Mutations of BRCA1, BRCA2, and PALB2 Genes in Breast Tumor Tissue: Relationship with the Effectiveness of Neoadjuvant Chemotherapy and Disease Prognosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Treatment
2.2. DNA Extraction
2.3. Microarrays Assay
2.4. Next-Generation Sequencing (NGS)
2.5. Statistics
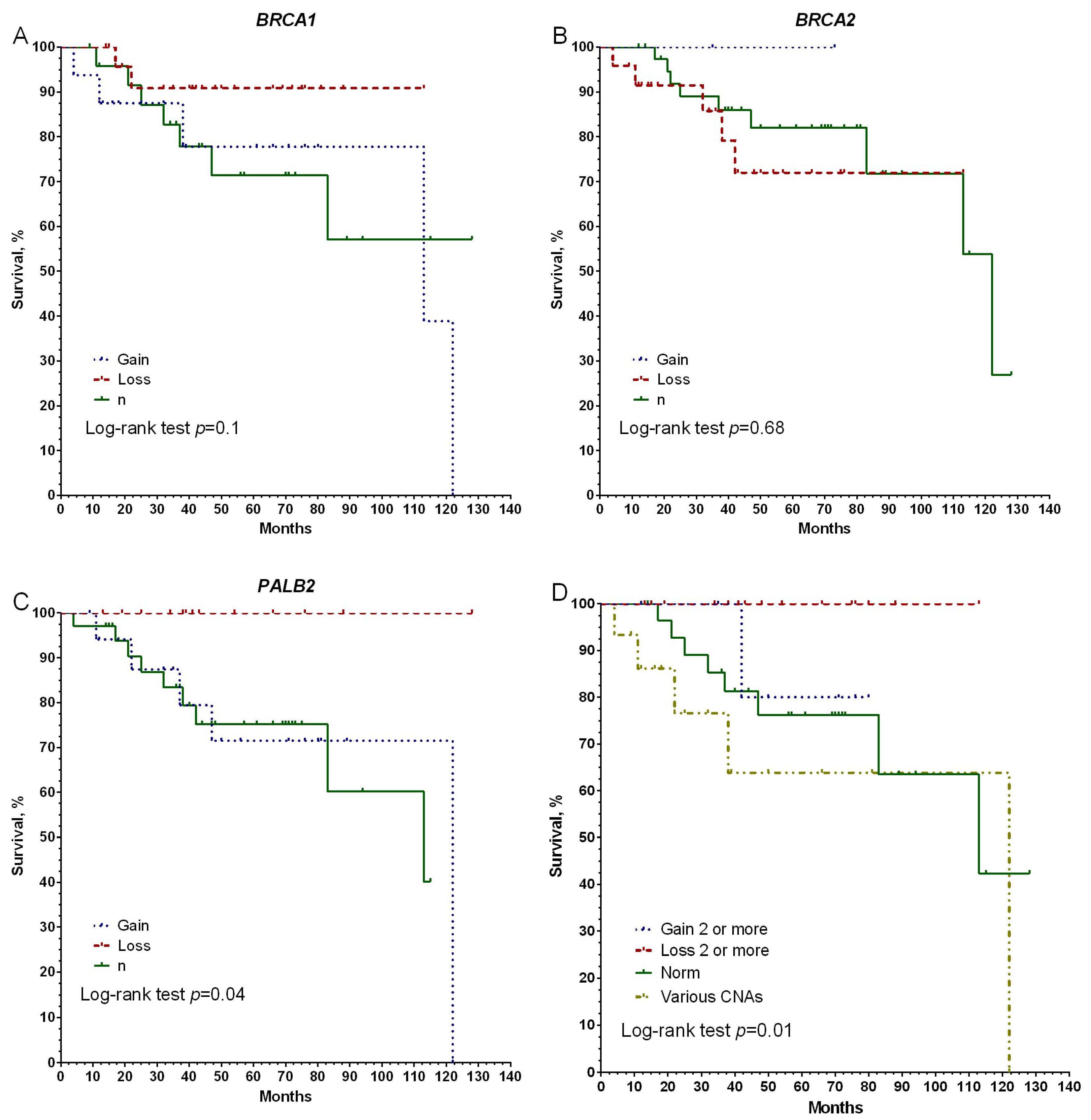
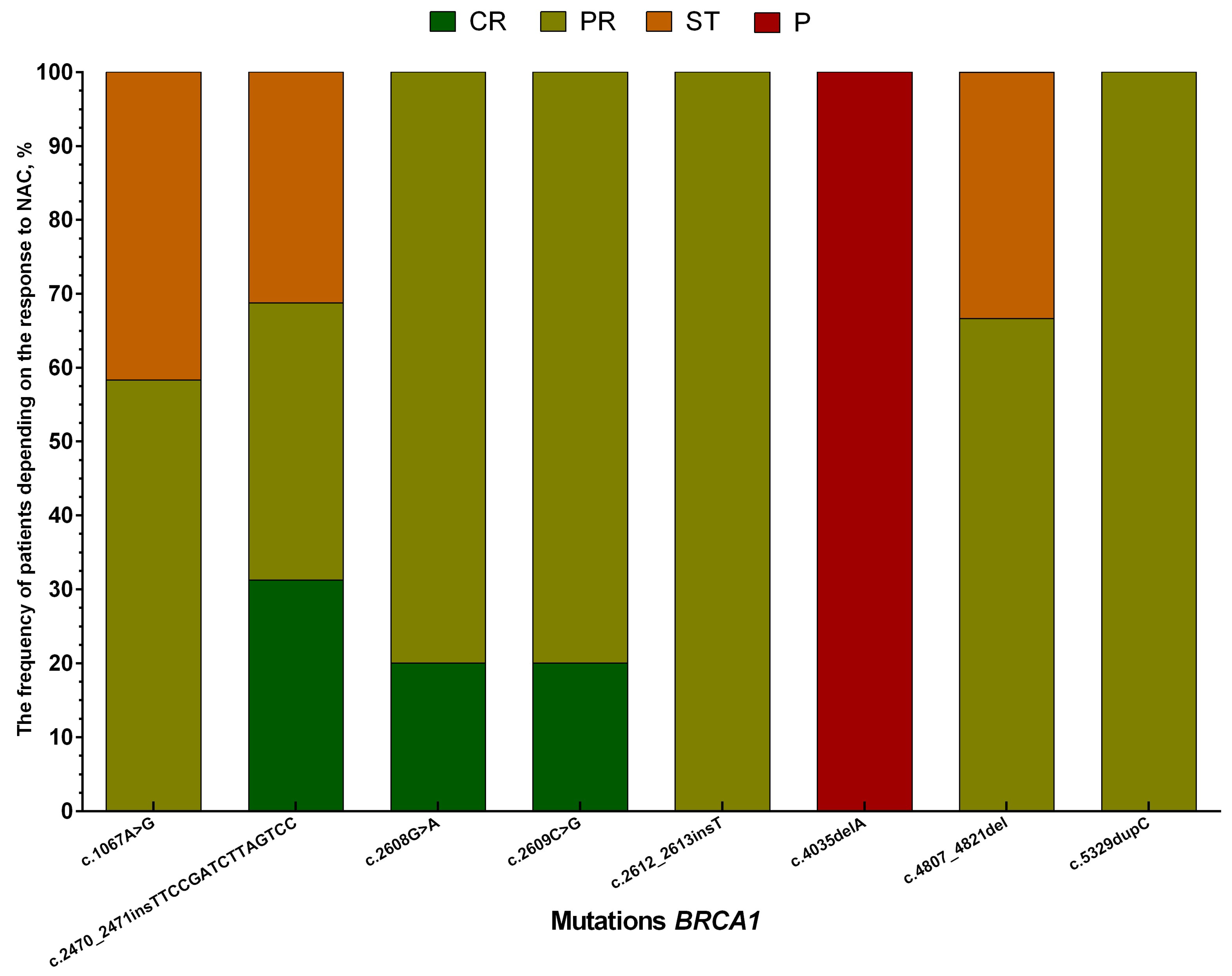
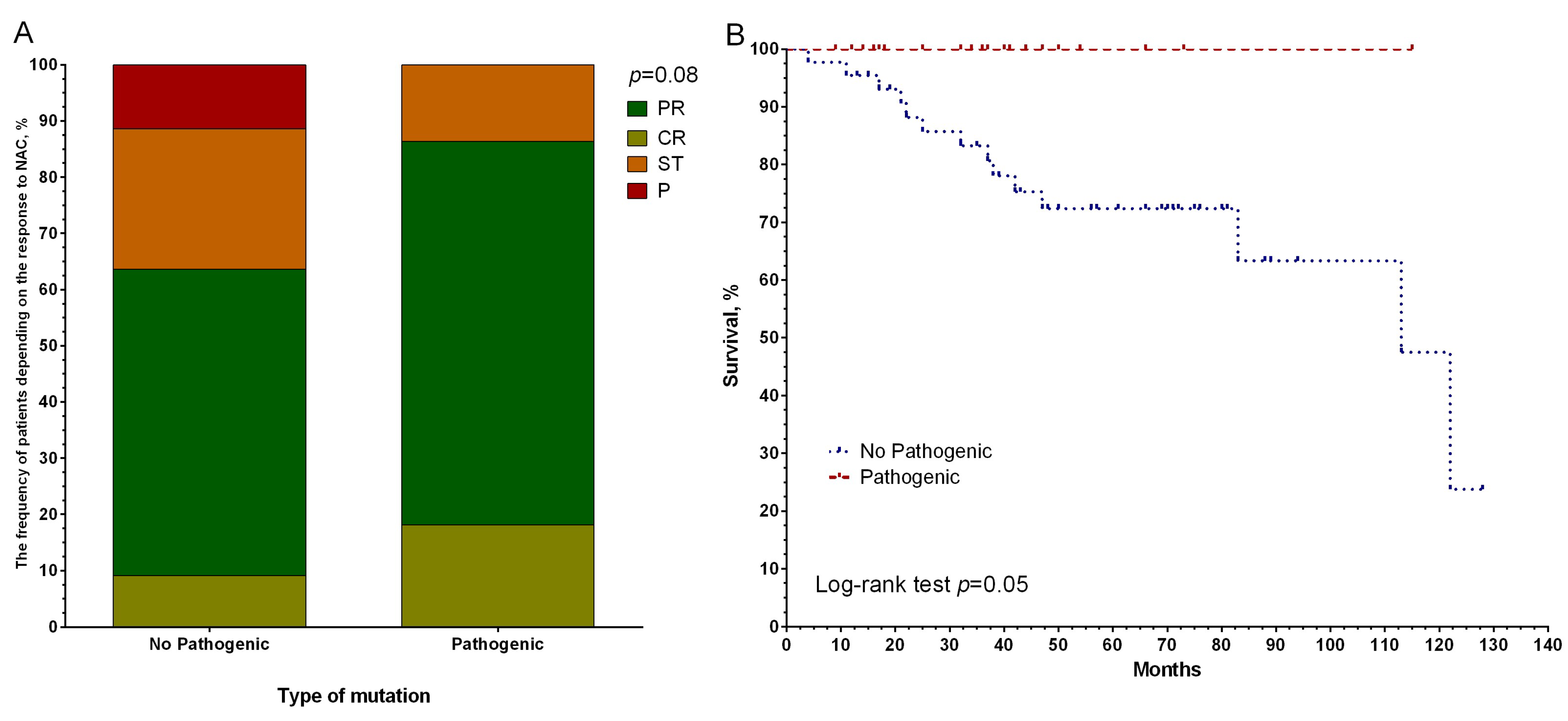
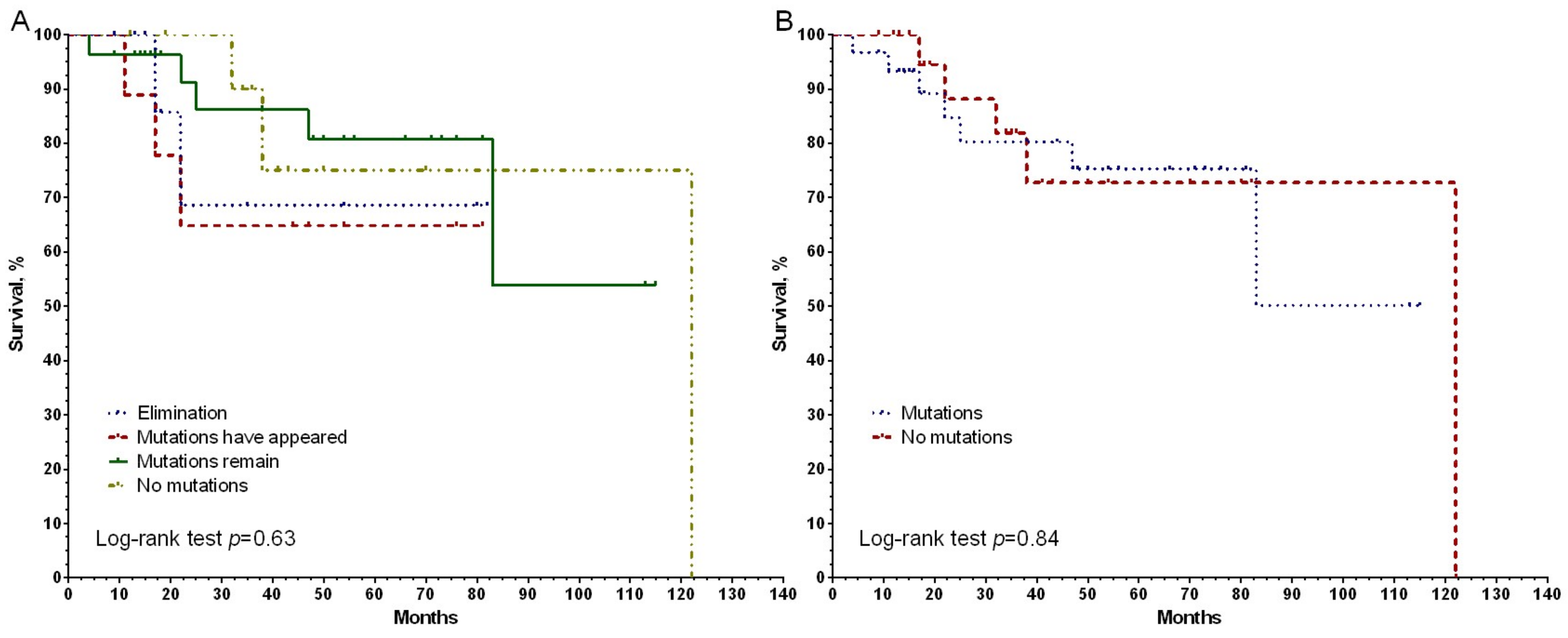
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Imyanitov, E. Features of drug sensitivity of hereditary tumors. Vopr. Onkol. 2016, 62, 221–226. [Google Scholar]
- Markowska, J.; Bar, J.; Mądry, R.; Słomska, I.; Mardas, M.; Grabowski, J. The expression of BRCA1, P53, KAI1, and Nm23 in ovaries of BRCA1 mutation carriers after prophylactic adnexectomy. Arch. Gynecol. Obstet. 2013, 288, 839–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alter, B.P. Fanconi anemia and the development of leukemia. Best Pract. Res. Clin. Haematol. 2014, 27, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Simard, J.; Tonin, P.; Durocher, F.; Morgan, K.; Rommens, J.; Gingras, S.; Samson, C.; Leblanc, J.-F.; Belanger, C.; Dion, F. Common origins of BRCA1 mutations in Canadian breast and ovarian cancer families. Nat. Genet. 1994, 8, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Byrski, T.; Huzarski, T.; Dent, R.; Gronwald, J.; Zuziak, D.; Cybulski, C.; Kladny, J.; Gorski, B.; Lubinski, J.; Narod, S. Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res. Treat. 2009, 115, 359–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.-A.; Mooij, T.M.; Roos-Blom, M.-J.; Jervis, S.; Van Leeuwen, F.E.; Milne, R.L.; Andrieu, N. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 2017, 317, 2402–2416. [Google Scholar] [CrossRef] [Green Version]
- Lord, C.J.; Ashworth, A. BRCAness revisited. Nat. Rev. Cancer 2016, 16, 110. [Google Scholar] [CrossRef]
- Tutt, A.; Tovey, H.; Cheang, M.C.U.; Kernaghan, S.; Kilburn, L.; Gazinska, P.; Owen, J.; Abraham, J.; Barrett, S.; Barrett-Lee, P. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: The TNT Trial. Nat. Med. 2018, 24, 628–637. [Google Scholar] [CrossRef] [Green Version]
- Kaklamani, V.G.; Jeruss, J.S.; Hughes, E.; Siziopikou, K.; Timms, K.M.; Gutin, A.; Abkevich, V.; Sangale, Z.; Solimeno, C.; Brown, K.L. Phase II neoadjuvant clinical trial of carboplatin and eribulin in women with triple negative early-stage breast cancer (NCT01372579). Breast Cancer Res. Treat. 2015, 151, 629–638. [Google Scholar] [CrossRef]
- Von Minckwitz, G.; Timms, K.; Untch, M.; Elkin, E.P.; Fasching, P.A.; Schneeweiss, A.; Salat, C.; Rezai, M.; Blohmer, J.U.; Zahm, D.M. Prediction of pathological complete response (PCR) by Homologous Recombination Deficiency (HRD) after carboplatin-containing neoadjuvant chemotherapy in patients with TNBC: Results from GeparSixto. J. Clin. Oncol. 2015, 33, 1004. [Google Scholar] [CrossRef]
- Wu, S.; Zhou, J.; Zhang, K.; Chen, H.; Luo, M.; Lu, Y.; Sun, Y.; Chen, Y. Molecular mechanisms of PALB2 function and its role in breast cancer management. Front. Oncol. 2020, 10, 301. [Google Scholar] [CrossRef] [Green Version]
- Couch, F.J.; Shimelis, H.; Hu, C.; Hart, S.N.; Polley, E.C.; Na, J.; Hallberg, E.; Moore, R.; Thomas, A.; Lilyquist, J. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017, 3, 1190–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cybulski, C.; Kluźniak, W.; Huzarski, T.; Wokołorczyk, D.; Kashyap, A.; Jakubowska, A.; Szwiec, M.; Byrski, T.; Dębniak, T.; Górski, B. Clinical outcomes in women with breast cancer and a PALB2 mutation: A prospective cohort analysis. Lancet Oncol. 2015, 16, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Antoniou, A.C.; Casadei, S.; Heikkinen, T.; Barrowdale, D.; Pylkäs, K.; Roberts, J.; Lee, A.; Subramanian, D.; De Leeneer, K.; Fostira, F. Breast-cancer risk in families with mutations in PALB2. N. Engl. J. Med. 2014, 371, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Castéra, L.; Harter, V.; Muller, E.; Krieger, S.; Goardon, N.; Ricou, A.; Rousselin, A.; Paimparay, G.; Legros, A.; Bruet, O. Landscape of pathogenic variations in a panel of 34 genes and cancer risk estimation from 5131 HBOC families. Genet. Med. 2018, 20, 1677–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heikkinen, T.; Kärkkäinen, H.; Aaltonen, K.; Milne, R.L.; Heikkilä, P.; Aittomäki, K.; Blomqvist, C.; Nevanlinna, H. The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clin. Cancer Res. 2009, 15, 3214–3222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiss, K.A.; Yu, S.; Judy, R.; Symecko, H.; Nathanson, K.L.; Domchek, S.M. Retrospective survival analysis of patients with advanced pancreatic ductal adenocarcinoma and germline BRCA or PALB2 mutations. JCO Precis. Oncol. 2018, 2, 1–9. [Google Scholar] [CrossRef]
- Kuemmel, S.; Harrach, H.; Schmutzler, R.K.; Kostara, A.; Ziegler-Löhr, K.; Dyson, M.H.; Chiari, O.; Reinisch, M. Olaparib for metastatic breast cancer in a patient with a germline PALB2 variant. NPJ Breast Cancer 2020, 6, 1–4. [Google Scholar] [CrossRef]
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef]
- Robson, M.; Tung, N.; Conte, P.; Im, S.-A.; Senkus, E.; Xu, B.; Masuda, N.; Delaloge, S.; Li, W.; Armstrong, A. OlympiAD final overall survival and tolerability results: Olaparib versus chemotherapy treatment of physician’s choice in patients with a germline BRCA mutation and HER2-negative metastatic breast cancer. Ann. Oncol. 2019, 30, 558–566. [Google Scholar] [CrossRef]
- Lacle, M.M.; Kornegoor, R.; Moelans, C.B.; Maes-Verschuur, A.H.; Van Der Pol, C.; Witkamp, A.J.; Van Der Wall, E.; Rueschoff, J.; Buerger, H.; Van Diest, P.J. Analysis of copy number changes on chromosome 16q in male breast cancer by multiplex ligation-dependent probe amplification. Mod. Pathol. 2013, 26, 1461–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, W.; Lu, K.; Wang, W.; Yao, J.; Hou, Y. PALB2 as a potential prognostic biomarker for colorectal cancer. Comput. Biol. Chem. 2020, 87, 107289. [Google Scholar] [CrossRef]
- Li, J.; Li, M.; Chen, P.; Ba, Q. High expression of PALB2 predicts poor prognosis in patients with advanced breast cancer. FEBS Open Bio 2018, 8, 56–63. [Google Scholar] [CrossRef]
- Schwartz, G.F.; Hortobagyi, G.N. Proceedings of the consensus conference on neoadjuvant chemotherapy in carcinoma of the breast, April 26–28, 2003, Philadelphia, Pennsylvania. Breast J. 2004, 10, 273–294. [Google Scholar] [CrossRef]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [Green Version]
- Duan, J.; Wainwright, M.S.; Comeron, J.M.; Saitou, N.; Sanders, A.R.; Gelernter, J.; Gejman, P.V. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet. 2003, 12, 205–216. [Google Scholar] [CrossRef]
- Löbrich, M.; Jeggo, P.A. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer 2007, 7, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Tsyganov, M.M.; Ibragimova, M.K.; Garbukov, E.Y.; Bragina, O.D.; Karchevskaya, A.A.; Usynin, E.A.; Litvyakov, N.V. Determination of BRCAness Phenotype in Breast Tumors for the Appointment of Neoadjuvant Chemotherapy Based on Platinum and Taxanes. Int. J. Mol. Sci. 2022, 24, 207. [Google Scholar] [CrossRef]
- Pylkäs, K.; Erkko, H.; Nikkilä, J.; Sólyom, S.; Winqvist, R. Analysis of large deletions in BRCA1, BRCA2 and PALB2 genes in Finnish breast and ovarian cancer families. BMC Cancer 2008, 8, 146. [Google Scholar] [CrossRef] [Green Version]
- Yazıcı, H.; Kılıç, S.; Akdeniz, D.; Şükrüoğlu, Ö.; Tuncer, Ş.B.; Avşar, M.; Kuru, G.; Çelik, B.; Küçücük, S.; Saip, P. Frequency of rearrangements versus small indels mutations in BRCA1 and BRCA2 genes in Turkish patients with high risk breast and ovarian cancer. Eur. J. Breast Health 2018, 14, 93–99. [Google Scholar]
- Tyulyandina, A.; Kekeeva, T.; Gorbunova, V.; Kolomiets, L.; Statsenko, G.; Saevets, V.; Khokhlova, S.V.; Tkachenko, S.; Koroleva, I.; Lisyanskaya, A.S. Non-interventional study OVATAR final report: Diagnostic and treatment approaches in Russian ovarian cancer population—BRCAm group analysis. J. Clin. Oncol. 2019, 15, e13111. [Google Scholar] [CrossRef]
- Han, S.-A.; Kim, S.-W.; Kang, E.; Park, S.K.; Ahn, S.-H.; Lee, M.H.; Nam, S.-J.; Han, W.; Bae, Y.T.; Kim, H.-A. The prevalence of BRCA mutations among familial breast cancer patients in Korea: Results of the Korean Hereditary Breast Cancer study. Fam. Cancer 2013, 12, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Simhadri, S.; Vincelli, G.; Huo, Y.; Misenko, S.; Foo, T.K.; Ahlskog, J.; Sørensen, C.S.; Oakley, G.G.; Ganesan, S.; Bunting, S.F. PALB2 connects BRCA1 and BRCA2 in the G2/M checkpoint response. Oncogene 2019, 38, 1585–1596. [Google Scholar] [CrossRef]
- Sy, S.M.; Huen, M.S.; Chen, J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. USA 2009, 106, 7155–7160. [Google Scholar] [CrossRef] [PubMed]
- Elsakov, P.; Kurtinaitis, J.; Petraitis, S.; Ostapenko, V.; Razumas, M.; Razumas, T.; Meskauskas, R.; Petrulis, K.; Luksite, A.; Lubiński, J. The contribution of founder mutations in BRCA1 to breast and ovarian cancer in Lithuania. Clin. Genet. 2010, 78, 373–376. [Google Scholar] [CrossRef]
- Caramelo, O.; Silva, C.; Caramelo, F.; Frutuoso, C.; Almeida-Santos, T. The effect of neoadjuvant platinum-based chemotherapy in BRCA mutated triple negative breast cancers-systematic review and meta-analysis. Hered. Cancer Clin. Pract. 2019, 17, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-J.; Xu, Y.; Lin, Y.; Zhu, H.-J.; Zhou, Y.-D.; Mao, F.; Zhang, X.-H.; Huang, X.; Zhong, Y.; Sun, Q. Platinum-based Neoadjuvant chemotherapy for breast Cancer with BRCA mutations: A meta-analysis. Front. Oncol. 2020, 10, 592998. [Google Scholar] [CrossRef]
- Gervas, P.; Klyuch, B.; Denisov, E.; Kiselev, A.; Molokov, A.; Pisareva, L.; Malinovskaya, E.; Choynzonov, E.; Cherdyntseva, N. New germline BRCA2 gene variant in the Tuvinian Mongol breast cancer patients. Mol. Biol. Rep. 2019, 46, 5537–5541. [Google Scholar] [CrossRef]
- Nguyen-Dumont, T.; MacInnis, R.J.; Steen, J.A.; Theys, D.; Tsimiklis, H.; Hammet, F.; Mahmoodi, M.; Pope, B.J.; Park, D.J.; Mahmood, K. Rare germline genetic variants and risk of aggressive prostate cancer. Int. J. Cancer 2020, 147, 2142–2149. [Google Scholar] [CrossRef]
- Yao, J.; Zhen, Y.; Fan, J.; Gong, Y.; Ye, Y.; Guo, S.; Liu, H.; Li, X.; Li, G.; Yang, P. Comprehensive characterization of CRC with germline mutations reveals a distinct somatic mutational landscape and elevated cancer risk in the Chinese population. Cancer Biol. Med. 2022, 19, 707. [Google Scholar] [CrossRef]
- Talens, F.; Teixeira, V.O.; Rosenberg, E.H.; Nederlof, P.M.; Debipersad, R.D.; Janatova, M.; Zemankova, P.; Kleibl, Z.; Duiker, E.W.; Wisman, G.B.A. Functional RAD51 based assay predicts in vivo PARP inhibitor response in ovarian cancer models beyond BRCA. Homol. Recomb. -Defic. Cancers Approaches Improv. Treat. Patient Sel. 2020, 1, 167–185. [Google Scholar]
- Southey, M.C.; Teo, Z.L.; Winship, I. PALB2 and breast cancer: Ready for clinical translation! Appl. Clin. Genet. 2013, 6, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dansonka-Mieszkowska, A.; Kluska, A.; Moes, J.; Dabrowska, M.; Nowakowska, D.; Niwinska, A.; Derlatka, P.; Cendrowski, K.; Kupryjanczyk, J. A novel germline PALB2 deletion in Polish breast and ovarian cancer patients. BMC Med. Genet. 2010, 11, 20. [Google Scholar] [CrossRef] [Green Version]
- Foulkes, W.D.; Ghadirian, P.; Akbari, M.R.; Hamel, N.; Giroux, S.; Sabbaghian, N.; Darnel, A.; Royer, R.; Poll, A.; Fafard, E. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007, 9, R83. [Google Scholar] [CrossRef]
- Erkko, H.; Xia, B.; Nikkilä, J.; Schleutker, J.; Syrjäkoski, K.; Mannermaa, A.; Kallioniemi, A.; Pylkäs, K.; Karppinen, S.-M.; Rapakko, K. A recurrent mutation in PALB2 in Finnish cancer families. Nature 2007, 446, 316–319. [Google Scholar] [CrossRef] [PubMed]
- Teo, Z.L.; Sawyer, S.D.; James, P.A.; Mitchell, G.; Trainer, A.H.; Lindeman, G.J.; Shackleton, K.; Cicciarelli, L.; Southey, M.C. The incidence of PALB2 c. 3113G> A in women with a strong family history of breast and ovarian cancer attending familial cancer centres in Australia. Fam. Cancer 2013, 12, 587–595. [Google Scholar] [CrossRef]
- Teo, Z.L.; Park, D.J.; Provenzano, E.; Chatfield, C.A.; Odefrey, F.A.; Nguyen-Dumont, T.; Dowty, J.G.; Hopper, J.L.; Winship, I.; Goldgar, D.E. Prevalence of PALB2 mutations in Australasian multiple-case breast cancer families. Breast Cancer Res. 2013, 15, R17. [Google Scholar] [CrossRef]
- Xia, B.; Sheng, Q.; Nakanishi, K.; Ohashi, A.; Wu, J.; Christ, N.; Liu, X.; Jasin, M.; Couch, F.J.; Livingston, D.M. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 2006, 22, 719–729. [Google Scholar] [CrossRef]
- Villarroel, M.C.; Rajeshkumar, N.; Garrido-Laguna, I.; De Jesus-Acosta, A.; Jones, S.; Maitra, A.; Hruban, R.H.; Eshleman, J.R.; Klein, A.; Laheru, D. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol. Cancer Ther. 2011, 10, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bleuyard, J.-Y.; Butler, R.M.; Esashi, F. Perturbation of PALB2 function by the T413S mutation found in small cell lung cancer. Wellcome Open Res. 2017, 2, 110. [Google Scholar] [CrossRef] [Green Version]
- Karachaliou, N.; Bracht, J.W.P.; Bruno, M.F.; Drozdowskyj, A.; Capitan, A.G.; Moran, T.; Carcereny, E.; Cobo, M.; Domine, M.; Chaib, I. Association of PALB2 messenger RNA expression with platinum-docetaxel efficacy in advanced non–small cell lung cancer. J. Thorac. Oncol. 2019, 14, 304–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padella, A.; Marconi, G.; Simonetti, G.; Di Rora, A.G.L.; Fontana, M.C.; Papayannidis, C.; Ottaviani, E.; Cavo, M.; Soverini, S.; Martinelli, G. Higher Expression of PALB2 Predict Poor Prognosis in AML Patients and Identifies Potential Targets of Synthetic Lethal Therapies. Blood 2018, 132, 1507. [Google Scholar] [CrossRef]
- Nykamp, K.; Anderson, M.; Powers, M.; Garcia, J.; Herrera, B.; Ho, Y.-Y.; Kobayashi, Y.; Patil, N.; Thusberg, J.; Westbrook, M. Sherloc: A comprehensive refinement of the ACMG–AMP variant classification criteria. Genet. Med. 2017, 19, 1105–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García, M.J.; Fernández, V.; Osorio, A.; Barroso, A.; LLort, G.; Lázaro, C.; Blanco, I.; Caldés, T.; De la Hoya, M.; y Cajal, T.R. Analysis of FANCB and FANCN/PALB2 fanconi anemia genes in BRCA1/2-negative Spanish breast cancer families. Breast Cancer Res. Treat. 2009, 113, 545–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, A.-Y.; Huang, J.; Hu, Z.; Li, W.-F.; Ma, Z.-L.; Tang, L.-L.; Zhang, B.; Su, F.-X.; Zhou, J.; Di, G.-H. The prevalence of PALB2 germline mutations in BRCA1/BRCA2 negative Chinese women with early onset breast cancer or affected relatives. Breast Cancer Res. Treat. 2009, 114, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.; de la Hoya, M.; Balmaña, J.; y Cajal, T.R.; Teulé, A.; Miramar, M.-D.; Esteban, E.; Infante, M.; Benítez, J.; Torres, A. Detection of a large rearrangement in PALB2 in Spanish breast cancer families with male breast cancer. Breast Cancer Res. Treat. 2012, 132, 307–315. [Google Scholar] [CrossRef] [Green Version]
- Thompson, E.R.; Gorringe, K.L.; Rowley, S.M.; Wong-Brown, M.W.; McInerny, S.; Li, N.; Trainer, A.H.; Devereux, L.; Doyle, M.A.; Li, J. Prevalence of PALB2 mutations in Australian familial breast cancer cases and controls. Breast Cancer Res. 2015, 17, 111. [Google Scholar] [CrossRef]
- Maxwell, K.N.; Hart, S.N.; Vijai, J.; Schrader, K.A.; Slavin, T.P.; Thomas, T.; Wubbenhorst, B.; Ravichandran, V.; Moore, R.M.; Hu, C. Evaluation of ACMG-guideline-based variant classification of cancer susceptibility and non-cancer-associated genes in families affected by breast cancer. Am. J. Hum. Genet. 2016, 98, 801–817. [Google Scholar] [CrossRef] [Green Version]
- Bergstrom, C.; Pence, C.; Berg, J.; Partain, N.; Sadeghi, N.; Mauer, C.; Pirzadeh-Miller, S.; Gao, A.; Li, H.; Unni, N. Clinicopathological features and outcomes in individuals with breast cancer and ATM, CHEK2, or PALB2 Mutations. Ann. Surg. Oncol. 2021, 28, 3383–3393. [Google Scholar] [CrossRef]



| Clinical and Pathological Parameter | Number of Patients, abs. (%) | |
|---|---|---|
| Age | ≤45 | 31 (47.0) |
| >45 | 35 (53.0) | |
| Menstrual status | Perimenopause | 35 (53.0) |
| Postmenopause | 31 (47.0) | |
| Histological type | Invasive ductal cancer | 45 (68.2) |
| Invasive lobular carcinoma | 21 (31.8) | |
| Size | T1 | 7 (10.6) |
| T2 | 53 (80.4) | |
| T3 | 3 (4.5) | |
| T4 | 3 (4.5) | |
| Lymphogenous metastasis | N0 | 25 (37.9) |
| N1 | 31 (47.0) | |
| N2 | 7 (10.6) | |
| N3 | 3 (4.5) | |
| Histological form | Unicentric | 31 (47.0) |
| Multicentric | 35 (53.0) | |
| Molecular subtype | Luminal B HER2-negative | 51 (77.3) |
| HER2+ | 5 (7.6) | |
| Triple-negative | 10 (15.2) | |
| NAC scheme | CAX | 11 (16.7) |
| AC | 12 (18.2) | |
| Taxotere | 21 (31.8) | |
| AT/ACT | 8 (12.1) | |
| CP | 14 (21.2) | |
| NAC effect | Complete regression | 8 (12.1) |
| Partial regression | 39 (59.1) | |
| Stabilization | 14 (21.2) | |
| Progression | 5 (7.6) | |
| Gene/CNA | BRCA1 | BRCA2 | PALB2 | |||
|---|---|---|---|---|---|---|
| Effect of NAC/Scheme of NAC | ||||||
| All Patients (abs. Number, %) | ||||||
| CR + PR | ST + P | CR + PR | ST + P | CR + PR | ST + P | |
| Gain | 11 (23.4) | 5 (26.3) | 1 (2.1) | 1 (5.3) | 15 (31.9) | 3 (15.8) |
| Loss | 21 (44.7) | 4 (21.1) | 15 (31.9) | 9 (47.4) | 12 (25.5) | 2 (10.5) |
| n | 15 (31.9) | 10 (52.6) | 31 (66) | 9 (47.4) | 20 (42.6) | 14 (73.7) |
| p-level | 0.16 | 0.19 | 0.07 | |||
| CAX (abs. number, %) | ||||||
| Gain | 3 (42.9) | 3 (75.0) | 0 (0) | 1 (25.0) | 2 (28.6) | 1 (25.0) |
| Loss | 2 (28.6) | 0 (0) | 4 (57.1) | 3 (75.0) | 2 (28.6) | 0 (0) |
| n | 2 (28.6) | 1 (25.0) | 3 (42.9) | 0 (0) | 3 (42.9) | 3 (75.0) |
| p-level | 0.44 | 0.16 | 0.44 | |||
| AC (abs. number, %) | ||||||
| Gain | 1 (11.1) | 1 (33.3) | 1 (11.1) | 0 (0) | 3 (33.3) | 2 (66.7) |
| Loss | 2 (22.2) | 0 (0) | 3 (33.3) | 1 (33.3) | 3 (33.3) | 0 (0) |
| n | 6 (66.7) | 2 (66.7) | 5 (55.6) | 2 (66.7) | 3 (33.3) | 1 (33.3) |
| p-level | 0.51 | 0.82 | 0.44 | |||
| Taxotere/AT/ACT (abs. number, %) | ||||||
| Gain | 6 (30.0) | 1 (11.1) | 0 (0) | 0 (0) | 8 (40.0) | 0 (0) |
| Loss | 7 (35.0) | 3 (33.3) | 4 (20.0) | 4 (44.4) | 3 (15.0) | 1 (11.1) |
| n | 7 (35.0) | 5 (55.6) | 16 (80.0) | 5 (55.6) | 9 (45.0) | 8 (88.9) |
| p-level | 0.45 | 0.39 | 0.05 | |||
| CP (abs. number, %) | ||||||
| Gain | 1 (9.1) | 0 (0) | 0 (0) | 0 (0) | 2 (18.2) | 0 (0) |
| Loss | 10 (90.9) | 1 (33.3) | 4 (36.4) | 1 (33.3) | 4 (36.4) | 1 (33.3) |
| n | 0 (0) | 2 (66.7) | 7 (63.6) | 2 (66.7) | 5 (45.5) | 2 (66.7) |
| p-level | 0.04 | 0.99 | 0.36 | |||
| № | Mutation | rs | Class | Type |
|---|---|---|---|---|
| 1 | c.1067A>G (p.Gln356Arg) | rs1799950 | Non-synonymous SNV | VUS |
| 2 | c.1114A>C (p.Asn372His) | rs144848 | Non-synonymous SNV | Little Clinical Significance |
| 3 | c.2077G>A (p.Asp693Asn) | rs4986850 | Non-synonymous SNV | Benign |
| 4 | c.2082C>T (p.Ser694=) | rs1799949 | Synonymous SNV | Benign |
| 5 | c.2311T>C (p.Leu771=) | rs16940 | Synonymous SNV | Benign |
| 6 | c.2470_2471insTTCCGATCTTAGTCC (p.Pro 824delinsLPILVP) | - | Non-frameshift insertion | VUS |
| 7 | c.2608G>A (p.Ala870Thr) | rs753256448 | Non-synonymous SNV | VUS |
| 8 | c.2609C>G (p.Ala870Gly) | rs1060502324 | Non-synonymous SNV | VUS |
| 9 | c.2612_2613insT (p.Phe872fs) | rs80357948 | Frameshift insertion | Pathogenic |
| 10 | c.2612C>T (p.Pro871Leu) | rs799917 | Non-synonymous SNV | Benign |
| 11 | c.3113A>G (p.Glu1038Gly) | rs16941 | Non-synonymous SNV | Benign |
| 12 | c.3396A>G (p.Lys1132=) | rs1801406 | Synonymous SNV | Little Clinical Significance |
| 13 | c.3548A>G (p.Lys1183Arg) | rs16942 | Non-synonymous SNV | Benign |
| 14 | c.4035delA (p.Glu1345fs) | rs80357711 | Frameshift deletion | Pathogenic |
| 15 | c.4308T>C (p.Ser1436=) | rs1060915 | Synonymous SNV | Benign |
| 16 | c.4563A>G (p.Leu1521=) | rs206075 | Synonymous SNV | Benign |
| 17 | c.4807_4821del (p.Pro1603_Val1607del) | rs80359888 | Non-frameshift deletion | VUS |
| 18 | c.4837A>G (p.Ser1613Gly) | rs1799966 | Non-synonymous SNV | Benign |
| 19 | c.4900A>G (p.Ser1634Gly) | rs1799966 | Non-synonymous SNV | Benign |
| 20 | c.4946T>C (p.Met1649Thr) | rs4986854 | Non-synonymous SNV | Benign |
| 21 | c.5019G>A (p.Met1673Ile) | rs1799967 | Non-synonymous SNV | Benign |
| 22 | c.5329dupC (p.Gln1777fs) | rs397507247, rs397507246, rs431825413; rs80357906 | Frameshift duplication | Pathogenic |
| 23 | c.6513G>C (p.Val2171=) | rs206076 | Synonymous SNV | Benign |
| 24 | c.7242A>G (p.Ser2414=) | rs1799955 | Synonymous SNV | Little Clinical Significance |
| 25 | c.999T>C (p.Ser333=) | - | Synonymous SNV | Benign |
| № | Mutation | rs | Class | Type |
|---|---|---|---|---|
| 1 | c.10076A>G (p.Glu3359Gly) | rs80358389 | Non-synonymous SNV | VUS |
| 2 | c.10234A>G (p.Ile3412Val) | rs1801426 | Non-synonymous SNV | Benign |
| 3 | c.1114A>C (p.Asn372His) | rs144848 | Non-synonymous SNV | Little Clinical Significance |
| 4 | c.125A>G (p.Tyr42Cys) | rs4987046 | Non-synonymous SNV | Benign |
| 5 | c.1295A>T (p.Glu432Val) | - | Non-synonymous SNV | VUS |
| 6 | c.1305A>G (p.Arg435=) | - | Synonymous SNV | VUS |
| 7 | c.1309_1310insAAATC | - | Frameshift insertion | VUS |
| 8 | c.1365A>G (p.Ser455=) | rs1801439 | Synonymous SNV | Benign |
| 9 | c.1514T>C (p.Ile505Thr) | rs28897708 | Non-synonymous SNV | Benign |
| 10 | c.2229T>C (p.His743=) | rs1801499 | Synonymous | Benign |
| 11 | c.-26G>A | rs1799943 | 5 prime UTR | VUS |
| 12 | c.2971A>G (p.Asn991Asp) | rs1799944 | Non-synonymous SNV | Benign |
| 13 | c.3396A>G (p.Lys1132=) | rs1801406 | Synonymous SNV | Little Clinical Significance |
| 14 | c.3807T>C (p.Val1269=) | rs543304 | Synonymous SNV | Likely Benign |
| 15 | c.3823_3824insAGCAGTTCC (p.Ile1275delinsKQFL) | - | Frameshift insertion | VUS |
| 16 | c.3824_3825del p.(Ile1275ArgfsTer4) | - | Frameshift deletion | Pathogenic |
| 17 | c.3824delT (p.Ile 1275fs) | - | Frameshift deletion | VUS |
| 18 | c.4068G>A (p.Leu1356=) | rs28897724 | Synonymous SNV | Benign |
| 19 | c.4258G>T (p.Asp1420Tyr) | rs28897727 | Non-synonymous SNV | Benign |
| 20 | c.4288_4289insGGAACTGAGT (p.Thr1430_1431Ala_delinsRNX) | - | Stop-gain | VUS |
| 21 | c.4563A>G (p.Leu1521=) | rs206075 | Synonymous SNV | Benign |
| 22 | c.5455C>T (p.Pro1819Ser) | rs80358768 | Non-synonymous SNV | Benign |
| 23 | c.5744C>T (p.Thr1915Met) | rs4987117 | Non-synonymous SNV | Benign |
| 24 | c.590C>G (p.Ser197Cys) | rs876659940 | Non-synonymous SNV | VUS |
| 25 | c.6513G>C (p.Val2171=) | rs206076 | Synonymous SNV | Benign |
| 26 | c.6577G>C (p.Glu2193Gln) | - | Non-synonymous SNV | VUS |
| 27 | c.7108_7109insCAT (p.K2370delinsTX) | - | Stop-gain | VUS |
| 28 | c.7110_7111insATATGTGGG (p.Lys2370delinsKICG) | - | Non-frameshift insertion | VUS |
| 29 | c.7242A>G (p.Ser2414=) | rs1799955 | Synonymous SNV | Little Clinical Significance |
| 30 | c.7397T>C (p.Val2466Ala) | rs169547 | Non-synonymous SNV | Benign |
| 31 | c.7544C>T (p.Thr2515Ile) | rs28897744 | Non-synonymous SNV | Benign |
| 32 | c.8208_8209insAG (p.Leu2737fs) | rs483353122 | Frameshift insertion | Pathogenic |
| 33 | c.865A>C (p.Asn289His) | rs766173 | Non-synonymous SNV | Benign |
| 34 | c.8673_8674delAA (p.Thr2891fs) | rs80359724 | Frameshift deletion | Pathogenic |
| 35 | c.8878C>G (p.Gln2960Glu) | - | Non-synonymous SNV | VUS |
| 36 | c.8878C>T p.(Gln2960Ter) | - | Non-synonymous SNV | Pathogenic |
| 37 | c.8881_8884del (p.Gly2961fs) | - | Frameshift deletion | VUS |
| 38 | c.8885T>A | - | Stop-gain | VUS |
| 39 | c.9038C>T (p.Thr3013Ile) | rs28897755 | Non-synonymous SNV | Benign |
| 40 | c.9090delA (p.T3030fs) | rs397507420 | Frameshift deletion | Pathogenic |
| 41 | c.9090dupA (p.Thr3030fs) | - | Frameshift duplication | VUS |
| 42 | c.9976A>T (p.Lys3326*) | rs11571833 | Stop-gain | Benign |
| № | Mutation | rs | Class | Type |
|---|---|---|---|---|
| 1 | c.1010T>C (p.Leu337Ser) | rs45494092 | Non-synonymous SNV | Benign |
| 2 | c.1288delC (p.Gln467fs) | - | Frameshift deletion | VUS |
| 3 | c.1675_1676insGAGTGAAAGGTAA ATCAAGATGTGTGCTCTTCCGACTCC (p.Q559delinsRVKGKSRCVLFRLQ) | - | Non-frameshift insertion | VUS |
| 4 | c.1676A>G (p.Gln559Arg) | rs152451 | Non-synonymous SNV | Benign |
| 5 | c.1686delG (p.Gly562fs) | - | Frameshift deletion | Pathogenic |
| 6 | c.1693delA (p.Ser565fs) | - | Frameshift deletion | VUS |
| 7 | c.1706_1707del (p.Lys569fs) | rs1060502759 | Frameshift deletion | Pathogenic |
| 8 | c.1706delA (p.Lys569fs) | - | Frameshift deletion | VUS |
| 9 | c.2014G>C (p.Glu672Gln) | rs45532440 | Non-synonymous SNV | Benign |
| 10 | c.212-58A>C | rs80291632 | Intronic | Benign |
| 11 | c.2552delA (p.Asn851fs) | - | Frameshift deletion | VUS |
| 12 | c.255dupA (p.Thr86fs) | - | Frameshift duplication | VUS |
| 13 | c.256delA (p.Thr86fs) | - | Frameshift deletion | VUS |
| 14 | c.2586+58C>T | rs249954 | Intronic | Benign |
| 15 | c.2587-38C>T | rs180177119 | Intronic | Likely benign |
| 16 | c.2993G>A (p.Gly998Glu) | rs45551636 | Non-synonymous SNV | Benign |
| 17 | c.3114-51T>A | rs249936 | Intronic | Benign |
| 18 | c.3300T>G (p.Thr1100=) | rs45516100 | Synonymous SNV | Benign |
| 19 | c.3351-53delT | rs35294437 | Intronic | VUS |
| 20 | c.3465dupT (p. Asp1156 Gln1157delinsX) | - | Stop-gain | VUS |
| 21 | c.747T>A (p.Pro249=) | - | Synonymous SNV | VUS |
| 22 | c.833T>A (p.Leu278Gln) | rs200843485 | Non-synonymous SNV | VUS |
| 23 | c.834A>T (p.Leu278=) | rs199919863 | Synonymous SNV | Likely benign |
| 24 | c.92_94del (p.31_32del) | - | Non-frameshift deletion | VUS |
| 25 | c.921delA (p. Lys307fs) | rs202151522 | Frameshift deletion | VUS |
| 26 | c.94C>G (p.Leu32Val) | rs151316635 | Non-synonymous SNV | Conflicting interpretations of pathogenicity |
| 27 | c.95_96insCGGAAG (p.Leu32 delinsLGR) | - | Non-frameshift insertion | VUS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsyganov, M.M.; Sorokovikova, S.S.; Lutzkaya, E.A.; Ibragimova, M.K. Mutations of BRCA1, BRCA2, and PALB2 Genes in Breast Tumor Tissue: Relationship with the Effectiveness of Neoadjuvant Chemotherapy and Disease Prognosis. Genes 2023, 14, 1554. https://doi.org/10.3390/genes14081554
Tsyganov MM, Sorokovikova SS, Lutzkaya EA, Ibragimova MK. Mutations of BRCA1, BRCA2, and PALB2 Genes in Breast Tumor Tissue: Relationship with the Effectiveness of Neoadjuvant Chemotherapy and Disease Prognosis. Genes. 2023; 14(8):1554. https://doi.org/10.3390/genes14081554
Chicago/Turabian StyleTsyganov, Matvey M., Sofia S. Sorokovikova, Elizaveta A. Lutzkaya, and Marina K. Ibragimova. 2023. "Mutations of BRCA1, BRCA2, and PALB2 Genes in Breast Tumor Tissue: Relationship with the Effectiveness of Neoadjuvant Chemotherapy and Disease Prognosis" Genes 14, no. 8: 1554. https://doi.org/10.3390/genes14081554
APA StyleTsyganov, M. M., Sorokovikova, S. S., Lutzkaya, E. A., & Ibragimova, M. K. (2023). Mutations of BRCA1, BRCA2, and PALB2 Genes in Breast Tumor Tissue: Relationship with the Effectiveness of Neoadjuvant Chemotherapy and Disease Prognosis. Genes, 14(8), 1554. https://doi.org/10.3390/genes14081554






