Micro RNA-175 Targets Claudin-1 to Inhibit Madin–Darby Canine Kidney Cell Adhesion
Abstract
:1. Introduction
2. Results
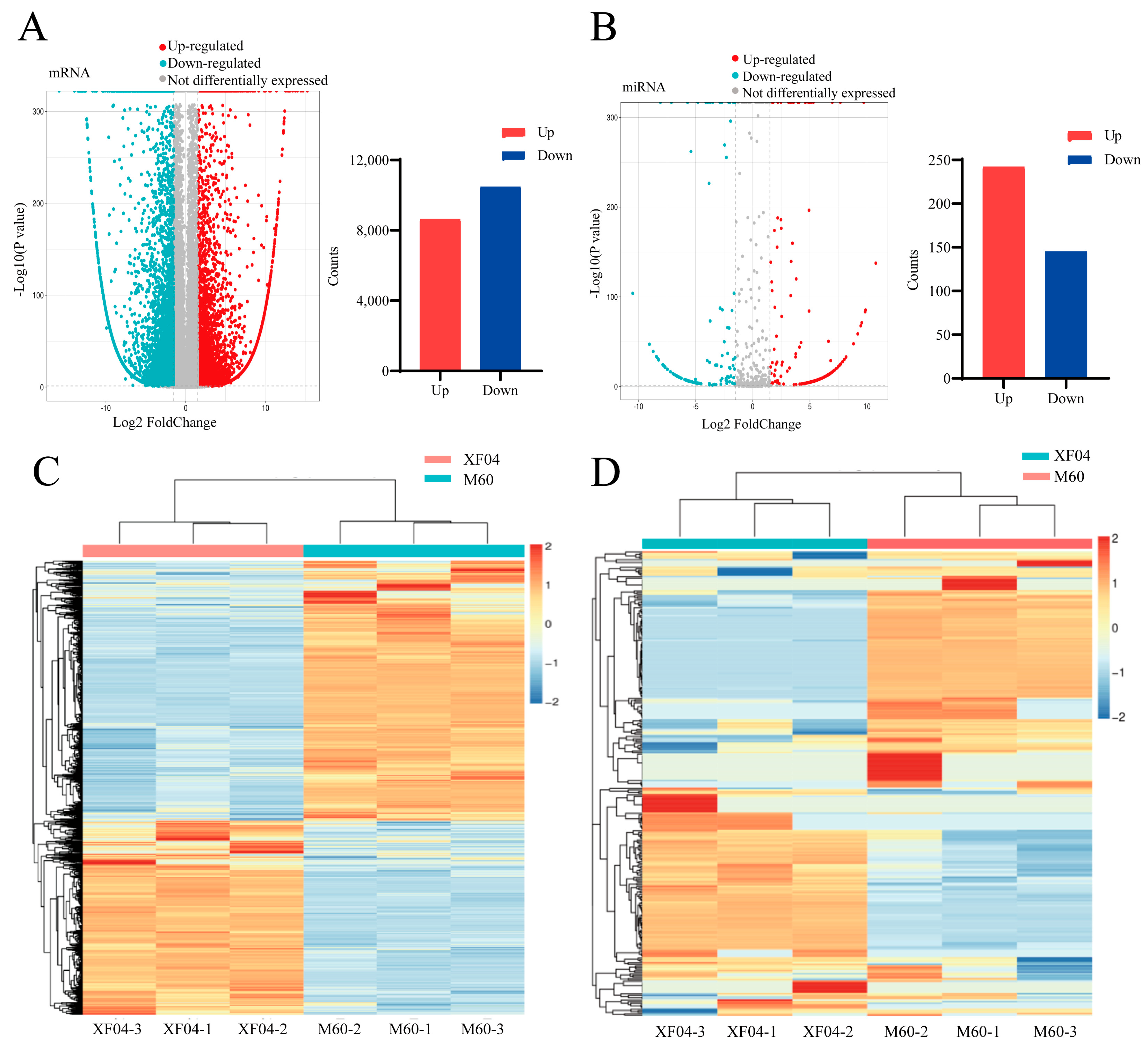
2.1. The Transcriptomic Analysis of Differentially Expressed mRNAs and miRNAs in Suspension and Adherent MDCK Cells
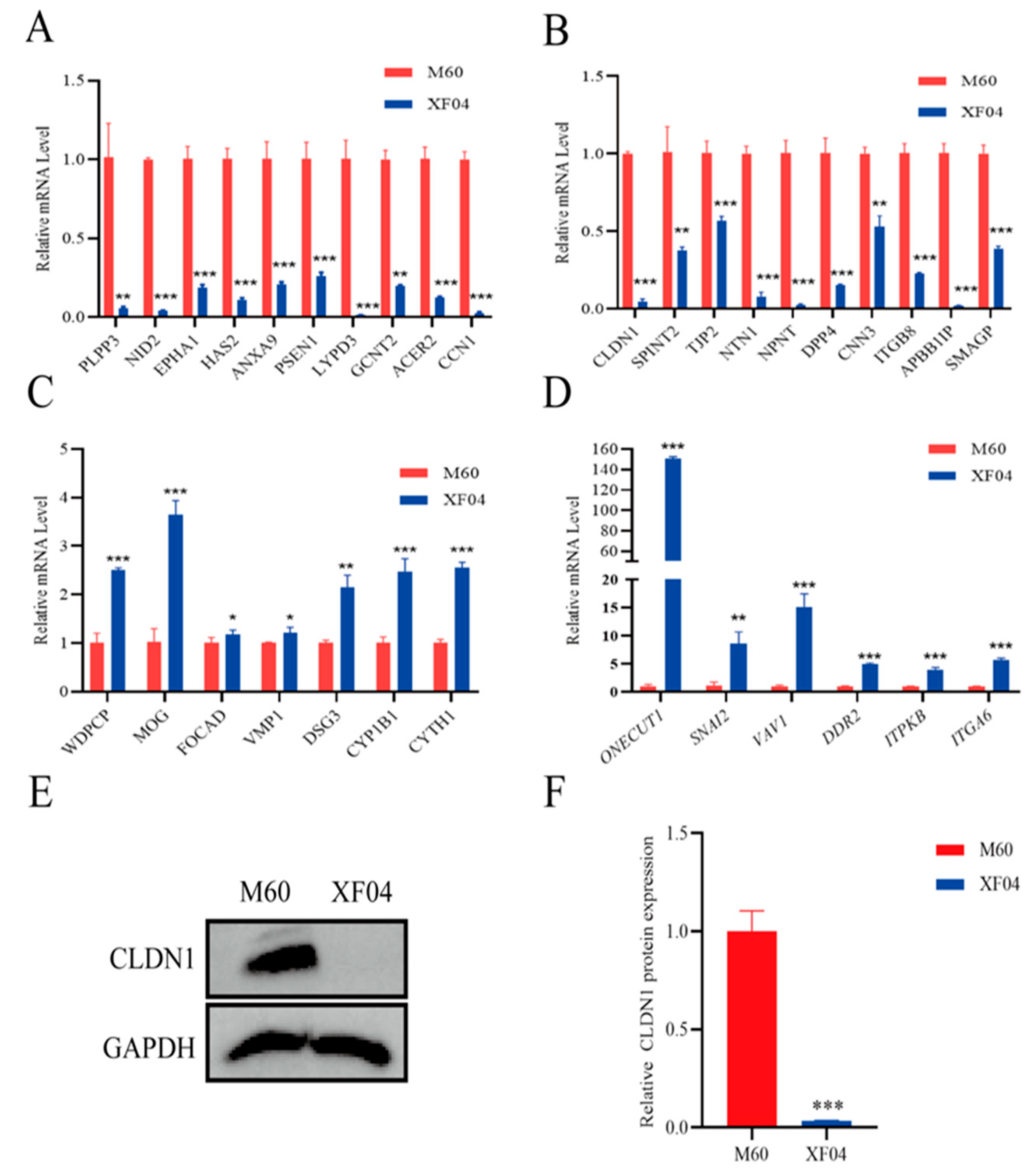
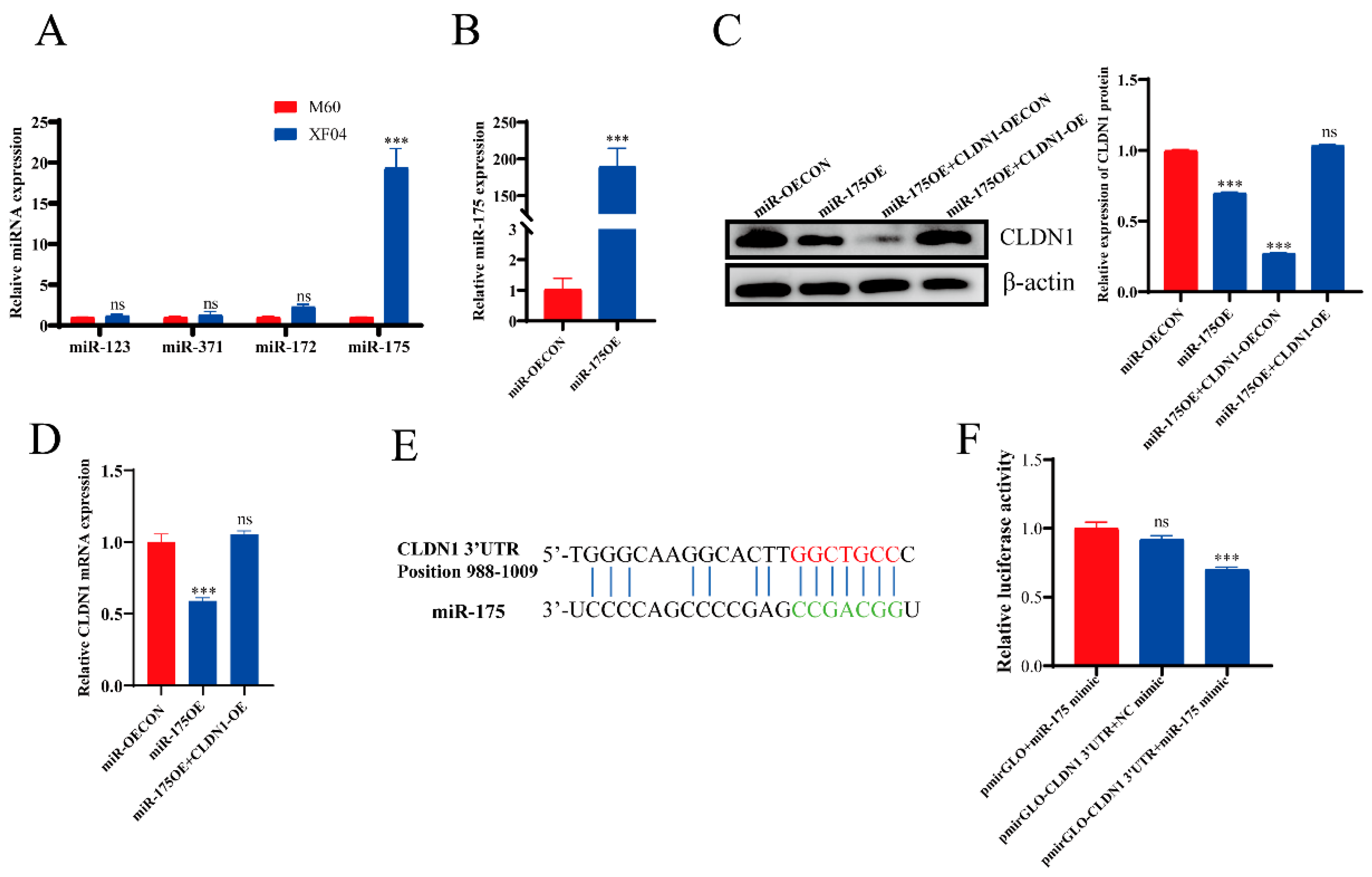
2.2. CLDN1 Expression Was Significantly Downregulated in the Suspension MDCK Cells
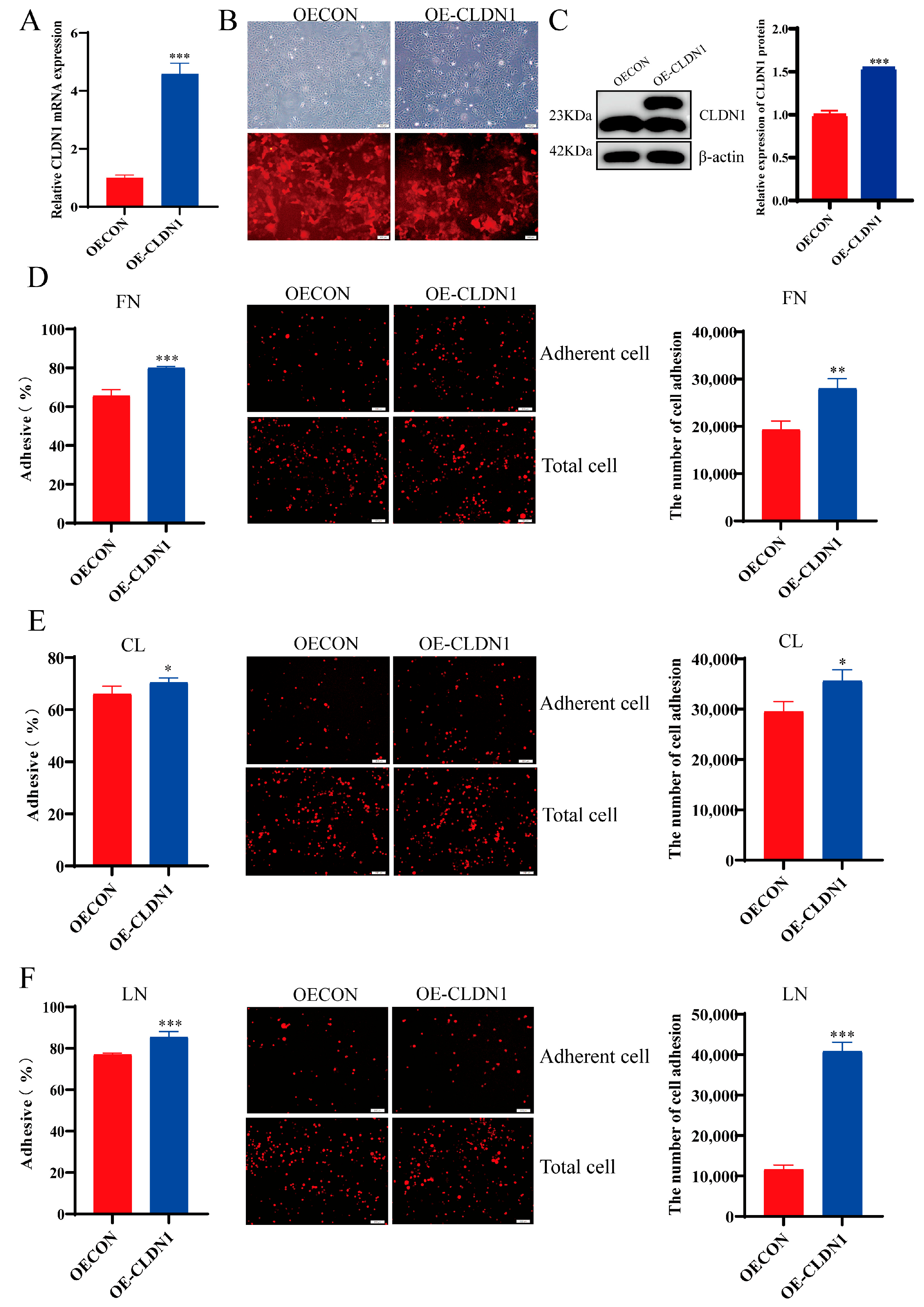
2.3. CLDN1 Promotes MDCK Cell Adhesion
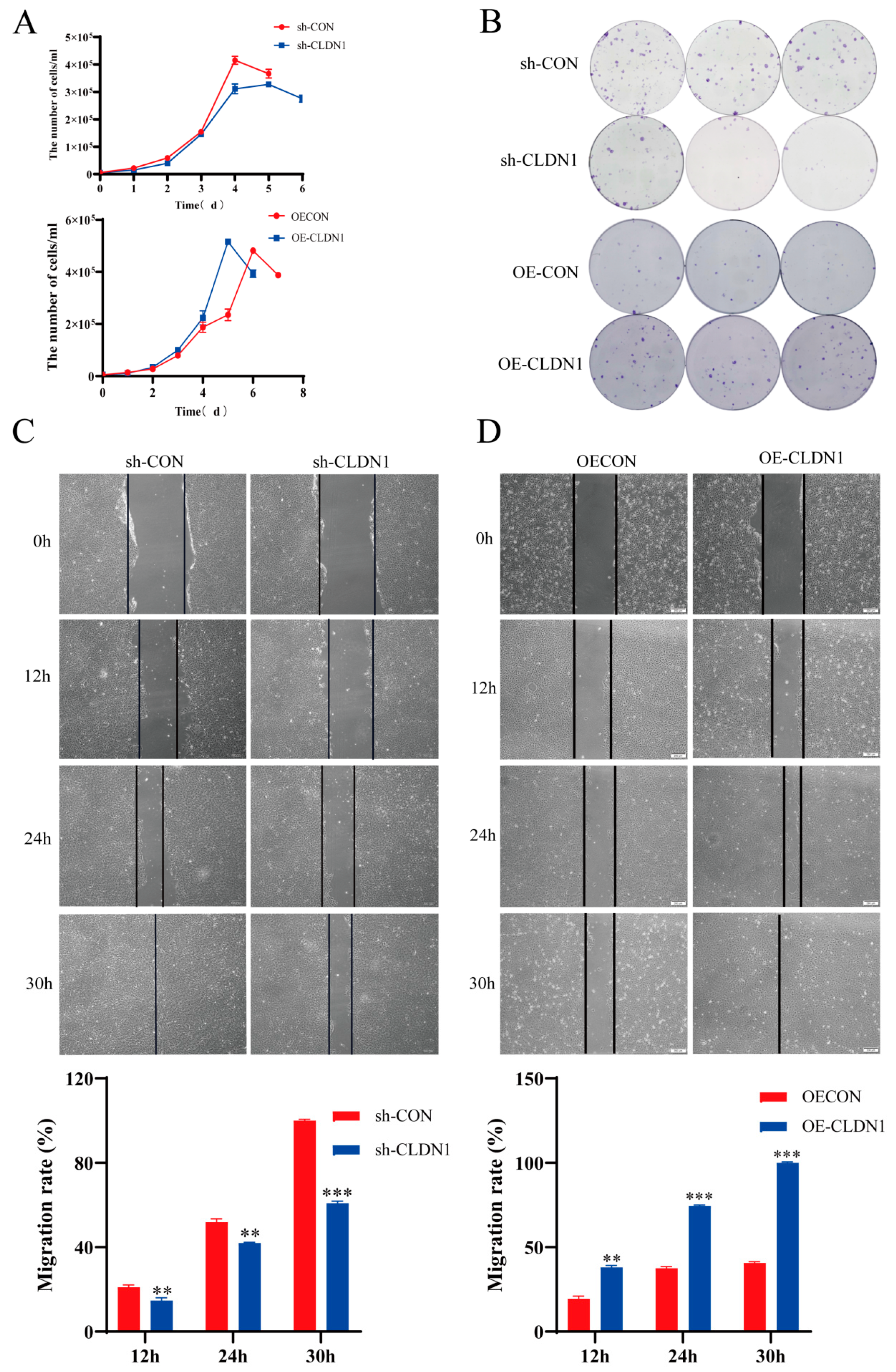
2.4. CLDN1 Promotes MDCK Cell Proliferation and Migration
2.5. miR-175 Directly Targets the CLDN1 3′-UTR
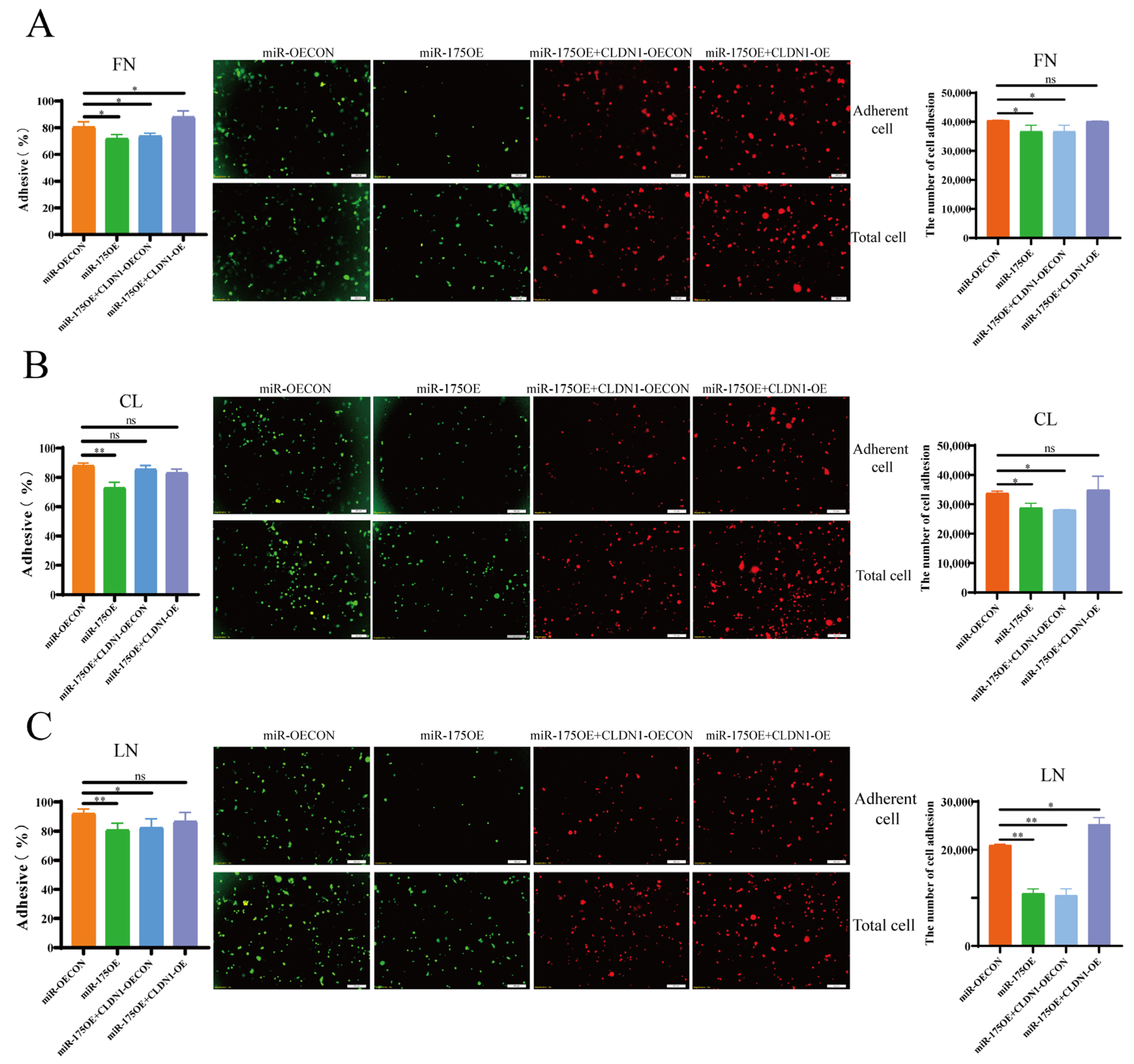
2.6. miR-175 Inhibits MDCK Cell Adhesion by Targeting CLDN1
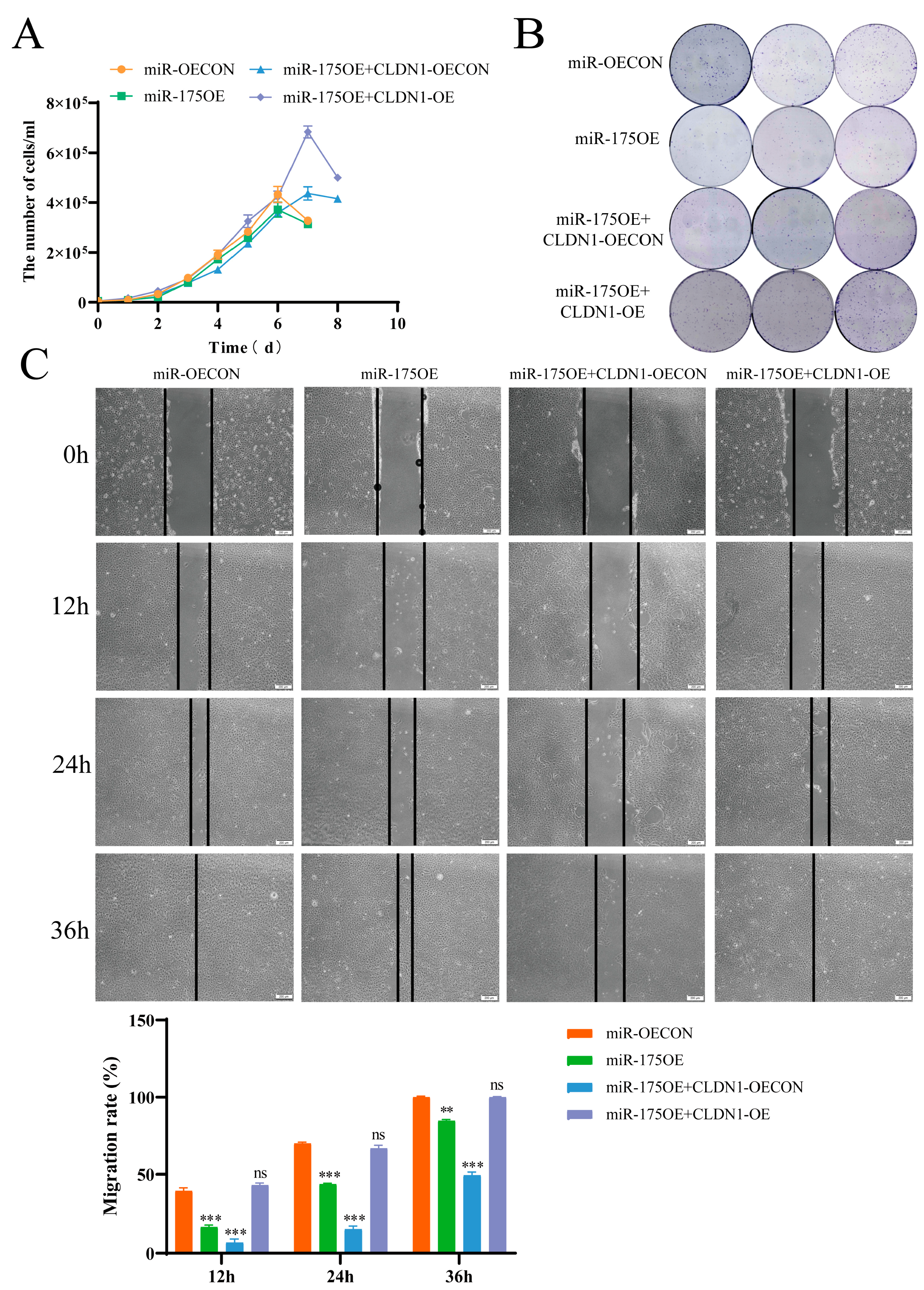
2.7. miR-175 Targets CLDN1 to Inhibit MDCK Cell Proliferation and Migration
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. mRNA Library Establishment
4.3. miRNA Library Establishment
4.4. Reverse Transcription and Polymerase Chain Reaction
4.5. Western Bloting
4.6. miR-175 Overexpression, CLDN1 Knockdown, Overexpression Cell Construction, and the Screening of Stable Cell Lines
4.7. Dual Luciferase Reporter Gene Test
4.8. Cell Proliferation Experiments
4.9. Cell Migration Assay
4.10. Cell Adhesion Assay
4.11. mRNA and miRNA Bioinformatics Analysis
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ren, Z.H.; Lu, Z.Z.; Wang, L. Rapid production of a H9N2 influenza vaccine from MDCK cells for protecting chicken against influenza virus infection. Microbiol. Biotechnol. 2015, 99, 2999–3013. [Google Scholar] [CrossRef] [PubMed]
- Itsuki, H.; Hitoshi, T.; Shimazaki, N. Suitability of NIID-MDCK cells as a substrate for cell-based influenza vaccine development from the perspective of adventitious virus susceptibility. Microbiol. Immunol. 2022, 66, 361–370. [Google Scholar] [CrossRef]
- Alexander, D.; Scott, A. Trivalent MDCK cell culture-derived influenza vaccine Optaflu® (Novartis Vaccines). Expert Rev. Vaccines 2009, 8, 679–688. [Google Scholar] [CrossRef]
- Pech, S.; Rehberg, M.; Janke, R.; Benndorf, D.; Genzel, Y.; Muth, T.; Sickmann, A.; Rapp, E. Tracking changes in adaptation to suspension growth for MDCK cells: Cell growth correlates with levels of metabolites, enzymes and proteins. Microbiol. Biotechnol. 2021, 105, 1861–1874. [Google Scholar] [CrossRef]
- Chu, C.; Lugovtsev, V.; Golding, H.; Betenbaugh, M.; Shiloach, J. Conversion of MDCK cell line to suspension culture by transfecting with human siat7e gene and its application for influenza virus production. Proc. Natl. Acad. Sci. USA 2009, 106, 14802–14807. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Obeng, G.; Park, E.J.; Appiah, M.G.; Kawamoto, E.; Gaowa, A.; Shimaoka, M. miRNA-200c-3p targets talin-1 to regulate integrin-mediated cell adhesion. Sci. Rep. 2021, 11, 21597. [Google Scholar] [CrossRef]
- Hunt, S.; Jones, A.V.; Hinsley, E.E.; Whawell, S.A.; Lambert, D.W. MicroRNA-124 suppresses oral squamous cell carcinoma motility by targeting ITGB1. FEBS Lett. 2010, 585, 187–192. [Google Scholar] [CrossRef]
- Hozaka, Y.; Seki, N.; Tanaka, T.; Asai, S.; Moriya, S.; Idichi, T.; Wada, M.; Tanoue, K.; Kawasaki, Y.; Mataki, Y.; et al. Molecular Pathogenesis and Regulation of the miR-29-3p-Family: Involvement of ITGA6 and ITGB1 in Intra-Hepatic Cholangiocarcinoma. Cancers 2021, 13, 2804. [Google Scholar] [CrossRef]
- Gong, C.; Yang, Z.; Wu, F.; Han, L.; Liu, Y.; Gong, W. miR-17 inhibits ovarian cancer cell peritoneal metastasis by targeting ITGA5 and ITGB1. Oncol. Rep. 2016, 36, 2177–2183. [Google Scholar] [CrossRef]
- Carolina, O.R.; María, C.O.; Rafael, S.F. Hsa-miR-183-5p Modulates Cell Adhesion by Repression of ITGB1 Expression in Prostate Cancer. Non-Coding RNA 2022, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Wei, F.; Zhang, J.; Li, B. miR-134 suppresses the migration and invasion of non-small cell lung cancer by targeting ITGB1. Oncol. Rep. 2017, 37, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Jin, X.; Li, Z.; Chen, X.; Zhao, J. miR-152-3p facilitates cell adhesion and hepatic metastases in colorectal cancer via targeting AQP11. Pathol. Res. Pract. 2023, 244, 154–189. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Haemmig, S.; Chen, J.; McCoy, M.; Cheng, H.S.; Zhou, H.; Pérez-Cremades, D.; Cheng, X.; Sun, X.; Haneo-Mejia, J.; et al. Endothelial cell-specific deletion of a microRNA accelerates atherosclerosis. Atherosclerosis 2022, 350, 9–18. [Google Scholar] [CrossRef]
- Süren, D.; Yildirim, M.; Sayïner, A.; Alïkanoğlu, A.S.; Atalay, I.; Gündüz, U.R.; Kaya, V.; Gündüz, Ş.; Oruç, M.T.; Sezer, C. Expression of claudin 1, 4 and 7 in thyroid neoplasms. Oncol. Lett. 2017, 13, 3722–3726. [Google Scholar] [CrossRef]
- Saurabh, D.; Rajat, S. Blood-Brain Barrier Permeability Is Exacerbated in Experimental Model of Hepatic Encephalopathy via MMP-9 Activation and Downregulation of Tight Junction Proteins. Mol. Neurobiol. 2017, 55, 3642–3659. [Google Scholar] [CrossRef]
- Wu, J.; Gao, F.; Xu, T.; Li, J.; Hu, Z.; Wang, C.; Long, Y.; He, X.; Deng, X.; Ren, D.; et al. CLDN1 induces autophagy to promote proliferation and metastasis of esophageal squamous carcinoma through AMPK/STAT1/ULK1 signaling. J. Cell. Physiol. 2019, 235, 2245–2259. [Google Scholar] [CrossRef]
- Takasawa, K.; Takasawa, A.; Akimoto, T.; Magara, K.; Aoyama, T.; Kitajima, H.; Murakami, T.; Ono, Y.; Kyuno, D.; Suzuki, H.; et al. Regulatory roles of claudin-1 in cell adhesion and microvilli formation. Biochem. Biophys. Res. Commun. 2021, 565, 36–42. [Google Scholar] [CrossRef]
- Kim, N.Y.; Pyo, J.S.; Kang, D.W.; Yoo, S.M. Loss of claudin-1 expression induces epithelial-mesenchymal transition through nuclear factor-κB activation in colorectal cancer. Pathol. Res. Pract. 2019, 215, 580–585. [Google Scholar] [CrossRef]
- Geoffroy, M.; Kleinclauss, A.; Kuntz, S.; Grillier-Vuissoz, I. Claudin 1 inhibits cell migration and increases intercellular adhesion in triple-negative breast cancer cell line. Mol. Biol. Rep. 2020, 47, 7643–7653. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Lauko, A.; Mu, Z.; Gutmann, D.H.; Naik, U.P.; Lathia, J.D. Junctional Adhesion Molecules in Cancer: A Paradigm for the Diverse Functions of Cell–Cell Interactions in Tumor Progression. Cancer Res. 2020, 80, 4878–4885. [Google Scholar] [CrossRef]
- Rodrigues, A.F.; Fernandes, P.; Laske, T.; Castro, R.; Alves, P.M.; Genzel, Y.; Coroadinha, A.S. Cell Bank Origin of MDCK Parental Cells Shapes Adaptation to Serum-Free Suspension Culture and Canine Adenoviral Vector Production. Int. J. Mol. Sci. 2020, 21, 6111. [Google Scholar] [CrossRef]
- Schröder, M.; Matischak, K.; Friedl, P. Serum- and protein-free media formulations for the Chinese hamster ovary cell line DUKXB11. J. Biotechnol. 2004, 18, 279–292. [Google Scholar] [CrossRef]
- Paillet, C.; Forno, G.; Kratje, R.; Etcheverrigaray, M. Suspension-Vero cell cultures as a platform for viral vaccine production. Vaccine 2009, 27, 6464–6467. [Google Scholar] [CrossRef]
- Malm, M.; Saghaleyni, R.; Lundqvist, M.; Giudici, M.; Chotteau, V.; Field, R.; Varley, P.G.; Hatton, D.; Grassi, L.; Svensson, T.; et al. Evolution from adherent to suspension: Systems biology of HEK293 cell line development. Sci. Rep. 2020, 10, 18996. [Google Scholar] [CrossRef]
- Dill, V.; Hoffmann, B.; Zimmer, A.; Beer, M.; Eschbaumer, M. Influence of cell type and cell culture media on the propagation of foot-and-mouth disease virus with regard to vaccine quality. Virol. J. 2018, 15, 46. [Google Scholar] [CrossRef]
- Zhang, J.; Qiu, Z.; Wang, S.; Liu, Z.; Qiao, Z.; Wang, J.; Duan, K.; Nian, X.; Ma, Z.; Yang, X. Suspended cell lines for inactivated virus vaccine production. Expert Rev. Vaccines 2023, 22, 468–480. [Google Scholar] [CrossRef]
- Krneta-Stankic, V.; Corkins, M.E.; Paulucci-Holthauzen, A. The Wnt/PCP formin Daam1 drives cell-cell adhesion during nephron development. Cell Rep. 2021, 36, 109–340. [Google Scholar] [CrossRef]
- Takuya, S. Regulation of the intestinal barrier by nutrients: The role of tight junctions. Anim. Sci. J. 2020, 91, e13357. [Google Scholar] [CrossRef]
- Xiao, K.; Song, Z.-H.; Jiao, L.-F.; Ke, Y.-L.; Hu, C.-H. Tight junction protein levels of occludin, claudin-1 and zonula occludens-1 (ZO-1). PLoS ONE 2014, 9, e104589. [Google Scholar] [CrossRef]
- You-Cheng, H.; Yu-Hsien, C.; Lin, K.Y.; Tzong-Der, W.; Lin, H.Y.; Varadharajan, T.; Hsin-Ling, Y. Antrodia camphorata inhibits metastasis and epithelial-to-mesenchymal transition via the modulation of claudin-1 and Wnt/β-catenin signaling pathways in human colon cancer cells. J. Ethnopharmacol. 2017, 208, 72–83. [Google Scholar] [CrossRef]
- Osada, T.; Gu, Y.-H.; Kanazawa, M.; Tsubota, Y.; Hawkins, B.T.; Spatz, M.; Milner, R.; del Zoppo, G.J. Interendothelial Claudin-5 Expression Depends on Cerebral Endothelial Cell–Matrix Adhesion by β1-Integrins. J. Cereb. Blood Flow Metab. 2011, 31, 1972–1985. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Lu, Q.; Chen, Y. Claudin-7 modulates cell-matrix adhesion that controls cell migration, invasion and attachment of human HCC827 lung cancer cells. Oncol. Lett. 2019, 17, 2890–2896. [Google Scholar] [CrossRef]
- Fuchs, M.T.; Foresti, M.; Vielmuth, F.; Waschke, J. Plakophilin-dependent Dsg3 oligomerization and binding properties are involved in desmosomal hyper-adhesion. FASEB J. 2019, 33, 662.38. [Google Scholar] [CrossRef]
- González-Mariscal, L.; Miranda, J.; Raya-Sandino, A.; Domínguez-Calderón, A.; Cuellar-Perez, F. ZO-2, a tight junction protein involved in gene expression, proliferation, apoptosis, and cell size regulation. Ann. N. Y. Acad. Sci. 2017, 1397, 35–53. [Google Scholar] [CrossRef]
- Yu, S.; Yan, C.; Wu, W.; He, S.; Liu, M.; Liu, J.; Yang, X.; Ma, J.; Lu, Y.; Jia, L. RU486 Metabolite Inhibits CCN1/Cyr61 Secretion by MDA-MB-231-Endothelial Adhesion. Front. Pharmacol. 2019, 10, 1296. [Google Scholar] [CrossRef]
- Villanueva, A.A.; Sanchez-Gomez, P.; Muñoz-Palma, E.; Puvogel, S.; Casas, B.S.; Arriagada, C.; Peña-Villalobos, I.; Lois, P.; Orellana, M.R.; Lubieniecki, F.; et al. The Netrin-1-Neogenin-1 signaling axis controls neuroblastoma cell migration via integrin-β1 and focal adhesion kinase activation. Cell Adhes. Migr. 2021, 15, 58–73. [Google Scholar] [CrossRef]
- Yumi, S. Possible involvement of crosstalk cell-adhesion mechanism by endometrial CD26/dipeptidyl peptidase IV and embryonal fibronectin in human blastocyst implantation. Mol. Hum. Reprod. 2006, 12, 491–495. [Google Scholar] [CrossRef]
- Khademi, R.; Malekzadeh, H.; Bahrami, S.; Saki, N.; Khademi, R.; Villa-Diaz, L.G. Regulation and Functions of α6-Integrin (CD49f) in Cancer Biology. Cancers 2023, 15, 3466. [Google Scholar] [CrossRef]
- Resnick, M.B.; Konkin, T.; Routhier, J. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: A tissue microarray study. Mod. Pathol. 2005, 18, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.-C.; Pan, S.-H.; Yang, S.-C.; Yu, S.-L.; Che, T.-F.; Lin, C.-W.; Tsai, M.-S.; Chang, G.-C.; Wu, C.-H.; Wu, Y.-Y.; et al. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am. J. Respir. Crit. Care Med. 2009, 179, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, S.; Kusumi, T.; Sato, F.; Odagiri, H.; Chiba, H.; Yoshihara, S.; Hakamada, K.; Sasaki, M.; Kijima, H. Decreased expression of claudin-1 correlates with recurrence status in breast cancer. Int. J. Mol. Med. 2007, 20, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, P.; Singh, A.B.; Deane, N.G.; No, Y.; Shiou, S.-R.; Schmidt, C.; Neff, J.; Washington, M.K.; Beauchamp, R.D. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J. Clin. Investig. 2005, 115, 1765–1776. [Google Scholar] [CrossRef]
- Suh, Y.; Yoon, C.-H.; Kim, R.-K.; Lim, E.-J.; Oh, Y.S.; Hwang, S.-G.; An, S.; Yoon, G.; Gye, M.C.; Yi, J.-M.; et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene 2013, 32, 4873–4882. [Google Scholar] [CrossRef]
- Oku, N.; Sasabe, E.; Ueta, E. Tight junction protein claudin-1 enhances the invasive activity of oral squamous cell carcinoma cells by promoting cleavage of laminin-5 gamma2 chain via matrix metalloproteinase (MMP)-2 and membrane-type MMP-1. Cancer Res. 2006, 66, 5251–5257. [Google Scholar] [CrossRef]
- Lemesle, M.; Geoffroy, M.; Alpy, F.; Tomasetto, C.-L.; Kuntz, S.; Grillier-Vuissoz, I. CLDN1 Sensitizes Triple-Negative Breast Cancer Cells to Chemotherapy. Cancers 2022, 14, 5026. [Google Scholar] [CrossRef]
- Cherradi, S.; Garambois, V.; Marines, J.; Andrade, A.F.; Fauvre, A.; Morand, O.; Fargal, M.; Mancouri, F.; Ayrolles-Torro, A.; Vezzo-Vié, N.; et al. Improving the response to oxaliplatin by targeting chemotherapy-induced CLDN1 in resistant metastatic colorectal cancer cells. Cell Biosci. 2023, 13, 72–75. [Google Scholar] [CrossRef]
- Akizuki, R.; Maruhashi, R.; Eguchi, H.; Kitabatake, K.; Tsukimoto, M.; Furuta, T.; Matsunaga, T.; Endo, S.; Ikari, A. Decrease in paracellular permeability and chemosensitivity to doxorubicin by claudin-1 in spheroid culture models of human lung adenocarcinoma A549 cells. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 769–780. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, J.; Jiang, Y. CLDN1 increases Drug Resistance of Non-Small Cell Lung Cancer by activating Autophagy via Up-Regulation of ULK1 phosphorylation. Med. Sci. Monit. 2017, 23, 2906–2916. [Google Scholar] [CrossRef]
- Sun, N.; Zhang, Y.; Dong, J.; Liu, G.; Liu, Z.; Wang, J.; Qiao, Z.; Zhang, J.; Duan, K.; Nian, X.; et al. Metabolomics profiling reveals differences in proliferation between tumorigenic and non-tumorigenic Madin-Darby canine kidney (MDCK) cells. PeerJ 2023, 11, e16077. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323–324. [Google Scholar] [CrossRef]
- Ding, R.; Qu, Y.; Wu, C.H.; Vijay-Shanker, K. Automatic gene annotation using GO terms from cellular component domain. BMC Med. Inform. Decis. Mak. 2018, 18, 119. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Kumar, L.; Futschik, M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 25–34. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Kivioja, T.; Vähärautio, A.; Karlsson, K.; Bonke, M.; Enge, M.; Linnarsson, S.; Taipale, J. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 2011, 9, 72–74. [Google Scholar] [CrossRef] [PubMed]
- Jan, W.K.; Marc, R. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA Targets. PLoS Biol. 2004, 2, e363. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Zhou, Z.; Lunetta, K.L.; Smith, A.K.; Wolf, E.J.; Stone, A.; A Schichman, S.; E McGlinchey, R.; Milberg, W.P.; Miller, M.W.; Logue, M.W. Correction for multiple testing in candidate-gene methylation studies. Epigenomics 2019, 11, 1089–1105. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Ma, F.; Wang, S.; Tang, T.; Ma, L.; Qiao, Z.; Ma, Z.; Wang, J.; Liu, Z. Micro RNA-175 Targets Claudin-1 to Inhibit Madin–Darby Canine Kidney Cell Adhesion. Genes 2024, 15, 1333. https://doi.org/10.3390/genes15101333
Li X, Ma F, Wang S, Tang T, Ma L, Qiao Z, Ma Z, Wang J, Liu Z. Micro RNA-175 Targets Claudin-1 to Inhibit Madin–Darby Canine Kidney Cell Adhesion. Genes. 2024; 15(10):1333. https://doi.org/10.3390/genes15101333
Chicago/Turabian StyleLi, Xiaoyun, Fangfang Ma, Siya Wang, Tian Tang, Liyuan Ma, Zilin Qiao, Zhongren Ma, Jiamin Wang, and Zhenbin Liu. 2024. "Micro RNA-175 Targets Claudin-1 to Inhibit Madin–Darby Canine Kidney Cell Adhesion" Genes 15, no. 10: 1333. https://doi.org/10.3390/genes15101333
APA StyleLi, X., Ma, F., Wang, S., Tang, T., Ma, L., Qiao, Z., Ma, Z., Wang, J., & Liu, Z. (2024). Micro RNA-175 Targets Claudin-1 to Inhibit Madin–Darby Canine Kidney Cell Adhesion. Genes, 15(10), 1333. https://doi.org/10.3390/genes15101333




