Circadian Rhythm Alteration of the Core Clock Genes and the Lipid Metabolism Genes Induced by High-Fat Diet (HFD) in the Liver Tissue of the Chinese Soft-Shelled Turtle (Trionyx sinensis)
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Diets
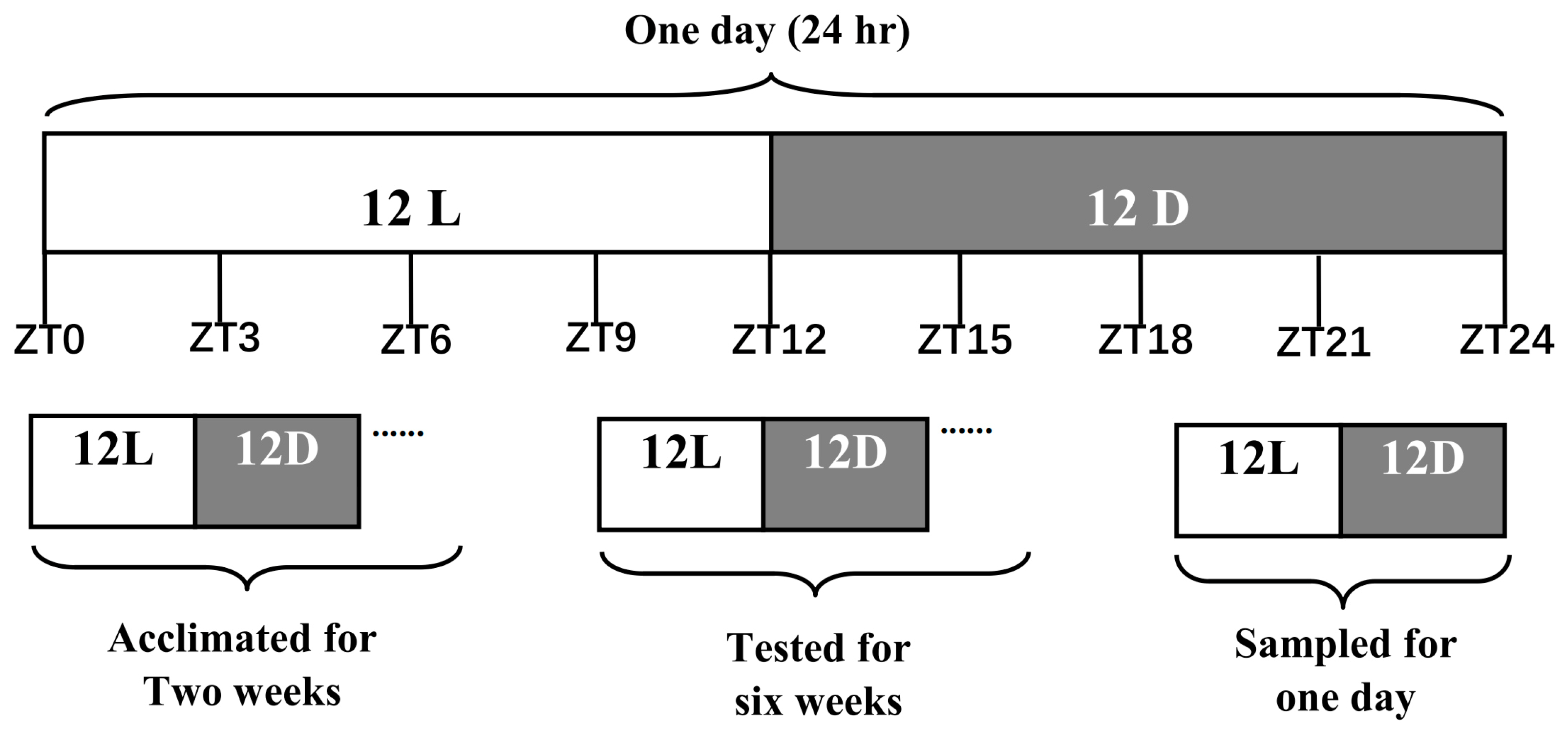
2.2. Animals and Experimental Design
2.3. Sample Collection
2.4. RNA Extraction and Expression by Quantitative Real-Time PCR (RT-qPCR) Analysis
2.5. Statistical Analysis
3. Results
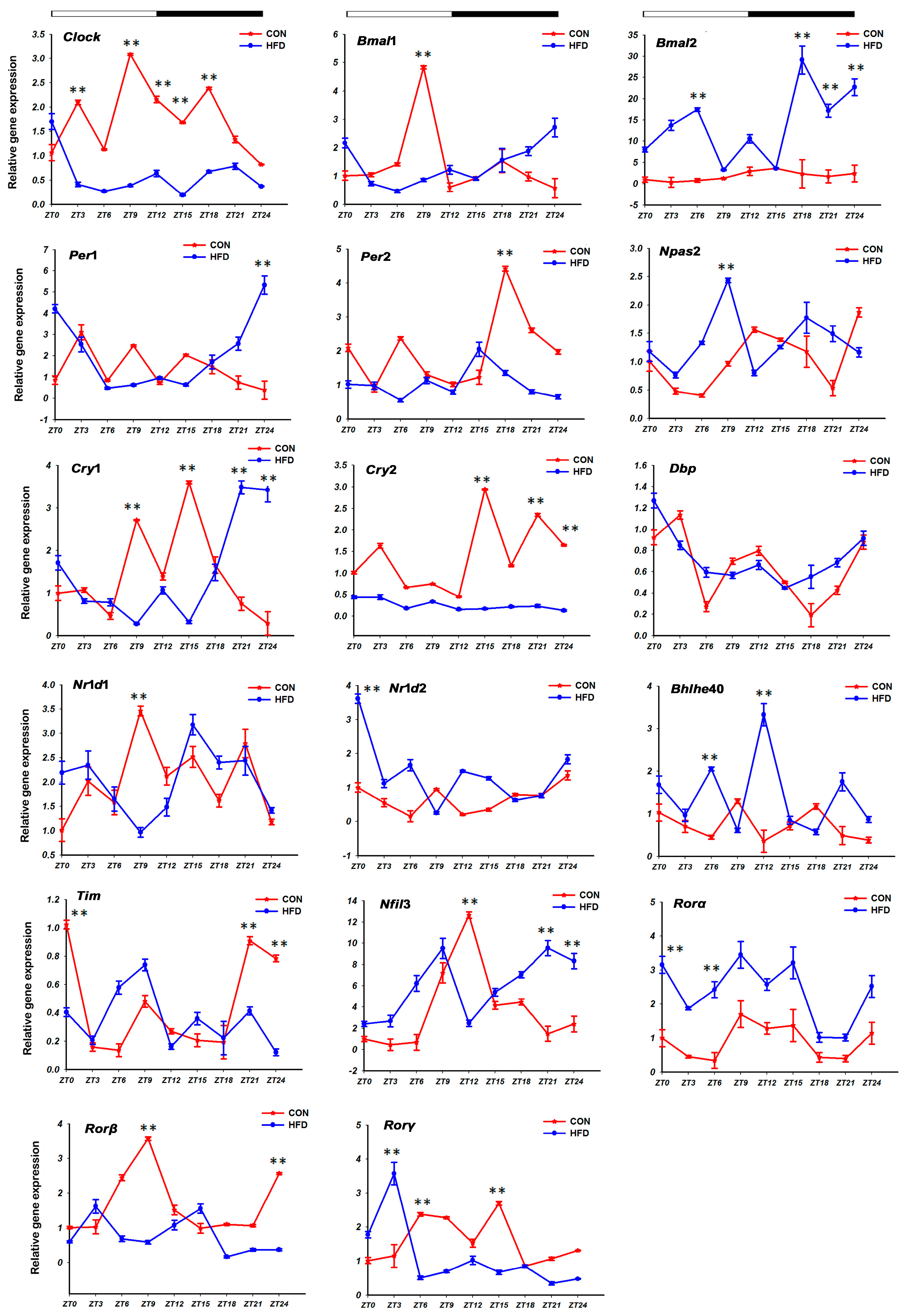
3.1. The HFD Altered Rhythmic mRNA Expression of the Core Clock Genes in Liver Tissue
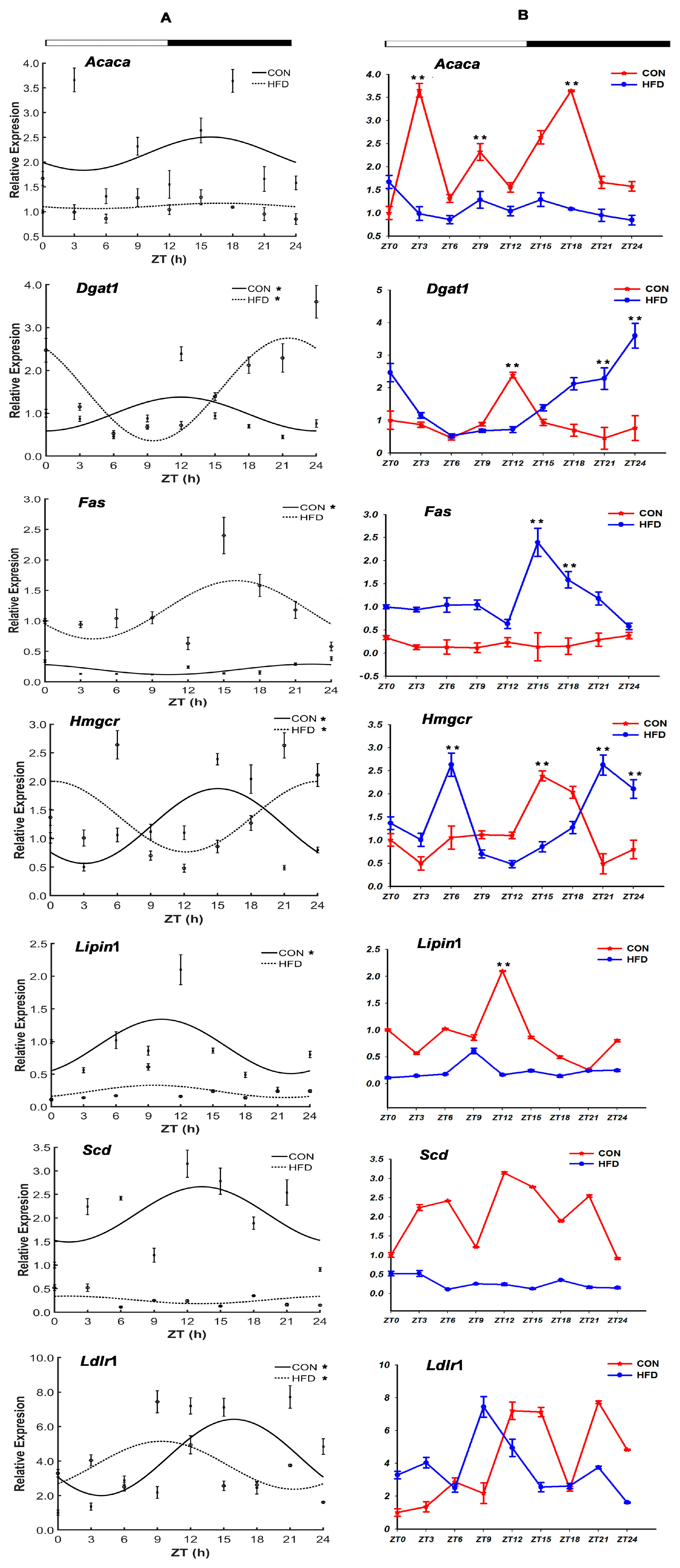
3.2. The HFD Altered Rhythmic mRNA Expression of the Lipid Metabolism Genes in the Liver Tissue
3.2.1. The Lipid Synthesis-Related Genes
3.2.2. The Lipid Oxygenolysis-Related Genes
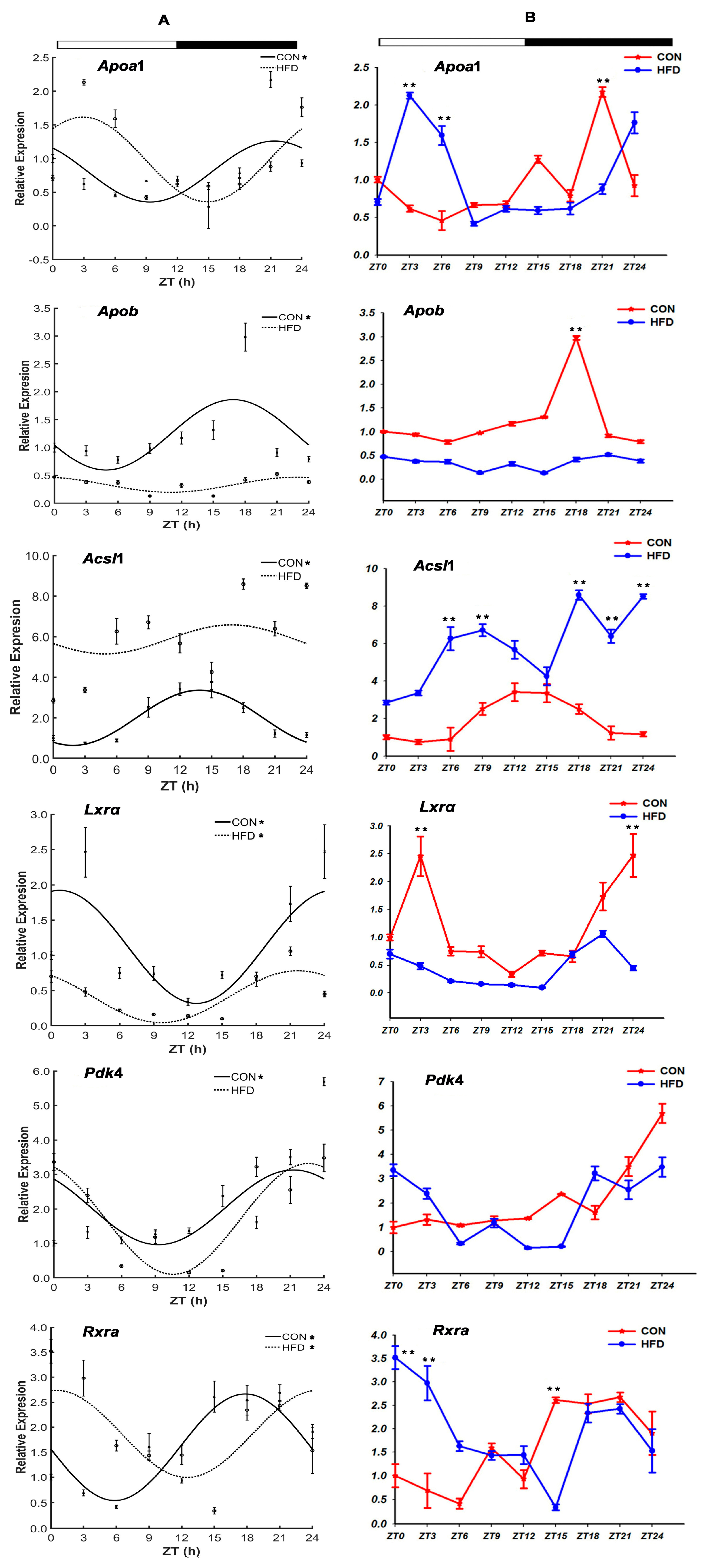
3.2.3. The Lipid Transport-Related Genes
3.3. The Correlation Analysis on Daily Expression between the Core Clock Genes and the Lipid Metabolism Genes in Liver Tissue
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jandot, A.; Calligaro, H.; Dkhissi-Benyahya, O. Endogenous functioning and light response of the retinal clock in vertebrates. Prog. Brain Res. 2022, 273, 49–69. [Google Scholar] [PubMed]
- Florian, A.; Daniel, M.; Benjamin, W.; Cédric, G.; Frédéric, G. Regulation of mammalian physiology by interconnected circadian and feeding rhythms. Front. Endocrinol. 2017, 8, 42. [Google Scholar]
- Tu, H.Q.; Li, S.; Xu, Y.L.; Zhang, Y.C.; Li, P.Y.; Liang, L.Y. Rhythmic cilia changes support scn neuron coherence in circadian clock. Science 2023, 380, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jiang, G.; Peng, Z.; Li, Y.; Wang, X.; Chu, W. The effect of high fat diet on daily rhythm of the core clock genes and muscle functional genes in the skeletal muscle of chinese soft-shelled turtle (Trionyx sinensis). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 213, 17–27. [Google Scholar] [CrossRef]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Bass, J. Circadian topology of metabolism. Nature 2012, 491, 348–356. [Google Scholar] [CrossRef]
- Bozek, K.; Relógio, A.; Kielbasa, S.M.; Heine, M.; Dame, C.; Kramer, A. Regulation of clock-controlled genes in mammals. PLoS ONE 2009, 4, e4882. [Google Scholar] [CrossRef]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U. The orphan nuclear receptor rev-erbalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Nakajima, Y. Bidirectional role of orphan nuclear receptor rorα in clock gene transcriptions demonstrated by a novel reporter assay system. FEBS Lett. 2004, 565, 122–126. [Google Scholar] [CrossRef]
- Reinke, H.; Asher, G. Circadian clock control of liver metabolic functions. Gastroenterology 2016, 150, 574–580. [Google Scholar] [CrossRef]
- Akhtar, R.A.; Reddy, A.B.; Maywood, E.S.; Clayton, J.D.; King, V.M. Circadian cycling of the mouse liver transcriptome, as revealed by cdna microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 2002, 12, 540–550. [Google Scholar] [CrossRef]
- Kornmann, B.; Schaad, O.; Reinke, H.; Saini, C.; Schibler, U. Regulation of circadian gene expression in liver by systemic signals and hepatocyte oscillators. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 319–330. [Google Scholar] [CrossRef]
- Acimovic, J.; Fink, M.; Pompon, D.; Bjorkhem, I.; Hirayama, J.; Sassone-Corsi, P. Crem modulates the circadian expression of cyp51, hmgcr and cholesterogenesis in the liver. Biochem. Biophys. Res. Commun. 2008, 376, 206–210. [Google Scholar] [CrossRef]
- Delezie, J.; Dumont, S.; Dardente, H.; Oudart, H.; Gréchez-Cassiau, A.; Klosen, P.; Teboul, M.; Delaunay, F.; Pévet, P.; Challet, E. The nuclear receptor rev-erbα is required for the daily balance of carbohydrate and lipid metabolism. Faseb J. 2012, 26, 3321–3335. [Google Scholar] [CrossRef]
- Nakahata, Y.; Kaluzova, M.; Grimaldi, B.; Sahar, S.; Hirayama, J.; Chen, D. The NAD+-Dependent Deacetylase SIRT1 Modulates CLOCK-Mediated Chromatin Remodeling and Circadian Control. Cell 2008, 134, 329–340. [Google Scholar] [CrossRef]
- Bellet, M.M.; Orozcosolis, R.; Sahar, S.; Eckelmahan, K.; Sassonecorsi, P. The time of metabolism: Nad+, sirt1, and the circadian clock. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 31–38. [Google Scholar] [CrossRef]
- Grimaldi, B.; Bellet, M.M.; Katada, S.; Astarita, G.; Hirayama, J.; Amin, R.H. Per2 controls lipid metabolism by direct regulation of pparγ. Cell Metab. 2010, 12, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Canaple, L.; Rambaud, J.; Dkhissi-Benyahya, O.; Rayet, B.; Tan, N.S.; Michalik, L. Reciprocal regulation of brain and muscle arnt-like protein 1 and peroxisome proliferator-activated receptor α defines a novel positive feedback loop in the rodent liver circadian clock. Mol. Endocrinol. 2006, 20, 1715–1727. [Google Scholar] [CrossRef] [PubMed]
- Kohsaka, A.; Bass, J. A sense of time: How molecular clocks organize metabolism. Trends Endocrinol. Metab. 2007, 18, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Downes, M.; Yu, R.T.; Bookout, A.L.; He, W.; Straume, M. Nuclear receptor expression links the circadian clock to metabolism. Cell 2006, 126, 801–810. [Google Scholar] [CrossRef]
- Nakamura, K.I.; Inoue, I.; Takahashi, S.; Komoda, T.; Katayama, S. Cryptochrome and period proteins are regulated by the clock/bmal1 gene: Crosstalk between the ppars/rxr-regulated and clock/bmal1-regulated systems. PPAR Res. 2008, 100, 348610. [Google Scholar] [CrossRef]
- Ming, C.C.; Chou, C.J.; Boozer, C.N. High-fat diet feeding reduces the diurnal variation of plasma leptin concentration in rats. Metab. Clin. Exp. 2000, 49, 503–507. [Google Scholar]
- Havel, P.J.; Townsend, R.; Chaump, L. High-fat meals reduce 24-h circulating leptin concentration in women. Diabetes 1999, 48, 334–341. [Google Scholar] [CrossRef]
- Maayan, B.; Zecharia, M.; Oren, F. High-fat diet delays and fasting advances the circadian expression of adiponectin signaling components in mouse liver. Endocrinology 2009, 1, 161–168. [Google Scholar]
- Li, H.H.; Pan, Y.X.; Liu, L.; Li, Y.L.; Shen, Y.D. Effects of high-fat diet on muscle textural properties, antioxidant status and autophagy of chinese soft-shelled turtle (Pelodiscus sinensis). Aquaculture 2019, 511, 734228. [Google Scholar] [CrossRef]
- Zhong, Y.W.; Pan, Y.X.; Liu, L.; Li, H.H.; Li, Y.L.; Jiang, J.; Xiang, J.; Zhang, J.S.; Chu, W.Y. Effects of high fat diet on lipid accumulation, oxidative stress and autophagy in the liver of chinese softshell turtle (Pelodiscus sinensis). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2020, 240, 110331. [Google Scholar] [CrossRef] [PubMed]
- China Fisheries Administration of Ministry of Agriculture and Rural Affairs; China National Aquatic Technology Promotion Station; China Fishery Society. 2020 China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2020. (In Chinese) [Google Scholar]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, Y.L.; Liu, L.; Wang, J.H.; Li, H.H.; Wu, P.; Zhang, J.S.; Chu, W.Y. Molecular characterization of myf5 and comparative expression patterns of myogenic regulatory factors in Siniperca chuatsi. Gene Expr. Patterns 2016, 20, 1–10. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef]
- Lazado, C.C.; Kumaratunga, H.; Nagasawa, K.; Babiak, I.; Giannetto, A.; Fernandes, J. Daily rhythmicity of clock gene transcripts in atlantic cod fast skeletal muscle. PLoS ONE 2014, 9, e99172. [Google Scholar] [CrossRef]
- Velarde, E.; Haque, R.; Iuvone, P.M.; Azpeleta, C.; Alonso-Gomez, A.L.; Delgado, M.J. Circadian clock genes of goldfish, Carassius auratus: cDNA cloning and rhythmic expression of period and cryptochrome transcripts in retina, liver, and gut. J. Biol. Rhythm. 2009, 24, 104–113. [Google Scholar] [CrossRef]
- Wu, P.; Bao, L.; Zhang, R.; Li, Y.; Liu, L.; Wu, Y. Impact of Short-Term Fasting on The Rhythmic Expression of the Core Circadian Clock and Clock-Controlled Genes in Skeletal Muscle of Crucian Carp (Carassius auratus). Genes 2018, 9, 526. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Chao, L.; Huang, Y.; Chen, S.; Qian, R. Effect of hyperlipidemia on the expression of circadian genes in apolipoprotein e knock-out atherosclerotic mice. Lipids Health Dis. 2009, 8, 60. [Google Scholar] [CrossRef]
- Hou, X.; Xu, S.; Maitland-Toolan, K.A.; Sato, K.; Jiang, B.; Ido, Y. Sirt1 regulates hepatocyte lipid metabolism through activating amp-activated protein kinase. J. Biol. Chem. 2008, 283, 20015–20026. [Google Scholar] [CrossRef]
- Kelley, D.E.; Bret, G.; Wing, R.R.; Jean-Aime, S. Skeletal muscle fatty acid metabolism in association with insulin resistance, obesity, and weight loss. AJP Endocrinol. Metab. 1999, 277, E1130–E1141. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, J.T.; Puigserver, P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl. Acad. Sci. USA 2007, 104, 12861–12866. [Google Scholar] [CrossRef]
- Zheng, S.; Liang, X.; Rogers, C.Q.; Rideout, D.; Min, Y. Involvement of adiponectin-sirt1-ampk signaling in the protective action of rosiglitazone against alcoholic fatty liver in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G364–G374. [Google Scholar]
- Ando, H.; Yanagihara, H.; Hayashi, Y.; Obi, Y.; Tsuruoka, S.; Takamura, T.; Kaneko, S.; Fujimura, A. Rhythmic messenger ribonucleic acid expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology 2005, 146, 5631–5636. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jie, X.; Yang, J.; Xie, M. Timed high-fat diet in the evening affects the hepatic circadian clock and pparα-mediated lipogenic gene expressions in mice. Genes Nutr. 2013, 8, 457–463. [Google Scholar] [CrossRef]
- Yanagihara, H.; Ando, H.; Hayashi, Y.; Obi, Y.; Fujimura, A. High-fat feeding exerts minimal effects on rhythmic mrna expression of clock genes in mouse peripheral tissues. Chronobiol. Int. 2009, 23, 905–914. [Google Scholar] [CrossRef]
- Huang, Y.; McNeil, G.P.; Jackson, R.F. Translational Regulation of the DOUBLETIME/CKIδ/ε Kinase by LARK Contributes to Circadian Period Modulation. PLoS Genet. 2014, 10, e1004536. [Google Scholar] [CrossRef] [PubMed]
- Llabre, J.E.; Trujillo, R.; Sroga, G.E.; Figueiro, M.G.; Vashishth, D. Circadian rhythm disruption with high-fat diet impairs glycemic control and bone quality. FASEB J. 2021, 9, 35. [Google Scholar] [CrossRef]
- Palmisano, B.T.; Stafford, J.M.; Pendergast, J.S. High-fat feeding does not disrupt daily rhythms in female mice because of protection by ovarian hormones. Front. Endocrinol. 2010, 8, 44. [Google Scholar] [CrossRef]
- Asher, G.; Gatfield, D.; Stratmann, M.; Reinke, H.; Dibner, C.; Kreppel, F. Sirt1 regulates circadian clock gene expression through per2 deacetylation. Cell 2008, 134, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Betancor, M.B.; Elsbeth, M.S.; Matteo, M.; Hervé, M.; Tocher, D.R.; Andrew, D. Daily rhythms in expression of genes of hepatic lipid metabolism in atlantic salmon (Salmo salar L.). PLoS ONE 2014, 9, e106739. [Google Scholar] [CrossRef]
- Martino, T.A.; Tata, N.; Belsham, D.D.; Chalmers, J.; Straume, M.; Lee, P. Disturbed diurnal rhythm alters gene expression and exacerbates cardiovascular disease with rescue by resynchronization. Hypertension 2007, 49, 1104–1113. [Google Scholar] [CrossRef]
- Parkes, H.A.; Preston, E.; Wilks, D.; Ballesteros, M.; Carpenter, L.; Wood, L. Overexpression of acyl-coa synthetase-1 increases lipid deposition in hepatic (hepg2) cells and rodent liver in vivo. Am. J. Physiol. Endocrinol. Metab. 2006, 291, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Bass, J. Obesity and metabolic syndrome in circadian clock mutant mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef]
- Lefebvre, P.; Chinetti, G.; Fruchart, J.C.; Staels, B. Sorting out the roles of pparα in energy metabolism and vascular homeostasis. J. Clin. Investig. 2006, 16, 571–580. [Google Scholar] [CrossRef]
- Sugden, M.C.; Caton, P.W.; Holness, M.J.; Miller, J.J. Peroxisome proliferator-activated receptors. Ref. Modul. Life Sci. 2021, 6, 229–235. [Google Scholar]
- Gilde, A.J. Peroxisome proliferator-activated receptor (ppar) α and pparβ/δ, but not pparγ, modulate the expression of genes involved in cardiac lipid metabolism. Circ. Res. 2003, 92, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Zhang, D.D.; Jiang, G.Z.; Li, X.F.; Zhang, C.N.; Zhou, M. Cloning and characterization of microsomal triglyceride transfer protein gene and its potential connection with peroxisome proliferator-activated receptor (PPAR) in blunt snout bream (Megalobrama amblycephala). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2015, 189, 23–33. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Lu, M.; Shi, H.; Yue, G.; Luo, H. Vaspin regulated cartilage cholesterol metabolism through mir155/lxrα and participated in the occurrence of osteoarthritis in rats. Life Sci. 2021, 269, 119096. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.H.; Wu, T.; Jin, L.; Yu, B.; Zhu, G.F.; Fu, Z.W. Chronobiology research on mammalian nutrition and metabolism. Chin. J. Cell Biol. 2011, 33, 190–196. (In Chinese) [Google Scholar]
- Gao, L.; Wang, K.; Cheng, M.; Zeng, Z.; Wang, T.; Wen, F. Circadian clock dysfunction of epithelial cells in pulmonary diseases. Int. J. Biochem. Cell Biol. 2021, 141, 106110. [Google Scholar] [CrossRef] [PubMed]
- Olaniyi, K.S.; Akintayo, C.O.; Oniyide, A.A.; Omoaghe, A.O.; Oyeleke, M.B.; Fafure, A.A. Acetate supplementation restores testicular function by modulating nrf2/ppar-γ in high fat diet-induced obesity in wistar rats. J. Diabetes Metab. Disord. 2021, 20, 1685–1696. [Google Scholar] [CrossRef]
- Shende, V.R.; Goldrick, M.M.; Suchitra, R.; Earnest, D.J.; Thomas, P. Expression and rhythmic modulation of circulating micrornas targeting the clock gene bmal1 in mice. PLoS ONE 2011, 6, e22586. [Google Scholar] [CrossRef]
 ”; white part means light, black part means dark; “CON” and “HFD” represent the CON group and the HFD group, respectively; ZT indicates zeitgeber time (h); “*” shows the gene with the characteristic of circadian rhythm in the CON group and the HFD group.
”; white part means light, black part means dark; “CON” and “HFD” represent the CON group and the HFD group, respectively; ZT indicates zeitgeber time (h); “*” shows the gene with the characteristic of circadian rhythm in the CON group and the HFD group.
 ”; white part means light, black part means dark; “CON” and “HFD” represent the CON group and the HFD group, respectively; ZT indicates zeitgeber time (h); “*” shows the gene with the characteristic of circadian rhythm in the CON group and the HFD group.
”; white part means light, black part means dark; “CON” and “HFD” represent the CON group and the HFD group, respectively; ZT indicates zeitgeber time (h); “*” shows the gene with the characteristic of circadian rhythm in the CON group and the HFD group.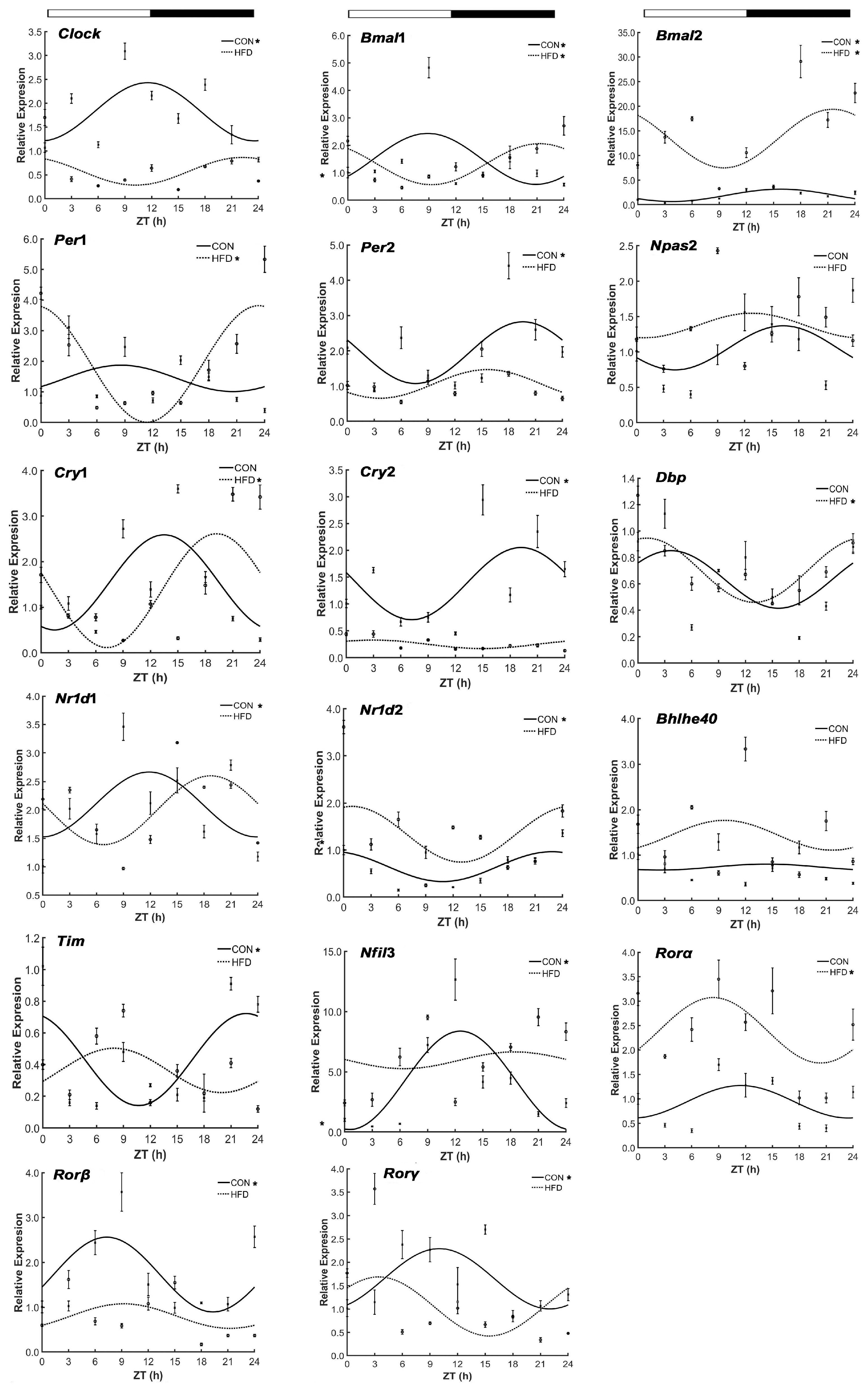

| Gene Name | Forward and Reverse Primers Sequence (5′-3′) | Annealing Temperature (°C) | Product Sizes (bp) | GeneBank Accession No. |
|---|---|---|---|---|
| Clock | F 5′ GTCATCGCTTAGTAGTCAGTCCTT 3′ | 57.0 | 187 | XM_014568700.2 |
| R 5′ TATCATTCGTGTTCTTTGCTCC 3′ | 57.3 | |||
| Bmal1 | F 5′ GATAAAGATGACCAACACGGAAGG 3′ | 61.9 | 338 | XM_014568878.2 |
| R 5′ TCACAGCCCACAACAAACAGAA 3′ | 61.3 | |||
| Bmal2 | F 5′ ACATTACTACCCTGTGGTTCCC 3′ | 57.4 | 287 | XM_006127687.3 |
| R 5′ GTCTCCAAGTCCTCCATTTCTG 3′ | 57.9 | |||
| Npas2 | F 5′ AGGCATTAGATGGCTTCGTTAT 3′ | 58.0 | 145 | XM_014578632.2 |
| R 5′ GAATGTTCTTGTTCTGGGAGGA 3′ | 58.3 | |||
| Tim | F 5′ TGGGAGCAGAGGCAGGAG 3′ | 58.8 | 248 | XM_025188709.1 |
| R 5′ CTGAACATGAGCGAGACGATTT 3′ | 59.6 | |||
| Cry1 | F 5′ GTTGGATTCACCACCTTGCTC 3′ | 59.3 | 300 | XM_025178935.1 |
| R 5′ GTGCTGTCCAAGGCTCGTAG 3′ | 58.2 | |||
| Cry2 | F 5′ CTGTTTATTGGCATCAGTCCCT 3′ | 58.6 | 154 | XM_006124501.2 |
| R 5′ CTCCTCTATTCCCTCATGTTTACG 3′ | 59.5 | |||
| Per1 | F 5′ TGCGTCAAGCAGGTCCAAG 3′ | 60.1 | 167 | XM_025181282.1 |
| R 5′ GAGACAGCCACGGCAAAGG 3′ | 61.2 | |||
| Per2 | F 5′ CACCTTCTTGTCCCTCTATCCA 3′ | 58.4 | 234 | XM_042853554.1 |
| R 5′ TCTTTGCCCACGAGTACCATG 3′ | 60.9 | |||
| Dbp | F 5′ ATGAACTTTGACCCTGACCCTG 3′ | 60.5 | 136 | XM_006132772.2 |
| R 5′ GGATTTTCCGTGCCTTCTTCAT 3′ | 62.3 | |||
| Nfil3 | F 5′ TCTGTGGTGGGCAGTAGTTGTA 3′ | 58.2 | 291 | XM_006131880.3 |
| R 5′ ATTCACTTGTAGCAGAGGAGGG 3′ | 57.9 | |||
| Bhlhe40 | F 5′ ACAGACAGTGGGTATGGAGGAG 3′ | 58.0 | 294 | XM_006132802.3 |
| R 5′ CAGCATAGGCAGATAGGCAGTT 3′ | 59.2 | |||
| Nr1d1 | F 5′ TCCTGAGCGGCGAGACCTAC 3′ | 62.5 | 256 | XM_025179480.1 |
| R 5′ GAGTCATCGGGGTGCTTCTTT 3′ | 60.7 | |||
| Nr1d2 | F 5′ CAATGGCTACCAGGGCAACA 3′ | 61.6 | 347 | XM_014579147.2 |
| R 5′ GCTTGGCAAACTCCACTACCTC 3′ | 60.7 | |||
| Rorα | F 5′ CATCGGGCTTCTTCCCTTATT 3′ | 60.2 | 207 | XM_025178325.1 |
| R 5′ TTACCTCCCTCTGCTTGTTCTG 3′ | 58.9 | |||
| Rorβ | F 5′ CTGCAAGGGTTTCTTTAGGAGG 3′ | 60.2 | 338 | XM_006137819.3 |
| R 5′ AGTAAGTGCCACCAGTTTCGTT 3′ | 58.4 | |||
| Rorγ | F 5′ CTACACCAGTCCCAACTTCACCA 3′ | 61.5 | 208 | XM_025182756.1 |
| R 5′ CCGTTCCCACATCTCCTCCA 3′ | 62.7 | |||
| Acaca | F 5′ CGTCCGAGAACCCCAAACTA 3′ | 60.0 | 264 | XM_006138625.3 |
| R 5′ CCAGCAACCCATCATCCAC 3′ | 58.8 | |||
| Dgat1 | F 5′ TTGCTGCCTCTGTTTTGTTTG 3′ | 59.1 | 167 | XM_014576010.1 |
| R 5′ TGACTGTCCTCTTTCGTTCCTTC 3′ | 60.2 | |||
| Fas | F 5′ CGTGGGCTTGGCTGCTATTC 3′ | 63.0 | 249 | XM_014580710.1 |
| R 5′ GGAGGACAACGGCTCTTACATT 3′ | 60.1 | |||
| Hmgcr | F 5′ TCATCAGTCTCGCTGGTCGTA 3′ | 58.9 | 235 | XM_014574160.1 |
| R 5′ GGAATGACTGCTTCACAGACCA 3′ | 59.9 | |||
| Lipin1 | F 5′ CACTGGGTGAACGAACGAGG 3′ | 61.0 | 191 | XM_006138555.2 |
| R 5′ GCAGGTCTGTTTCCAAAGGCT 3′ | 60.9 | |||
| Scd | F 5′ GAGGTTTTACAAGCCTTCCGTG 3′ | 60.8 | 291 | XM_014579917.1 |
| R 5′ TCGCTGGTGGCGTAGTCGT 3′ | 62.1 | |||
| Ldlr1 | F 5′ CCAACGCTCAGCAGAAAACC 3′ | 60.7 | 166 | XM_014577039.1 |
| R 5′ GGTTTGCCGAACTGGTCTTG 3′ | 60.4 | |||
| Cpt1a | F 5′ GAGCAGGGATACAGGGAAGAGG 3′ | 61.8 | 193 | XM_006131643.1 |
| R 5′ CATTCTCCCAAAGGTGTCCAAC 3′ | 60.9 | |||
| Sirt1 | F 5′ AGTAGACTTCCCAGACCTTCCAG 3′ | 58.6 | 208 | XM_006125276.3 |
| R 5′ AACCTGTTCCAGCGTATCTATGT 3′ | 57.7 | |||
| Pparα | F 5′ AGAGGAGGATGATCTCAGAAACC 3′ | 58.3 | 186 | XM_014575807.1 |
| R 5′ GATGCTGGTGAAAGGGTGTCTG 3′ | 60.8 | |||
| Pparβ | F 5′ GGAGGACCAGACCGTTTGCC 3′ | 63.8 | 167 | XM_006115662.2 |
| R 5′ CCGTAATGAAATCCCGATGCTA 3′ | 61.8 | |||
| Pparγ | F 5′ GTGGAGACAAGGCTTCTGGATT 3′ | 59.9 | 216 | XM_006117601.2 |
| R 5′ GCATTCGCCCAAACCTGATA 3′ | 60.9 | |||
| Rxra | F 5′ AAGGACCGAAATGAGAACGAGG 3′ | 62.3 | 221 | XM_014580328.1 |
| R 5′ ATTCGCTTTGCCCATTCCAC 3′ | 62.3 | |||
| Lxrα | F 5′ AGACCCTCATAACCGTGAAGCA 3′ | 61.2 | 215 | XM_006135352.2 |
| R 5′ TGATGCTTTCAGTCTCTGGATTGTA 3′ | 61.0 | |||
| Pdk4 | F 5′ CAGTCCGAAATAGACACCACGAT 3′ | 60.8 | 253 | XM_014569249.2 |
| R 5′ TTCACCACTCCCACCACATCAC 3′ | 62.5 | |||
| Acsl1 | F 5′ ATACAGGCAAGTCTGGGAGGAA 3′ | 60.4 | 128 | XM_006128490.2 |
| R 5′ TCAGTTTGTCCATAGCCTTCGT 3′ | 59.1 | |||
| Apoa1 | F 5′ GCTGGCTCCCTACTACACGC 3′ | 59.9 | 223 | XM_006121675.2 |
| R 5′ CAGGACCTCCATCTTCTGCTTG 3′ | 61.3 | |||
| Apob | F 5′ GACTGAACAGCCCATTAGCCA 3′ | 60.1 | 160 | XM_014577946.2 |
| R 5′ GTGACTTGTGCCATCATACCGT 3′ | 59.8 | |||
| Rpl19 | F 5′ TCGTATGCCCGAGAAGGTGA 3′ | 61.2 | 180 | XM_006126213.1 |
| R 5′ GCCTTGAGTTTGTGGATGTGCT 3′ | 61.6 |
| Gene Name | Mesor | Amplitude | Acro(p) | Peak of Expression/ZT(h) | ANOVA(p) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CON | HFD | CON | HFD | CON | HFD | CON | HFD | CON | HFD | |
| Clock | 1.82 | 0.57 | 0.61 | 0.29 | 0.12 | 0.34 | 11.57 | 22.20 | <0.05 | <0.05 |
| Bmal1 | 1.51 | 1.31 | 0.93 | 0.75 | 0.28 | 0.03 | 8.89 | 21.30 | <0.05 | <0.05 |
| Bmal2 | 1.91 | 13.43 | 1.25 | 5.95 | 0.01 | 0.28 | 15.80 | 21.53 | <0.05 | <0.05 |
| Npas2 | 1.06 | 1.37 | 0.31 | 0.17 | 0.42 | 0.75 | 16.26 | 12.77 | <0.05 | <0.05 |
| Tim | 0.43 | 0.36 | 0.29 | 0.14 | 0.12 | 0.33 | 22.69 | 7.96 | <0.05 | <0.05 |
| Cry1 | 1.55 | 1.36 | 1.04 | 1.25 | 0.04 | 0.02 | 13.49 | 21.89 | n.s. | <0.05 |
| Cry2 | 1.38 | 0.25 | 0.67 | 0.08 | 0.19 | 0.33 | 19.20 | 2.97 | <0.05 | n.s. |
| Per1 | 1.44 | 1.91 | 0.43 | 1.90 | 0.61 | 0.01 | 8.59 | 23.36 | <0.05 | <0.05 |
| Per2 | 1.95 | 1.06 | 0.87 | 0.40 | 0.20 | 0.11 | 19.60 | 15.62 | <0.05 | n.s. |
| Dbp | 0.63 | 0.70 | 0.22 | 0.24 | 0.31 | 0.03 | 3.68 | 0.99 | <0.05 | <0.05 |
| Nfil3 | 4.29 | 5.95 | 4.08 | 0.70 | 0.02 | 0.89 | 12.55 | 18.42 | <0.05 | <0.05 |
| Bhlhe40 | 0.74 | 1.44 | 0.07 | 0.33 | 0.92 | 0.73 | 14.40 | 9.67 | <0.05 | <0.05 |
| Nr1d1 | 2.10 | 2.00 | 0.57 | 0.61 | 0.22 | 0.13 | 11.87 | 18.75 | <0.05 | n.s. |
| Nr1d2 | 0.65 | 1.34 | 0.32 | 0.59 | 0.15 | 0.36 | 22.87 | 0.90 | <0.05 | <0.05 |
| Rorα | 0.94 | 2.40 | 0.33 | 0.67 | 0.29 | 0.24 | 11.46 | 8.39 | n.s. | <0.05 |
| Rorβ | 1.73 | 0.80 | 0.83 | 0.27 | 0.13 | 0.51 | 7.31 | 9.23 | <0.05 | <0.05 |
| Rorγ | 1.65 | 1.06 | 0.64 | 0.63 | 0.05 | 0.39 | 10.01 | 3.43 | <0.05 | <0.05 |
| Gene Name | Mesor | Amplitude | Acro(p) | Peak of Expression/ZT(h) | ANOVA(p) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CON | HFD | CON | HFD | CON | HFD | CON | HFD | CON | HFD | |
| Acaca | 2.17 | 1.12 | 0.34 | 0.05 | 0.77 | 0.91 | 15.88 | 16.86 | <0.05 | <0.05 |
| Dgat1 | 0.98 | 1.56 | 0.39 | 1.20 | 0.26 | 0.00 | 11.97 | 21.52 | <0.05 | <0.05 |
| Fas | 0.20 | 1.18 | 0.09 | 0.48 | 0.11 | 0.13 | 22.31 | 16.04 | <0.05 | n.s. |
| Hmgcr | 1.22 | 1.38 | 0.65 | 0.62 | 0.03 | 0.18 | 15.04 | 0.17 | <0.05 | <0.05 |
| Lipin1 | 0.92 | 0.24 | 0.42 | 0.09 | 0.15 | 0.37 | 10.23 | 9.59 | <0.05 | <0.05 |
| Scd | 2.08 | 0.26 | 0.59 | 0.08 | 0.22 | 0.52 | 13.32 | 1.21 | n.s. | n.s. |
| Ldlr1 | 4.21 | 3.76 | 2.21 | 1.39 | 0.17 | 0.16 | 15.93 | 9.37 | <0.05 | <0.05 |
| Gene Name | Mesor | Amplitude | Acro(p) | Peak of Expression/ZT(h) | ANOVA(p) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CON | HFD | CON | HFD | CON | HFD | CON | HFD | CON | HFD | |
| Cpt1a | 2.10 | 1.79 | 0.94 | 0.33 | 0.01 | 0.71 | 14.54 | 1.92 | <0.05 | <0.05 |
| Sirt1 | 0.93 | 1.24 | 0.28 | 0.41 | 0.21 | 0.23 | 23.46 | 1.29 | <0.05 | n.s. |
| Pparα | 0.97 | 0.89 | 0.38 | 0.54 | 0.10 | 0.02 | 19.47 | 1.07 | <0.05 | <0.05 |
| Pparβ | 2.23 | 1.01 | 1.92 | 0.75 | 0.01 | 0.09 | 19.31 | 1.07 | <0.05 | <0.05 |
| Pparγ | 1.60 | 3.95 | 0.86 | 1.64 | 0.17 | 0.12 | 18.36 | 10.30 | <0.05 | <0.05 |
| Gene Name | Mesor | Amplitude | Acro(p) | Peak of Expression/ZT(h) | ANOVA(p) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CON | HFD | CON | HFD | CON | HFD | CON | HFD | CON | HFD | |
| Apoa1 | 0.81 | 0.98 | 0.45 | 0.63 | 0.14 | 0.03 | 21.37 | 2.90 | <0.05 | n.s. |
| Apob | 1.23 | 0.33 | 0.63 | 0.14 | 0.11 | 0.03 | 16.87 | 22.94 | <0.05 | n.s. |
| Acsl1 | 2.00 | 5.87 | 1.36 | 0.71 | 0.00 | 0.77 | 13.86 | 16.88 | <0.05 | <0.05 |
| Lxrα | 1.27 | 0.41 | 0.65 | 0.37 | 0.15 | 0.01 | 14.06 | 21.66 | <0.05 | <0.05 |
| Pdk4 | 4.29 | 5.95 | 4.08 | 0.70 | 0.02 | 0.89 | 12.55 | 18.42 | <0.05 | <0.05 |
| Rxra | 1.60 | 1.86 | 1.06 | 0.87 | 0.01 | 0.06 | 17.85 | 0.49 | <0.05 | <0.05 |
| Gene | Gene | Correlation Coefficient r | Gene | Gene | Correlation Coefficient r | Gene | Gene | Correlation Coefficient r |
|---|---|---|---|---|---|---|---|---|
| Clock | Bmal1 | 0.71 | Clock | Per2 | 0.51 | Bmal2 | Nfil3 | 0.54 |
| Clock | Nr1d1 | 0.63 | Nr1d2 | Tim | 0.69 | Bmal2 | Per2 | 0.50 |
| Clock | Nfil3 | 0.56 | Bmal1 | Tim | 0.81 | Per2 | Cry2 | 0.54 |
| Bmal1 | Nr1d1 | 0.63 | Nfil3 | Dgat1 | 0.84 | Bmal2 | Hmgcr | 0.64 |
| Per2 | Fas | 0.84 | Bmal2 | Ldlr1 | 0.72 | Per2 | Hmgcr | 0.80 |
| Cry2 | Fas | 0.61 | Cry2 | Pparβ | 0.55 | Nfil3 | Sirt1 | 0.80 |
| Nfil3 | Cpt1a | 0.56 | Bmal2 | Pparβ | 0.51 | Bmal2 | Stirt | 0.72 |
| Per2 | Cpt1a | 0.63 | Tim | Apoa1 | 0.55 | Cry2 | Pparγ | 0.64 |
| Nr1d1 | Cpt1a | 0.58 | Cry2 | Apoa1 | 0.67 | Bmal2 | Acsl1 | 0.75 |
| Bmal2 | Cpt1a | 0.78 | Nr1d2 | Lxrα | 0.51 | Per2 | Acsl1 | 0.57 |
| Cry2 | Rxra | 0.64 | Nfil3 | Lxrα | −0.56 | Clock | Acsl1 | 0.53 |
| Bmal2 | Rxra | 0.59 | Clock | Lxrα | 0.50 | Nr1d2 | Pdk4 | 0.55 |
| Per2 | Rxra | 0.54 | Bmal1 | Pparα | 0.52 | Cry2 | Lipin1 | −0.54 |
| Clock | Tim | −0.50 | Nr1d1 | Pparα | −0.54 |
| Gene | Gene | Correlation Coefficient r | Gene | Gene | Correlation Coefficient r | Gene | Gene | Correlation Coefficient r |
|---|---|---|---|---|---|---|---|---|
| Bmal1 | Cry1 | 0.81 | Bmal1 | Per1 | 0.84 | Per1 | Dbp | 0.79 |
| Bmal1 | Dbp | 0.58 | Per1 | Cry1 | 0.74 | Cry1 | Dgat1 | 0.80 |
| Cry1 | Hmgcr | 0.64 | Bmal1 | Dgat1 | 0.88 | Dbp | Dgat1 | 0.51 |
| Bmal2 | Hmgcr | 0.50 | Per1 | Dgat1 | 0.88 | Per1 | Pparγ | 0.50 |
| Dbp | Pparα | 0.77 | Bmal1 | Pparα | 0.53 | Dbp | Rxra | 0.70 |
| Per1 | Pparα | 0.73 | Cry1 | Pparα | 0.62 | Per1 | Rxra | 0.50 |
| Cry1 | Lxrα | 0.72 | Bmal1 | Lxrα | 0.54 | Bmal2 | Pparβ | −0.52 |
| Per1 | Lxrα | 0.53 | Clock | Lxrα | 0.56 | Bmal2 | Ldlr1 | −0.56 |
| Bmal2 | Rorα | −0.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Liu, L.; Deng, S.; Zou, L.; He, Y.; Zhu, X.; Li, H.; Hu, Y.; Chu, W.; Wang, X. Circadian Rhythm Alteration of the Core Clock Genes and the Lipid Metabolism Genes Induced by High-Fat Diet (HFD) in the Liver Tissue of the Chinese Soft-Shelled Turtle (Trionyx sinensis). Genes 2024, 15, 157. https://doi.org/10.3390/genes15020157
Liu L, Liu L, Deng S, Zou L, He Y, Zhu X, Li H, Hu Y, Chu W, Wang X. Circadian Rhythm Alteration of the Core Clock Genes and the Lipid Metabolism Genes Induced by High-Fat Diet (HFD) in the Liver Tissue of the Chinese Soft-Shelled Turtle (Trionyx sinensis). Genes. 2024; 15(2):157. https://doi.org/10.3390/genes15020157
Chicago/Turabian StyleLiu, Li, Lingli Liu, Shiming Deng, Li Zou, Yong He, Xin Zhu, Honghui Li, Yazhou Hu, Wuying Chu, and Xiaoqing Wang. 2024. "Circadian Rhythm Alteration of the Core Clock Genes and the Lipid Metabolism Genes Induced by High-Fat Diet (HFD) in the Liver Tissue of the Chinese Soft-Shelled Turtle (Trionyx sinensis)" Genes 15, no. 2: 157. https://doi.org/10.3390/genes15020157
APA StyleLiu, L., Liu, L., Deng, S., Zou, L., He, Y., Zhu, X., Li, H., Hu, Y., Chu, W., & Wang, X. (2024). Circadian Rhythm Alteration of the Core Clock Genes and the Lipid Metabolism Genes Induced by High-Fat Diet (HFD) in the Liver Tissue of the Chinese Soft-Shelled Turtle (Trionyx sinensis). Genes, 15(2), 157. https://doi.org/10.3390/genes15020157






