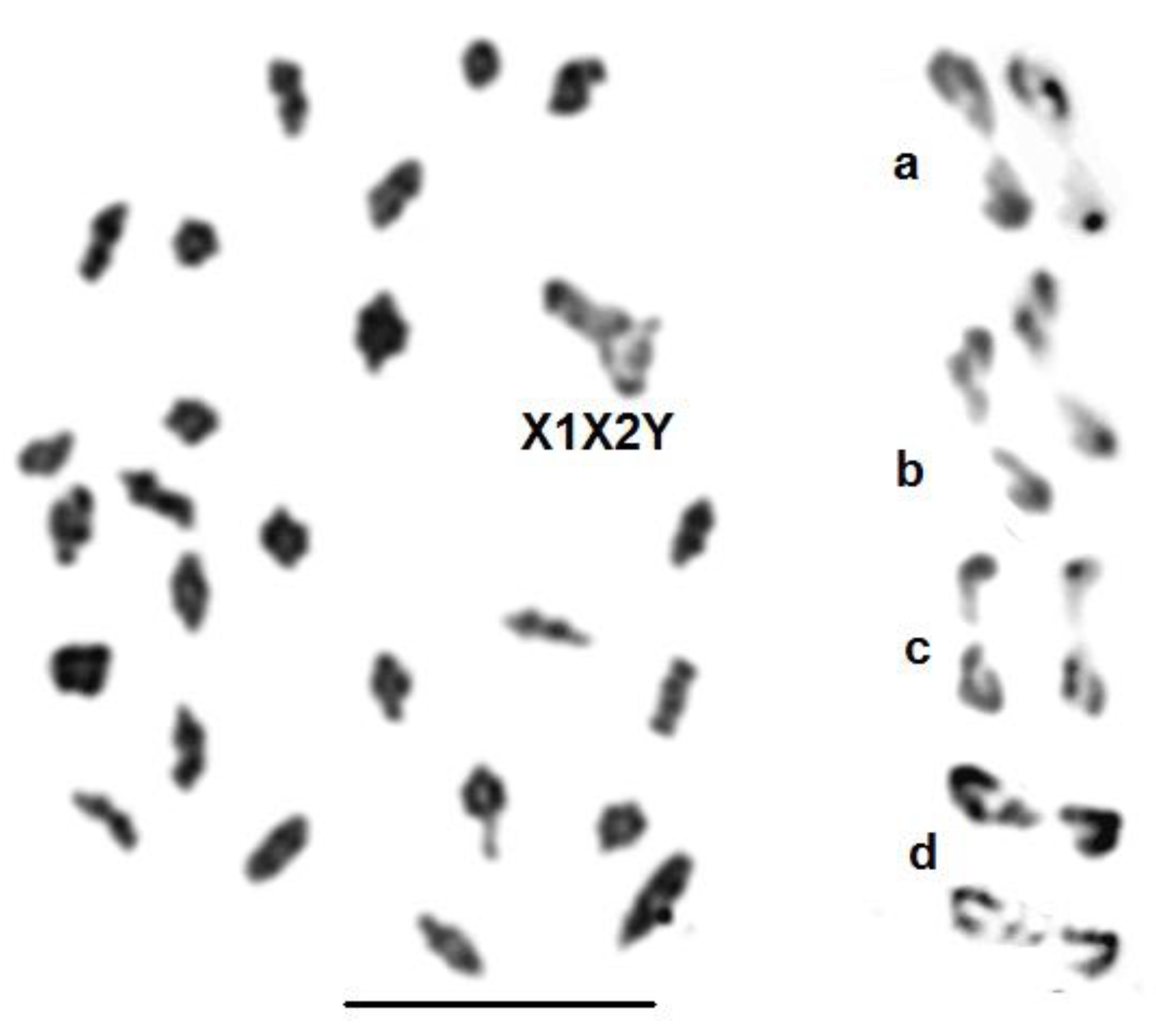
3.4. Metaphase I
4. Discussion
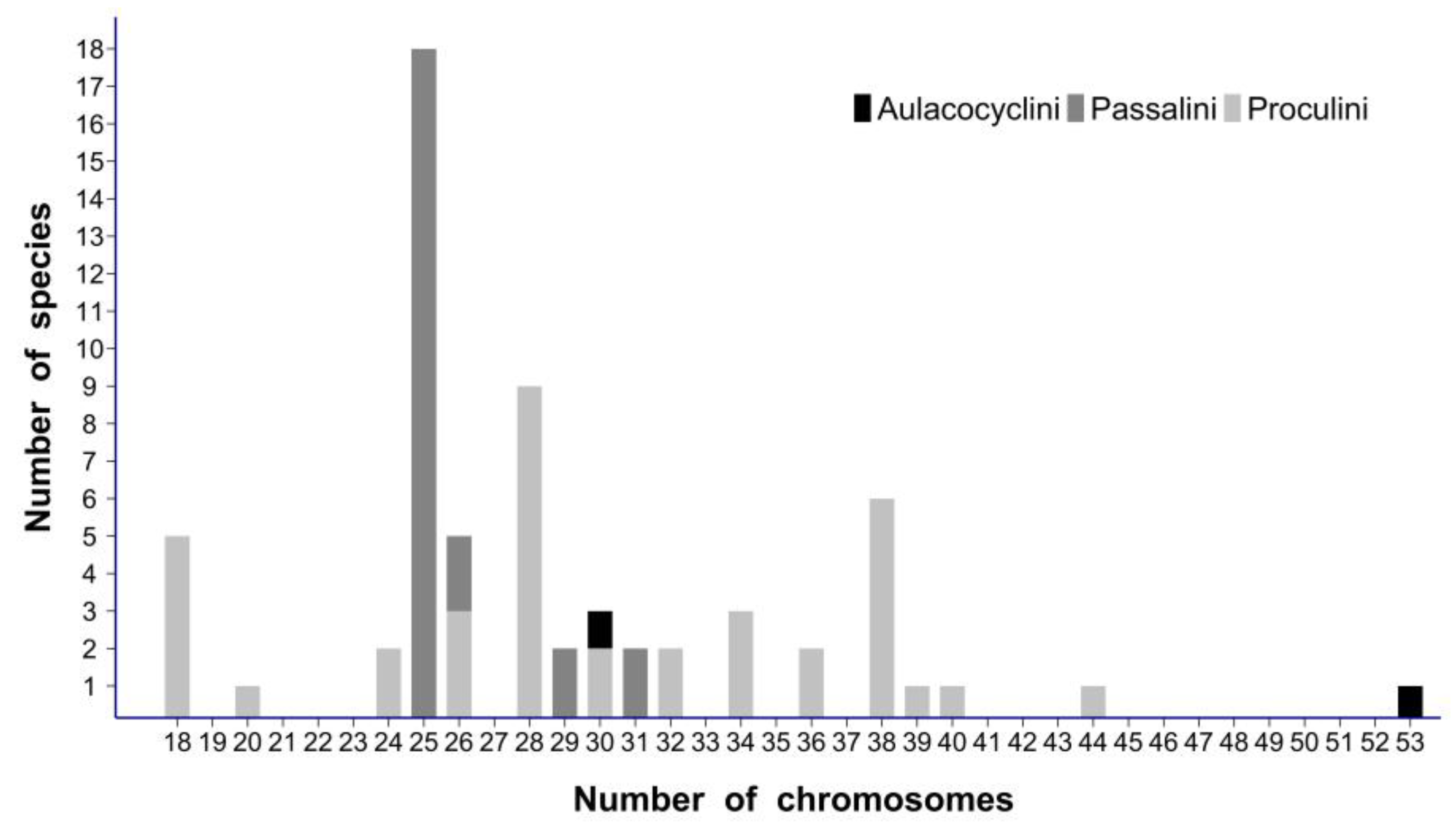
4.1. Formation of Multiple Acrocentric Autosomes by Centromere Fission of Ancestral Metacentric Chromosomes: A Mechanism of Recent Karyotype Evolution Characteristic of New Caledonian Passalidae?
4.2. X Chromosome Fission in New Caledonian Passalidae: An Exceptional Event?
4.3. X Chromosome Fission Associated with Multiple Centromere Fissions of Autosomes: A Rare, but Characteristic Pattern of Chromosome Evolution in Beetles
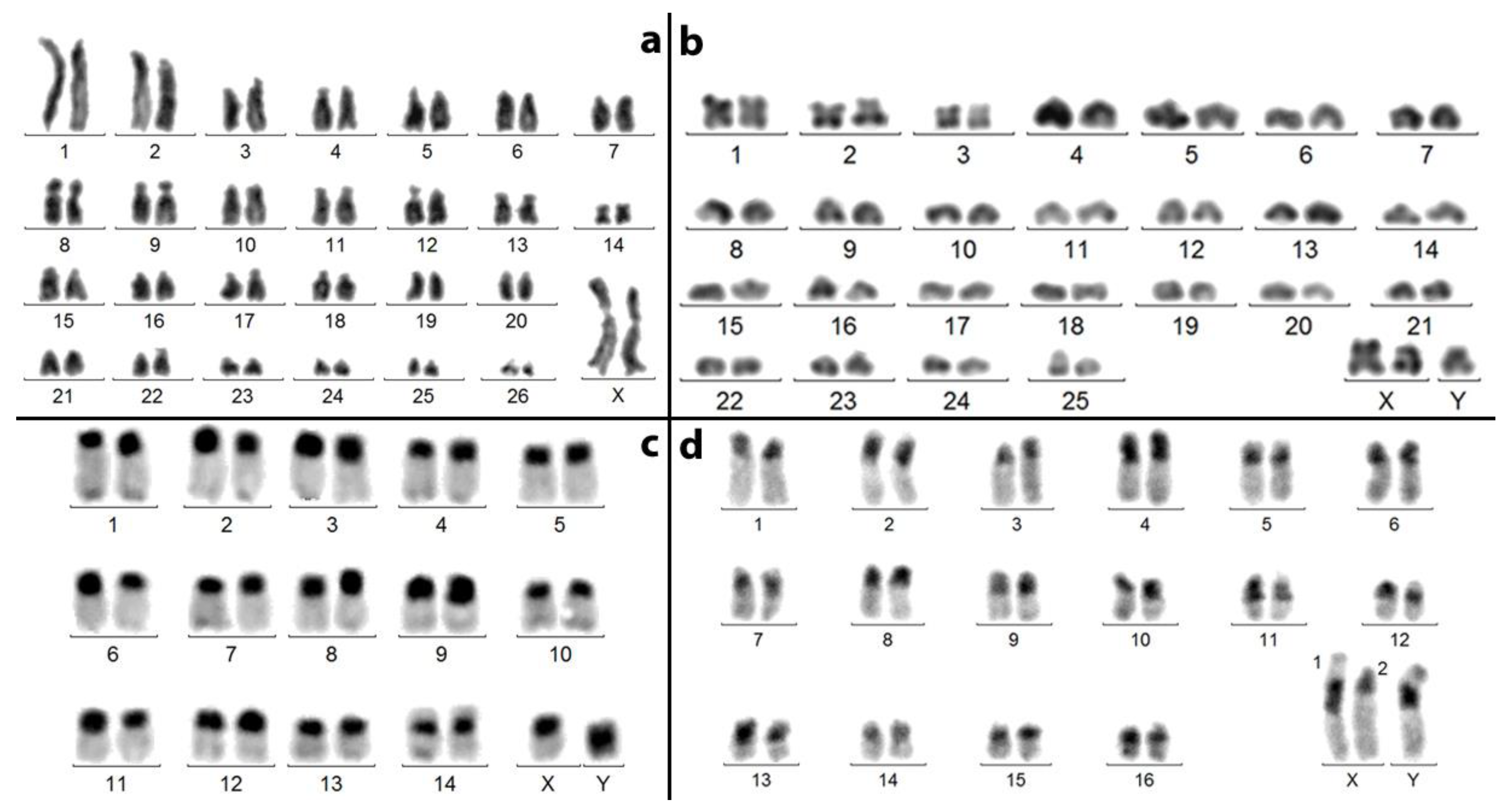
- The 2 n = 53/54 karyotype of V. xatarti (Vesperidae) almost exclusively composed of acrocentric chromosomes (Figure 6a) [25] looks very similar to that of A. tricuspis (Figure 6b). The sex chromosome composition remains uncertain (only somatic cells were available) and the formula is either 53,X/54,XX or 53,neoX/54neoX neoX. The large phylogenetic distance between these two species indicates that this chromosome similarity is a convergence.
- Amongst Chrysomelidae, a similar evolution probably occurred in three subfamilies. (A) In several species of the neotropical genus Botanochara (Cassidinae), increases in chromosome numbers from 2 n = 27 to 51 parallel the accumulation of acrocentric chromosomes and are associated with multiple sex chromosomes [26,35]. (B) In genus Aulacophora (Galerucinae) from India, increases in chromosome numbers from 2 n = 30 to 59 also parallel the formation of multiple sex chromosomes [36]. In genus Isotes, the 2 n male formula increases from 23,X in Isotes multipunctata to 37,X in Isotes tetraspilota [13]. (C) In genera Pachybrachis and Cryptocephalus (Cryptocephalinae), most of the species studied share either a 30,XY (16/35) or a 16,XY (7/35) male karyotype [37]. Our unpublished studies indicate that in the 30,XY karyotype of Cryptocephalus globicollis Suffrian, 1847 (Figure 6c, http://insect-cytogenetics.fr/ accessed on 17 January 2023), all autosomes but pair 14 are acrocentric, which is compatible with a derivation from the 16,XY karyotypes by repeated centric fissions of non-acrocentric chromosomes.
- Amongst Tenebrionidae, some species of genus Blaps, such as Blaps mucronata Latreille, 1804 (Figure 6d, http://insect-cytogenetics.fr/ accessed on 17 January 2023), display a 2 n = 35,neoX1neoX2neoY male karyotype, in which all 32 autosomes are acrocentric (Figure 6d), whereas the number of autosomes is limited to 16 and 18 (non-acrocentric?) in other species, such as Blaps judaeorum and Blaps cribosa [38]. In this way, the same evolution by multiple centric fissions of autosomes may have occurred. There is also an increase in the number of sex chromosomes in this genus [13], but this is associated with an X-autosome translocation [33]. The 36,neoX1neoX2neoX3neoY/38,neoX1neoX1neoX2neoX2neoX3neoX3 male and female karyotypes of Gnaptor spinnimanus Pallas, 1781 [33] probably originated from the same process.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- White, M.J.D. Animal Cytology and Evolution, 3rd ed.; Cambridge University Press: Cambridge, UK, 1977; 468p. [Google Scholar]
- Siqueira Castro, M.; Recco-Pimentel, S.M.; Tadeu Rocha, G. Karyotypic characterization of Ramphastidae (Piciformes, Aves). Genet. Mol. Biol. 2002, 25, 147–150. [Google Scholar] [CrossRef]
- Dutrillaux, B. Chromosomal evolution in primates: Tentative phylogeny from Microcebus murinus (Prosimian) to man. Hum. Genet. 1979, 48, 251–314. [Google Scholar] [CrossRef]
- Dutrillaux, B.; Couturier, J.; Muleris, M.; Lombard, M.; Chauvier, G. Chromosomal phylogeny of forty-two species or subspecies of cercopithecoids (Primates Catarrhini). Ann. Génét. 1982, 25, 96–109. [Google Scholar] [PubMed]
- Takagi, N.; Sasaki, M. A Phylogenetic study of bird karyotypes. Chromosoma 1974, 46, 91–120. [Google Scholar] [CrossRef]
- Burt, D.W. Origin and evolution of avian microchromosomes. Cytogenet. Gen. Res. 2002, 96, 97–112. [Google Scholar] [CrossRef]
- Ferguson-Smith, M.A.; Trifonov, V. Mammalian karyotype evolution. Nat. Rev. Genet. 2007, 8, 950–962. [Google Scholar] [CrossRef]
- Rochi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity 2012, 108, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Richard, F.; Dutrillaux, B. Origin of human chromosome 21 and its Consequences: A 50-million-year-old story. Chromosome Res. 1998, 6, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Boucher, S. Évolution et phylogénie des Coléoptères Passalidae. Les taxons du groupe famille. La tribu néotropicale des Proculini et son complexe Veturius. Ann. Soc. Entomol. Fr. 2005, 41, 239–604. [Google Scholar] [CrossRef] [Green Version]
- Boucher, S. Les Passalides de l’archipel du Vanuatu. Remarques faunistiques et biogéographiques; comparaison avec la Nouvelle-Calédonie. Ann. Soc. Entomol. Fr. 1991, 27, 361–374. [Google Scholar] [CrossRef]
- Serrano, J.; Galián, J.; Reyes-Castillo, P. Karyotype evolution and phylogeny of Mexican Passalidae (Coleoptera: Polyphaga: Scarabaeoidea). J. Zool. Syst. Evol. Res. 1998, 36, 159–167. [Google Scholar] [CrossRef]
- Smith, S.G.; Virkki, N. Animal Cytogenetics, Vol. 3: Insecta 5: Coleoptera; Gebrüder Borstraeger: Berlin, Germany, 1978; pp. 1–234. [Google Scholar]
- Petitpierre, E. Cytogenetics, Cytotaxonomy and Genetics of Chrysomelidae. In Biology of Chrysomelidae; Jolivet, P., Petitpierre, E., Hsiao, T.H., Eds.; Series Entomologica 42; Kluwer Acad. Publ.: Dordrecht, The Netherlands, 1988; pp. 131–159. [Google Scholar]
- Dutrillaux, A.-M.; Dutrillaux, B. Chromosome analysis of 82 species of Scarabaeoidea (Coleoptera), with special focus on NOR localization. Cytogenet. Genome Res. 2012, 136, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Dutrillaux, A.-M.; Dutrillaux, B. Chromosome study of Lucanidae (Coleoptera), with emphasis on recurrent X-autosome rearrangements. Trends Entomol. 2022, 18, 73–82. [Google Scholar]
- Virkki, N.; Reyes-Castillo, P. Cytotaxonomy of Passalidae (Coleoptera). An. Esc. Nac. Cienc. Biol. Mex. 1972, 19, 49–83. [Google Scholar]
- Mesa, A.; Martins, V. Karyotypical diversity among Brazilian species of the genus Passalus (Coleoptera, Scarabaeoidea, Passalidae). Rev. Bras. Gen. 1986, 9, 735–740. [Google Scholar]
- Boucher, S.; Dutrillaux, B.; Dutrillaux, A.-M. Parthenogenetic reproduction demonstrated in the diploid Spasalus puncticollis (Lepeletier & Serville 1825), n. stat., from the Antilles (Coleoptera, Scarabaeoidea, Passalidae). Comptes Rendus Biol. 2015, 338, 738–744. [Google Scholar] [CrossRef]
- Shaffer, E.L. Mitochondria and other cytoplasmic structures in the spermatogenesis of Passalus cornutus. Biol. Bull. 1917, 32, 407–435. [Google Scholar] [CrossRef]
- Dutrillaux, A.-M.; Xie, H.; Dutrillaux, B. Nucleolus and chromosome relationships at pachynema in four Scarabaeoidea (Coleoptera) species with various combinations of NOR and sex chromosomes. Chromosome Res. 2007, 15, 417–427. [Google Scholar] [CrossRef]
- Mesa, A.; Ferreira, A.; Martins, V. The chromosomes of an Australian Passalid, Aulacocyclus edentulus MacL. (Coleoptera, Passalidae, Aulacocyclinae). J. Aust. Ent. Soc. 1978, 17, 385–388. [Google Scholar] [CrossRef]
- Dutrillaux, A.-M.; Sigwalt, D.; Dutrillaux, B. (Ovo-) viviparity in the darkling beetle, Alegoria castelnaui (Tenebrioninae: Ulomini), from Guadeloupe. Eur. J. Entomol. 2010, 107, 481–485. [Google Scholar] [CrossRef] [Green Version]
- ISCN (1978). An International System for human Cytogenetic Nomenclature. Cytogenet. Cell Genet. 1978, 21, 309–404. [Google Scholar]
- Dutrillaux, A.-M.; Moulin, S.; Dutrillaux, B. Présence d’un caryotype très original à 53-54 chromosomes chez Vesperus xatarti Mulsant 1839 (Coleoptera: Cerambycidae; Vesperinae). Ann. Soc. Entomol. Fr. 2006, 43, 81–86. [Google Scholar] [CrossRef] [Green Version]
- Julio, D.M.; Fernandez, F.R.; Costa, C.; Almeida, M.C.; Cella, D.M. Mechanisms of karyotype differentiation in Cassidinae sensu lato (Coleoptera, Polyphaga, Chrysomelidae) based on seven species of the Brazilian fauna and overview of the cytogenetic data. Micron 2010, 41, 26–38. [Google Scholar] [CrossRef]
- Dutrillaux, A.-M.; Dutrillaux, B. Y-chromosome disomy and trisomy in Scarabaeid and Cerambycid beetles. Cytogenet. Genome Res. 2011, 132, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Carrel, L.; Willard, H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 2005, 434, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Loda, A.; Heard, E. Xist RNA in action: Past, present, and future. PLoS Genet. 2019, 15, e1008333. [Google Scholar] [CrossRef] [Green Version]
- Couturier, J.; Dutrillaux, B. Replication studies and demonstration of position effect in rearrangements involving the Human X chromosome. In Cytogenetics of the Mammalian X Chromosome, Part A, Basic Mechanisms of X Chromosome Behavior; Sandberg, A.A., Ed.; Alan R Liss Inc.: New York, NY, USA, 1983; pp. 375–403. [Google Scholar]
- Gupta, V.; Parisi, M.; Sturgill, D.; Nuttall, R.; Doctolero, M.; Dudko, O.K.; Malley, J.D.; Eastman, P.S.; Oliver, B. Global analysis of X chromosome dosage compensation. J. Biol. 2006, 5, 3. [Google Scholar]
- Prince, E.G.; Kirkland, D.; Demuth, J.P. Hyperexpression of the X chromosome in both sexes results in extensive female bias of X-linked genes in the flour beetle. Genome Biol. Evol. 2010, 2, 336–346. [Google Scholar] [CrossRef] [Green Version]
- Dutrillaux, B.; Dutrillaux, A.-M. Why are X-autosome rearrangements so frequent in beetles? A study of 50 cases. Genes 2023, 14, 150. [Google Scholar] [CrossRef] [PubMed]
- Proença, S.J.R.; Galián, J. Chromosome evolution in the genus Cicindela: Physical mapping and activity of rDNA in the tiger beetle species Cicindela littoralis and C. flexuosa. J. Zool. Syst. Evol. Res. 2003, 41, 227–232. [Google Scholar] [CrossRef]
- Panzera, F.; Mazzella, M.C.; De Vaio, E.S. Cytological studies on three species of neotropical cassidines (Coleoptera, Chrysomelidae). Genetica 1983, 62, 61–68. [Google Scholar] [CrossRef]
- Manna, G.K.; and Lahiri, M. Chromosome complement and meiosis in forty-six species of Coleoptera. Chromosome Inf. Serv. 1972, 13, 9–11. [Google Scholar]
- Alegre, C.; Petitpierre, E. New contribution to the study of the European Cryptocephalinae (Coleoptera, Chrysomelidae). Experientia 1986, 42, 580–583. [Google Scholar] [CrossRef]
- Wahrman, J.; Nezer, R.; Freund, O. Multiple sex chromosome mechanisms with “segregation bodies”. Chromosomes Today 1973, 4, 434. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dutrillaux, B.; Dutrillaux, A.-M.; Salazar, K.; Boucher, S. Multiple Chromosome Fissions, Including That of the X Chromosome, in Aulacocyclus tricuspis Kaup (Coleoptera, Passalidae) from New Caledonia: Characterization of a Rare but Recurrent Pathway of Chromosome Evolution in Animals. Genes 2023, 14, 1487. https://doi.org/10.3390/genes14071487
Dutrillaux B, Dutrillaux A-M, Salazar K, Boucher S. Multiple Chromosome Fissions, Including That of the X Chromosome, in Aulacocyclus tricuspis Kaup (Coleoptera, Passalidae) from New Caledonia: Characterization of a Rare but Recurrent Pathway of Chromosome Evolution in Animals. Genes. 2023; 14(7):1487. https://doi.org/10.3390/genes14071487
Chicago/Turabian StyleDutrillaux, Bernard, Anne-Marie Dutrillaux, Karen Salazar, and Stéphane Boucher. 2023. "Multiple Chromosome Fissions, Including That of the X Chromosome, in Aulacocyclus tricuspis Kaup (Coleoptera, Passalidae) from New Caledonia: Characterization of a Rare but Recurrent Pathway of Chromosome Evolution in Animals" Genes 14, no. 7: 1487. https://doi.org/10.3390/genes14071487





