First Contiguous Genome Assembly of Japanese Lady Bell (Adenophora triphylla) and Insights into Development of Different Leaf Types
Abstract
1. Introduction
2. Results and Discussion
2.1. Estimation of Genome Size
2.2. Genome Assembly
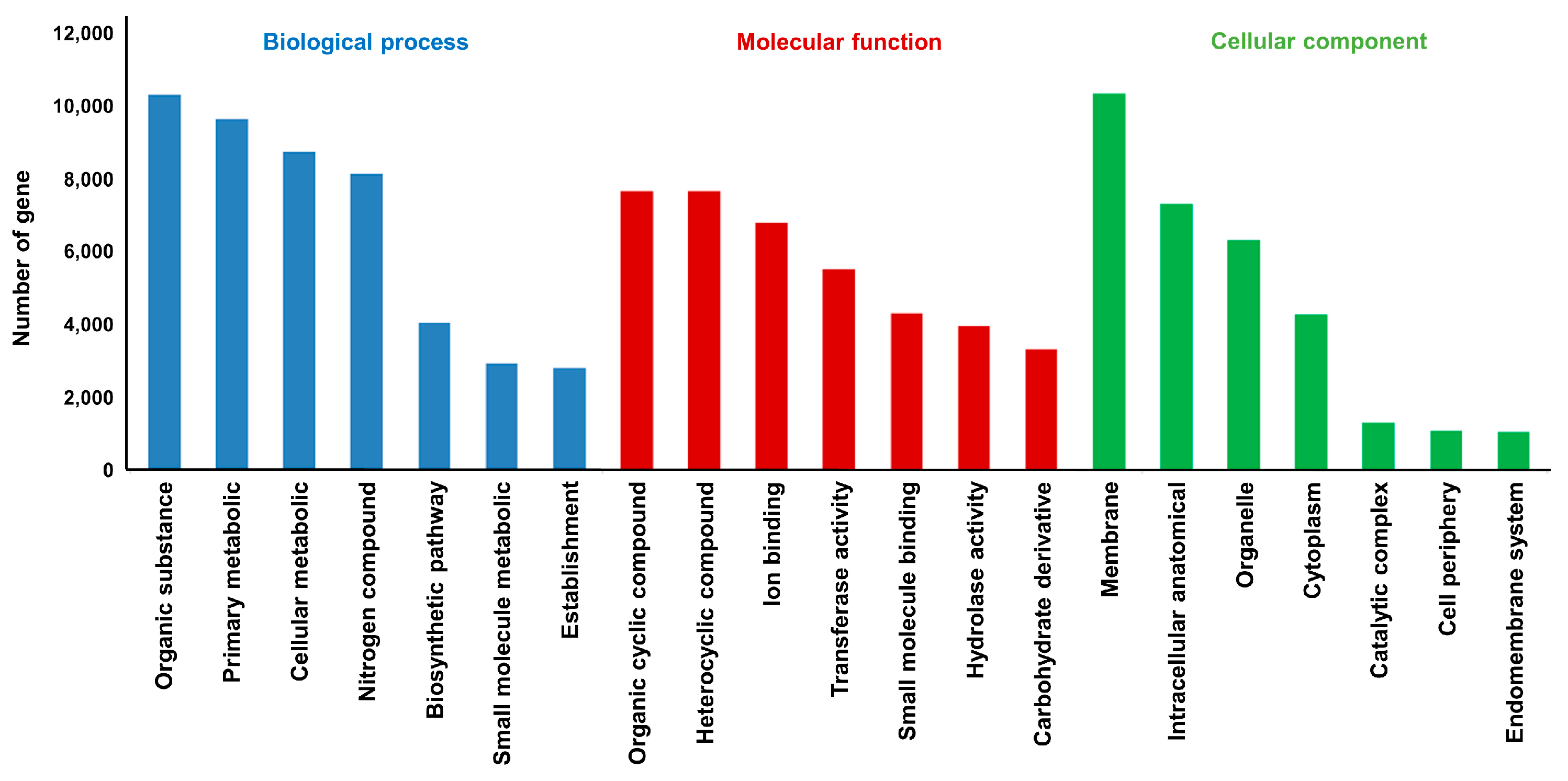
2.3. Genome Annotation
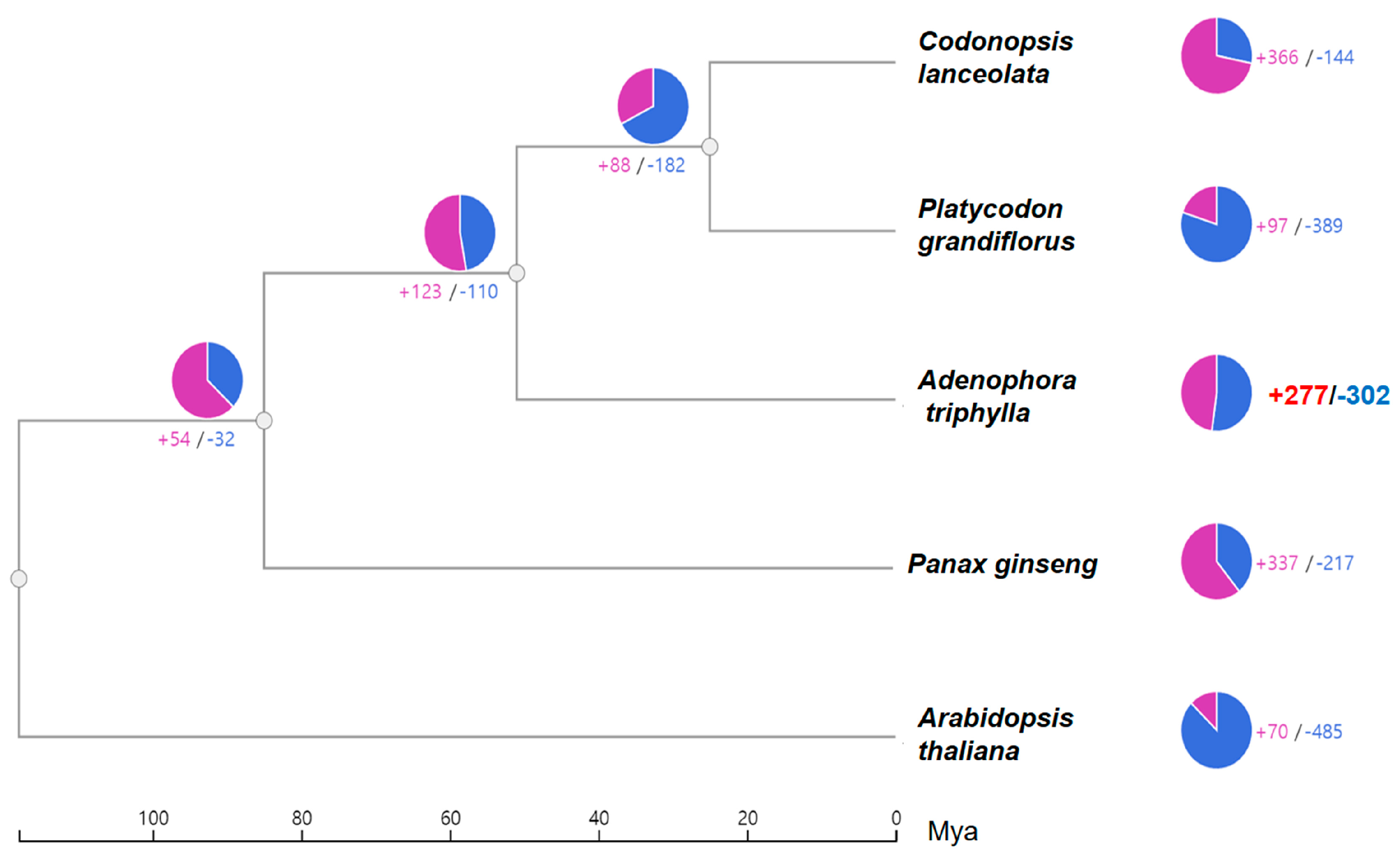
2.4. Genome Comparison
2.5. β-Amyrin Synthase Genes in A. triphylla
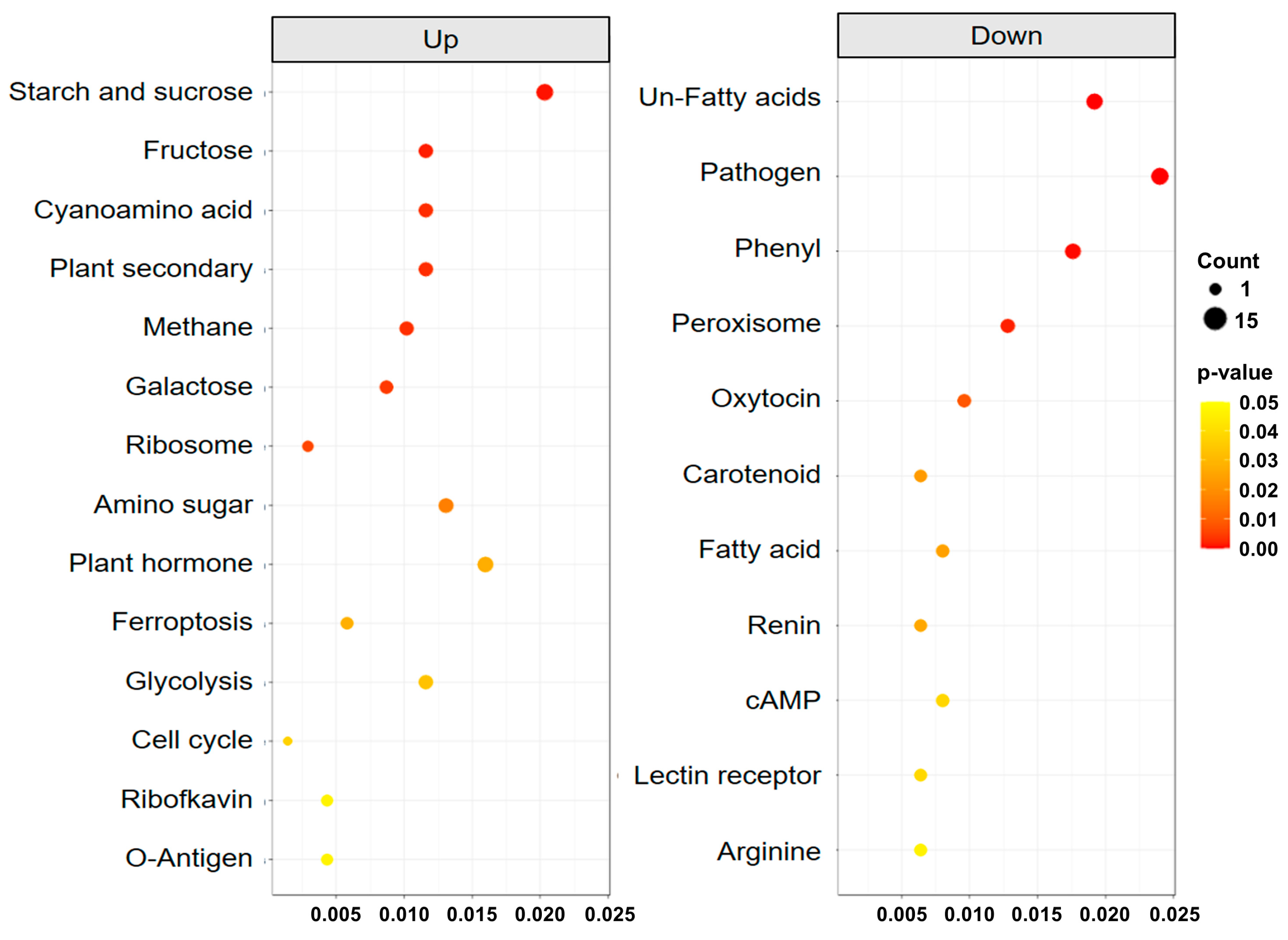
2.6. Analysis of Transcriptomes Involved in the Development of Different Leaf Types
3. Materials and Methods
3.1. Plant Materials and DNA Preparation
3.2. Library Preparation, Sequencing, and Genome Size Estimation
3.3. Genome Assembly
3.4. Gene Annotation
3.5. Genome Comparisons
3.6. RNA Sequencing and Transcriptome Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, S.-H. Ethyl Acetate Fraction of Adenophora triphylla var. japonica inhibits migration of lewis lung carcinoma cells by suppressing macrophage polarization toward an M2 phenotype. J. Pharmacopunct. 2019, 22, 253. [Google Scholar] [CrossRef] [PubMed]
- Ham, Y.-A.; Choi, H.-J.; Chung, M.-J.; Ham, S.-S. Component analysis and antioxidant activity of Adenophora triphylla. J. Korean Soc. Food Sci. Nutr. 2009, 38, 274–279. [Google Scholar] [CrossRef]
- Lee, S.-E.; Lee, E.-H.; Lee, T.-J.; Kim, S.-W.; Kim, B.-H. Anti-obesity effect and action mechanism of Adenophora triphylla root ethanol extract in C57BL/6 obese mice fed a high-fat diet. Biosci. Biotechnol. Biochem. 2013, 77, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.-J.; Kim, S.-H.; Oh, H.-T.; Chung, M.-J.; Cui, C.-B.; Ham, S.-S. Effects of Adenophora triphylla ethylacetate extract on mRNA levels of antioxidant enzymes in human HepG2 cells. J. Korean Soc. Food Sci. Nutr. 2008, 37, 1238–1243. [Google Scholar] [CrossRef][Green Version]
- Chung, M.J.; Lee, S.; Park, Y.I.; Kwon, K.H. Antioxidative and neuroprotective effects of extract and fractions from Adenophora triphylla. J. Korean Soc. Food. Sci. Nutr. 2016, 45, 1580–1588. [Google Scholar] [CrossRef]
- Park, D. The edible wild vegetable containing biological active substances. Euliiglob. Korean 2010, 231–234. [Google Scholar]
- Kim, J.-H.; Hong, J.-Y.; Shin, S.-R.; Yoon, K.-Y. Comparison of antioxidant activity in wild plant (Adenophora triphylla) leaves and roots as a potential source of functional foods. Int. J. Food Sci. Nutr. 2009, 60, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Chen, S.; Chen, W.; Zhang, P.; Su, Z.; Zhang, L.; Xu, M.; Guo, L. A chromosome-level reference genome of Chinese balloon flower (Platycodon grandiflorus). Front. Genet. 2022, 13, 869784. [Google Scholar] [CrossRef]
- Kim, J.; Kang, S.-H.; Park, S.-G.; Yang, T.-J.; Lee, Y.; Kim, O.T.; Chung, O.; Lee, J.; Choi, J.-P.; Kwon, S.-J. Whole-genome, transcriptome, and methylome analyses provide insights into the evolution of platycoside biosynthesis in Platycodon grandiflorus, a medicinal plant. Hortic. Res. 2020, 7, 112. [Google Scholar] [CrossRef]
- Jang, W.; Kang, J.-N.; Jo, I.-H.; Lee, S.-M.; Park, G.-H.; Kim, C.-K. The chromosome-level genome assembly of lance asiabell (Codonopsis lanceolata), a medicinal and vegetable plant of the Campanulaceae family. Front. Genet. 2023, 14, 1100819. [Google Scholar] [CrossRef]
- Dumschott, K.; Schmidt, M.H.; Chawla, H.S.; Snowdon, R.; Usadel, B. Oxford Nanopore sequencing: New opportunities for plant genomics? J. Exp. Bot. 2020, 71, 5313–5322. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Das, A.; Kainer, D.; Schalamun, M.; Morales-Suarez, A.; Schwessinger, B.; Lanfear, R. The draft nuclear genome assembly of Eucalyptus pauciflora: A pipeline for comparing de novo assemblies. GigaScience 2020, 9, giz160. [Google Scholar] [CrossRef]
- Tsukaya, H. Leaf shape diversity with an emphasis on leaf contour variation, developmental background, and adaptation. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2018; pp. 48–57. [Google Scholar]
- Fritz, M.A.; Rosa, S.; Sicard, A. Mechanisms underlying the environmentally induced plasticity of leaf morphology. Front. Genet. 2018, 9, 478. [Google Scholar] [CrossRef] [PubMed]
- Kierzkowski, D.; Runions, A.; Vuolo, F.; Strauss, S.; Lymbouridou, R.; Routier-Kierzkowska, A.-L.; Wilson-Sanchez, D.; Jenke, H.; Galinha, C.; Mosca, G. A growth-based framework for leaf shape development and diversity. Cell 2019, 177, 1405–1418. e1417. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, N.; Runions, A.; Tsiantis, M. Leaf shape diversity: From genetic modules to computational models. Annu. Rev. Plant Biol. 2021, 72, 325–356. [Google Scholar] [CrossRef]
- Ajayi, O.E.; Yoon, S.Y.; Moon, S.; Kim, K.H.; Kim, J.H.; Chung, J.-W.; Jang, K.-I.; Hyun, T.K. Variability in Phytochemical Contents and Biological Activities among Adenophora triphylla Genotypes. Appl. Sci. 2023, 13, 11184. [Google Scholar] [CrossRef]
- Kim, D.; Kim, K.-Y. Adenophora triphylla var. japonica inhibits Candida biofilm formation, increases susceptibility to antifungal agents and reduces infection. Int. J. Mol. Sci. 2021, 22, 12523. [Google Scholar] [CrossRef]
- Asano, N.; Nishida, M.; Miyauchi, M.; Ikeda, K.; Yamamoto, M.; Kizu, H.; Kameda, Y.; Watson, A.A.; Nash, R.J.; Fleet, G.W. Polyhydroxylated pyrrolidine and piperidine alkaloids from Adenophora triphylla var. japonica (Campanulaceae). Phytochemistry 2000, 53, 379–382. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, X.; Jeong, H.K.; Wei, H.; Jeong, B.R. Growth and physiological responses of Adenophora triphylla (Thunb.) A. DC. plug seedlings to day and night temperature regimes. Agronomy 2018, 8, 173. [Google Scholar] [CrossRef]
- Yu, H.; Liu, M.; Yin, M.; Shan, T.; Peng, H.; Wang, J.; Chang, X.; Peng, D.; Zha, L.; Gui, S. Transcriptome analysis identifies putative genes involved in triterpenoid biosynthesis in Platycodon grandiflorus. Planta 2021, 254, 34. [Google Scholar] [CrossRef]
- Zhang, D.; Li, W.; Xia, E.-h.; Zhang, Q.-j.; Liu, Y.; Zhang, Y.; Tong, Y.; Zhao, Y.; Niu, Y.-c.; Xu, J.-h. The medicinal herb Panax notoginseng genome provides insights into ginsenoside biosynthesis and genome evolution. Mol. Plant 2017, 10, 903–907. [Google Scholar] [CrossRef] [PubMed]
- Seki, H.; Tamura, K.; Muranaka, T. P450s and UGTs: Key players in the structural diversity of triterpenoid saponins. Plant Cell Physiol. 2015, 56, 1463–1471. [Google Scholar] [CrossRef] [PubMed]
- Busta, L.; Serra, O.; Kim, O.T.; Molinas, M.; Peré-Fossoul, I.; Figueras, M.; Jetter, R. Oxidosqualene cyclases involved in the biosynthesis of triterpenoids in Quercus suber cork. Sci. Rep. 2020, 10, 8011. [Google Scholar] [CrossRef]
- Lee, D.-J.; Choi, J.-W.; Kang, J.-N.; Lee, S.-M.; Park, G.-H.; Kim, C.-K. Chromosome-Scale Genome Assembly and Triterpenoid Saponin Biosynthesis in Korean Bellflower (Platycodon grandiflorum). Int. J. Mol. Sci. 2023, 24, 6534. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Kang, M.; Kim, Y.S. A triterpenoid saponin from Adenophora triphylla var. japonica suppresses the growth of human gastric cancer cells via regulation of apoptosis and autophagy. Tumor Biol. 2014, 35, 12021–12030. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.-X.; Liu, M.-Y.; Fu, C.-X.; Liu, M.-Y. Chromosome Studies of 10 Chinese Species of Adenophora Fisch. J. Syst. Evol. 1987, 25, 180. [Google Scholar]
- Sugiura, T. Studies on the chromosome numbers in Campanulaceae I. Campanuloideae-Campanuleae. Cytologia 1942, 12, 418–434. [Google Scholar] [CrossRef][Green Version]
- Ou, S.; Chen, J.; Jiang, N. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res. 2018, 46, e126. [Google Scholar] [CrossRef]
- Guiglielmoni, N.; Houtain, A.; Derzelle, A.; Van Doninck, K.; Flot, J.-F. Overcoming uncollapsed haplotypes in long-read assemblies of non-model organisms. BMC Bioinform. 2021, 22, 303. [Google Scholar] [CrossRef]
- Zhang, T.; Xing, W.; Wang, A.; Zhang, N.; Jia, L.; Ma, S.; Xia, Q. Comparison of Long-Read Methods for Sequencing and Assembly of Lepidopteran Pest Genomes. Int. J. Mol. Sci. 2022, 24, 649. [Google Scholar] [CrossRef]
- Ma, B.; Kuang, L.; Xin, Y.; He, N. New insights into long terminal repeat retrotransposons in mulberry species. Genes 2019, 10, 285. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-H.; Wang, X.-F.; Lu, T.; Li, M.-R.; Jiang, P.; Zhao, J.; Liu, S.-T.; Fu, X.-Q.; Wendel, J.F.; Van de Peer, Y. Reshuffling of the ancestral core-eudicot genome shaped chromatin topology and epigenetic modification in Panax. Nat. Commun. 2022, 13, 1902. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, S.J.; James, D.E.; Mann, M. Protein phosphorylation: A major switch mechanism for metabolic regulation. Trends Endocrinol. Metab. 2015, 26, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Cross, T.G.; Scheel-Toellner, D.; Henriquez, N.V.; Deacon, E.; Salmon, M.; Lord, J.M. Serine/threonine protein kinases and apoptosis. Exp. Cell Res. 2000, 256, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.I.; Xia, H.; Aguilera-Aguirre, L.; Hage, A.; van Tol, S.; Shan, C.; Xie, X.; Sturdevant, G.L.; Robertson, S.J.; McNally, K.L. Envelope protein ubiquitination drives entry and pathogenesis of Zika virus. Nature 2020, 585, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Polito, L.; Bortolotti, M.; Battelli, M.G.; Bolognesi, A. Xanthine oxidoreductase: A leading actor in cardiovascular disease drama. Redox Biol. 2021, 48, 102195. [Google Scholar] [CrossRef]
- He, J.-Y.; Ma, N.; Zhu, S.; Komatsu, K.; Li, Z.-Y.; Fu, W.-M. The genus Codonopsis (Campanulaceae): A review of phytochemistry, bioactivity and quality control. J. Nat. Med. 2015, 69, 1–21. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, H.; Kim, J.; Seol, G.H.; Lee, K.-W. Lancemaside A, a major triterpene saponin of Codonopsis lanceolata enhances regulation of nitric oxide synthesis via eNOS activation. BMC Complement. Med. Ther. 2019, 19, 110. [Google Scholar] [CrossRef]
- Shin, B.-K.; Kwon, S.W.; Park, J.H. Chemical diversity of ginseng saponins from Panax ginseng. J. Ginseng Res. 2015, 39, 287–298. [Google Scholar] [CrossRef]
- Ren, J.; Bi, Y.; Sowers, J.R.; Hetz, C.; Zhang, Y. Endoplasmic reticulum stress and unfolded protein response in cardiovascular diseases. Nat. Rev. Cardiol. 2021, 18, 499–521. [Google Scholar] [CrossRef] [PubMed]
- Garavito, M.F.; Narváez-Ortiz, H.Y.; Zimmermann, B.H. Pyrimidine metabolism: Dynamic and versatile pathways in pathogens and cellular development. J. Genet. Genom. 2015, 42, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Pien, S.; Wyrzykowska, J.; Fleming, A.J. Novel marker genes for early leaf development indicate spatial regulation of carbohydrate metabolism within the apical meristem. Plant J. 2001, 25, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Wick, R. Porechop: An Adapter Trimmer for Oxford Nanopore Reads. 2018. Available online: https://github.com/rrwick/Porechop (accessed on 14 December 2022).
- Marçais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef]
- Ranallo-Benavidez, T.R.; Jaron, K.S.; Schatz, M.C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 2020, 11, 1432. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Liu, H.; Wu, S.; Li, A.; Ruan, J. SMARTdenovo: A de novo assembler using long noisy reads. Gigabyte 2021, 2021, gigabyte15. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Hu, J.; Fan, J.; Sun, Z.; Liu, S. NextPolish: A fast and efficient genome polishing tool for long-read assembly. Bioinformatics 2020, 36, 2253–2255. [Google Scholar] [CrossRef]
- Roach, M.J.; Schmidt, S.A.; Borneman, A.R. Purge Haplotigs: Allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinform. 2018, 19, 460. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, Z.; Sun, Z.; Hu, B.; Ayoola, A.O.; Liang, F.; Li, J.; Sandoval, J.R.; Cooper, D.N.; Ye, K. An efficient error correction and accurate assembly tool for noisy long reads. bioRxiv 2023. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Jayakodi, M.; Lee, S.C.; Choi, B.S.; Jang, W.; Lee, J.; Kim, H.H.; Waminal, N.E.; Lakshmanan, M.; van Nguyen, B. Genome and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnol. J. 2018, 16, 1904–1917. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2015. Available online: http://www.repeatmasker.org (accessed on 12 January 2023).
- Kang, S.-H.; Kim, B.; Choi, B.-S.; Lee, H.O.; Kim, N.-H.; Lee, S.J.; Kim, H.S.; Shin, M.J.; Kim, H.-W.; Nam, K. Genome assembly and annotation of soft-shelled adlay (Coix lacrymajobi Variety mayuen), a cereal and medicinal crop in the Poaceae Family. Front. Plant Sci. 2020, 11, 630. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Sun, J.; Lu, F.; Luo, Y.; Bie, L.; Xu, L.; Wang, Y. OrthoVenn3: An integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 2023, 51, W397–W403. [Google Scholar] [CrossRef]
- Mendes, F.K.; Vanderpool, D.; Fulton, B.; Hahn, M.W. CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics 2020, 36, 5516–5518. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner; Lawrence Berkeley National Lab. (LBNL): Berkeley, CA, USA, 2014. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.; Haas, B.; Yassour, M.; Levin, J.; Thompson, D.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Zaharia, M.; Bolosky, W.; Curtis, K.; Fox, A.; Patterson, D.; Shenker, S.; Stoica, I.; Karp, R.; Sittler, T. Faster and more accurate sequence alignment with SNAP. arXiv 2011, arXiv:1111.5572. [Google Scholar] [CrossRef]
- Stanke, M.; Diekhans, M.; Baertsch, R.; Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008, 24, 637–644. [Google Scholar] [CrossRef]

| Parameter | Value |
|---|---|
| Genome assembly (Pipeline 2) | |
| Number of sequences | 8432 contigs |
| Total length of sequences | 2,477,772,497 bp |
| N50 length | 403,696 bp |
| Smallest sequence | 18,168 bp |
| Longest sequence | 3,658,388 bp |
| Average length | 293,853 bp |
| Complete BUSCO (version 5.0) | 95.6% |
| Gene annotation | |
| Number of protein-coding genes | 57,729 |
| Total length of protein-coding genes | 55,297,059 bp |
| Smallest gene length | 102 bp |
| Longest gene length | 16,275 bp |
| Average gene length | 958 bp |
| Repeat content | 79.2% |
| GC content | 43.6% |
| Functionally annotated | 88.4% |
| Complete BUSCO (version 5.0) | 75.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, J.-N.; Lee, S.-M.; Choi, J.-W.; Lee, S.-S.; Kim, C.-K. First Contiguous Genome Assembly of Japanese Lady Bell (Adenophora triphylla) and Insights into Development of Different Leaf Types. Genes 2024, 15, 58. https://doi.org/10.3390/genes15010058
Kang J-N, Lee S-M, Choi J-W, Lee S-S, Kim C-K. First Contiguous Genome Assembly of Japanese Lady Bell (Adenophora triphylla) and Insights into Development of Different Leaf Types. Genes. 2024; 15(1):58. https://doi.org/10.3390/genes15010058
Chicago/Turabian StyleKang, Ji-Nam, Si-Myung Lee, Ji-Weon Choi, Seung-Sik Lee, and Chang-Kug Kim. 2024. "First Contiguous Genome Assembly of Japanese Lady Bell (Adenophora triphylla) and Insights into Development of Different Leaf Types" Genes 15, no. 1: 58. https://doi.org/10.3390/genes15010058
APA StyleKang, J.-N., Lee, S.-M., Choi, J.-W., Lee, S.-S., & Kim, C.-K. (2024). First Contiguous Genome Assembly of Japanese Lady Bell (Adenophora triphylla) and Insights into Development of Different Leaf Types. Genes, 15(1), 58. https://doi.org/10.3390/genes15010058






