Identification of Quantitative Trait Loci and Candidate Genes Controlling Seed Dormancy in Eggplant (Solanum melongena L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Materials and Phenotypic Identification
2.2. DNA Extraction and Mixing Pool Sequencing
2.3. Analysis of Seed-Dormancy-Related QTLs
2.4. Candidate Gene Prediction
2.5. Analysis of Candidate Genes’ Expression Levels
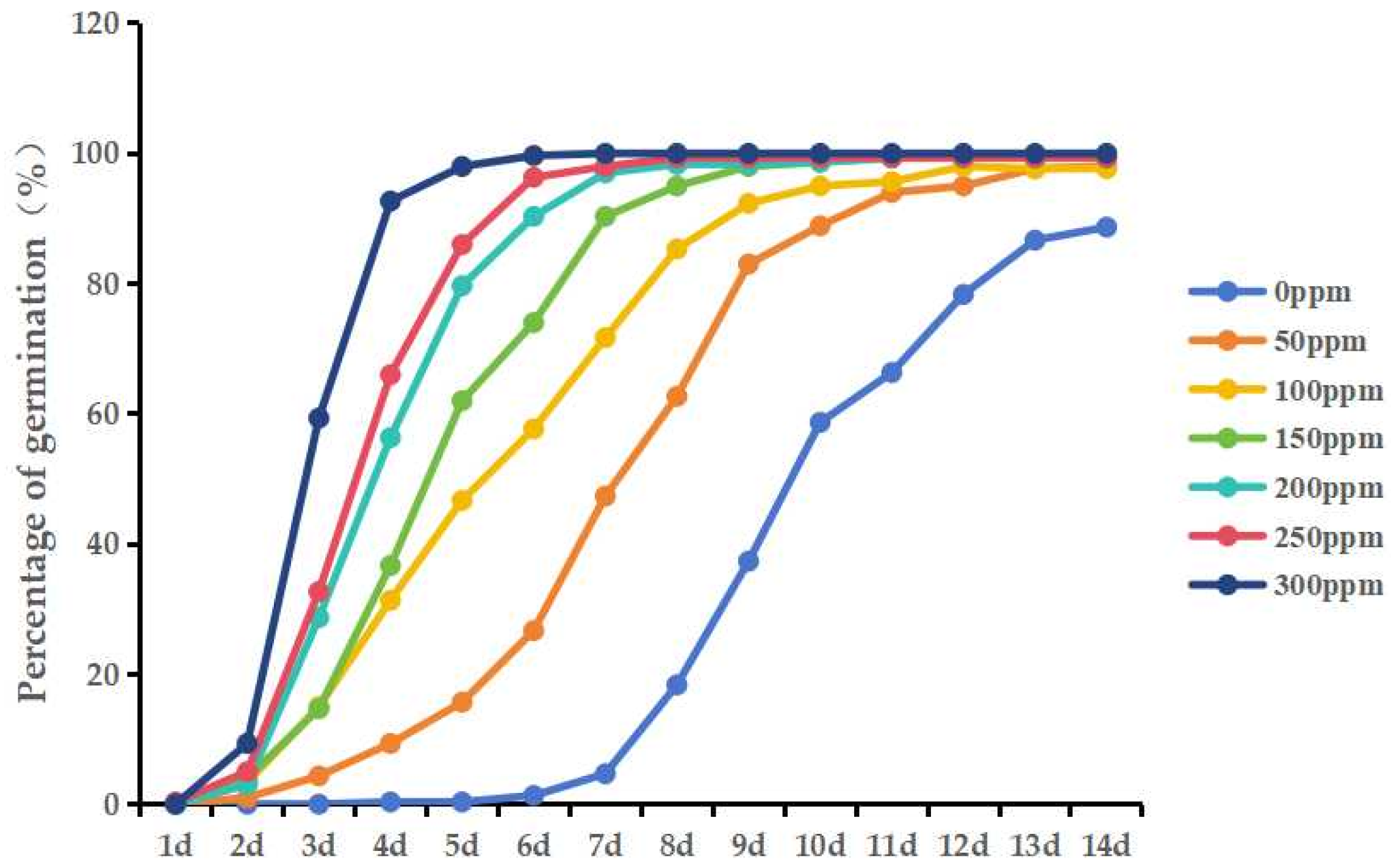
2.6. Treatment of Seeds with Different Concentrations of GA3
3. Results
3.1. Statistical Analysis of the Seed Dormancy Characteristics
3.2. Parental and Extreme Mixed Pool Sequencing
3.3. Detection of QTL Sites with Major Effects
3.4. Candidate Gene Prediction
3.5. Verification of Quantitative Expression
3.6. Effects of Different Concentrations of GA3 on the Germination Percentage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Forti, C.; Ottobrino, V.; Bassolino, L.; Toppino, L.; Rotino, G.L.; Pagano, A.; Macovei, A.; Balestrazzi, A. Molecular dynamics of pre-germinative metabolism in primed eggplant (Solanum melongena L.) seeds. Hortic. Res. 2020, 7, 87. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S. Seed dormancy and germination. Curr. Biol. 2017, 27, 874–878. [Google Scholar] [CrossRef]
- Baskin, J.M.; Hidayati, S.N.; Baskin, C.C.; Walck, J.L.; Huang, Z.Y.; Chien, C.T. Evolutionary considerations of the presence of both morphophysiological and physiological seed dormancy in the highly advanced euasterids II order Dipsacales. Seed Sci. Res. 2006, 16, 233–242. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Cao, Y.J.; Wang, C.M.; Zhai, H.Q.; Wan, J.M.; Yoshimura, A. Detection and analysis of QTL for seed dormancy in rice (Oryza sativa L.) using RIL and CSSL population. Yi Chuan Xue Bao 2003, 30, 453–458. (In Chinese) [Google Scholar]
- Liu, F.; Zhang, H.; Ding, L.; Soppe, W.J.J.; Xiang, Y. REVERSAL OF RDO5 1, a Homolog of Rice Seed Dormancy4, Interacts with bHLH57 and Controls ABA Biosynthesis and Seed Dormancy in Arabidopsis. Plant Cell 2020, 32, 1933–1948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Miao, X.L.; Xia, X.C.; He, Z.H. Cloning of seed dormancy genes (TaSdr) associated with tolerance to pre-harvest sprouting in common wheat and development of a functional marker. Theor. Appl. Genet. 2014, 127, 855–866. [Google Scholar] [CrossRef]
- Hisano, H.; Hoffie, R.E.; Abe, F.; Munemori, H.; Matsuura, T.; Endo, M.; Mikami, M.; Nakamura, S.; Kumlehn, J.; Sato, K. Regulation of germination by targeted mutagenesis of grain dormancy genes in barley. Plant Biotechnol. J. 2022, 20, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Barral, N.; Rodríguez-Gacio, M.D.C.; Matilla, A.J. Delay of Germination-1 (DOG1): A Key to Understanding Seed Dormancy. Plants 2020, 9, 480. [Google Scholar] [CrossRef]
- Graeber, K.; Linkies, A.; Müller, K.; Wunchova, A.; Rott, A.; Leubner-Metzger, G. Cross-species approaches to seed dormancy and germination: Conservation and biodiversity of ABA-regulated mechanisms and the Brassicaceae DOG1 genes. Plant Mol. Biol. 2010, 73, 67–87. [Google Scholar] [CrossRef]
- Bentsink, L.; Jowett, J.; Hanhart, C.J.; Koornneef, M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 17042–17047. [Google Scholar] [CrossRef] [PubMed]
- Zha, P.; Liu, S.R.; Li, Y.; Ma, T.T.; Yang, L.W.; Jing, Y.J.; Lin, R.C. The Evening Complex and the Chromatin-Remodeling Factor PICKLE Coordinately Control Seed Dormancy by Directly Repressing DOG1 in Arabidopsis. Plant Commun. 2019, 1, 100011. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Berdeja, A.; Stitzer, M.C.; Taylor, M.A.; Okada, M.; Ezcurra, E.; Runcie, D.E.; Schmitt, J. Functional variants of DOG1 control seed chilling responses and variation in seasonal life-history strategies in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2020, 117, 2526–2534. [Google Scholar] [CrossRef] [PubMed]
- Rikiishi, K.; Maekawa, M. Seed maturation regulators are related to the control of seed dormancy in wheat (Triticum aestivum L.). PLoS ONE 2014, 9, e107618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.P.; Liu, H.H.; Shen, S.X.; Zhang, X.E. Improvement of eggplant seed germination and seedling emergence at low temperature by seed priming with incorporation SA into KNO3 solution. Front. Agric. China 2011, 5, 534–537. [Google Scholar] [CrossRef]
- Muthusamy, A.; Kudwa, P.P.; Prabhu, V.; Mahato, K.K.; Babu, V.S.; Rao, M.R.; Gopinath, P.M.; Satyamoorthy, K. Influence of helium-neon laser irradiation on seed germination in vitro and physico-biochemical characters in seedlings of brinjal (Solanum melongena L.) var. Mattu Gulla. Photochem. Photobiol. 2012, 88, 1227–1235. [Google Scholar] [CrossRef]
- Gonzalez, L.M.R. Germination response of eggplant (Solanum melongena L.) seeds to different vinegar concentration as seed priming agents. Int. J. Sci. Res. Publ. 2015, 5, 2250–3153. [Google Scholar]
- Ali, M.; Hayat, S.; Ahmad, H.; Ghani, M.I.; Amin, B.; Atif, M.J.; Cheng, Z.H. Priming of Solanum melongena L. seeds enhances germination, alters antioxidant enzymes, modulates ROS, and improves early seedling growth: Indicating aqueous garlic extract as seed-priming bio-stimulant for eggplant production. Appl. Sci. 2019, 9, 2203. [Google Scholar] [CrossRef]
- Subramaniam, R.; Kumar, V.S. Allele mining, amplicon sequencing and computational prediction of Solanum melongena L. FT/TFL1 gene homologs uncovers putative variants associated to seed dormancy and germination. PLoS ONE 2023, 18, e0285119. [Google Scholar] [CrossRef]
- Wei, Q.Z.; Wang, J.L.; Wang, W.H.; Hu, T.H.; Hu, H.J.; Bao, C.L. A high-quality chromosome-level genome assembly reveals genetics for important traits in eggplant. Hortic. Res. 2020, 7, 153. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.Y.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Fenner, M. The effects of the parent environment on seed germinability. Seed Sci. Res. 1991, 1, 75–84. [Google Scholar] [CrossRef]
- Baskin, J.M.; Baskin, C.C. A classification system for seed dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef]
- Seo, M.; Hanada, A.; Kuwahara, A.; Endo, A.; Okamoto, M.; Yamauchi, Y.; North, H.; Marion-Poll, A.; Sun, T.P.; Koshiba, T.; et al. Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006, 48, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Chen, Q.; Wu, Y.R.; Liu, R.J.; Zhang, H.W.; Wang, P.F.; Li, Y.L.; Wang, S.F.; Tang, S.Y.; Liu, C.Y.; et al. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 2016, 85, 348–361. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Footitt, S. Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J. Exp. Bot. 2017, 68, 843–856. [Google Scholar] [CrossRef]
- Melan, M.A.; Enriquez, A.; Peterman, T.K. The LOX1 Gene of Arabidopsis Is Temporally and Spatially Regulated in Germinating Seedlings. Plant Physiol. 1994, 105, 385–393. [Google Scholar] [CrossRef]
- Xu, F.; Tang, J.Y.; Wang, S.X.; Cheng, X.; Wang, H.R.; Ou, S.J.; Gao, S.P.; Li, B.S.; Qian, Y.W.; Gao, C.X.; et al. Antagonistic control of seed dormancy in rice by two bHLH transcription factors. Nat. Genet. 2022, 54, 1972–1982. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.P.; Chen, W.Q.; Xu, Z.Y.; Chen, M.; Yu, D. Functions of WRKYs in plant growth and development. Trends Plant Sci. 2023, 28, 630–645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Xie, Z.; Zou, X.L.; Casaretto, J.; Ho, T.H.D.; Shen, Q.J. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004, 134, 1500–1513. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Yang, G.; Komatsu, S.; Shen, Q.J. Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant Physiol. 2006, 46, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Zhang, Z.L.; Zou, X.L.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.X.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Zentella, R.; Zhang, Z.L.; Park, M.; Thomas, S.G.; Endo, A.; Murase, K.; Fleet, C.M.; Jikumaru, Y.; Nambara, E.; Kamiya, Y.; et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007, 19, 3037–3057. [Google Scholar] [CrossRef]
- Zhou, C.; Lin, Q.; Lan, J.; Zhang, T.; Liu, X.; Miao, R.; Mou, C.; Nguyen, T.; Wang, J.; Zhang, X.; et al. WRKY Transcription Factor OsWRKY29 Represses Seed Dormancy in Rice by Weakening Abscisic Acid Response. Front. Plant Sci. 2020, 11, 691. [Google Scholar] [CrossRef]
- Wang, H.M.; Hou, Y.X.; Wang, S.; Tong, X.H.; Tang, L.Q.; Abolore, A.A.; Zhang, J.; Wang, Y.F. WRKY72 Negatively Regulates Seed Germination Through Interfering Gibberellin Pathway in Rice. Rice Sci. 2021, 28, 1–5. [Google Scholar] [CrossRef]
- Ali, F.; Qanmber, G.; Li, F.; Wang, Z. Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res. 2021, 35, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Sajeev, N.; Koornneef, M.; Bentsink, L. A commitment for life: Decades of unraveling the molecular mechanisms behind seed dormancy and germination. Plant Cell 2024, 12, 328. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Kumar, R.; Rehal, P.K.; Toora, P.K.; Ayele, B.T. Molecular Mechanisms Underlying Abscisic Acid/Gibberellin Balance in the Control of Seed Dormancy and Germination in Cereals. Front. Plant Sci. 2018, 9, 668. [Google Scholar] [CrossRef] [PubMed]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 Gene Family: Functional Evolution and Molecular Mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef] [PubMed]



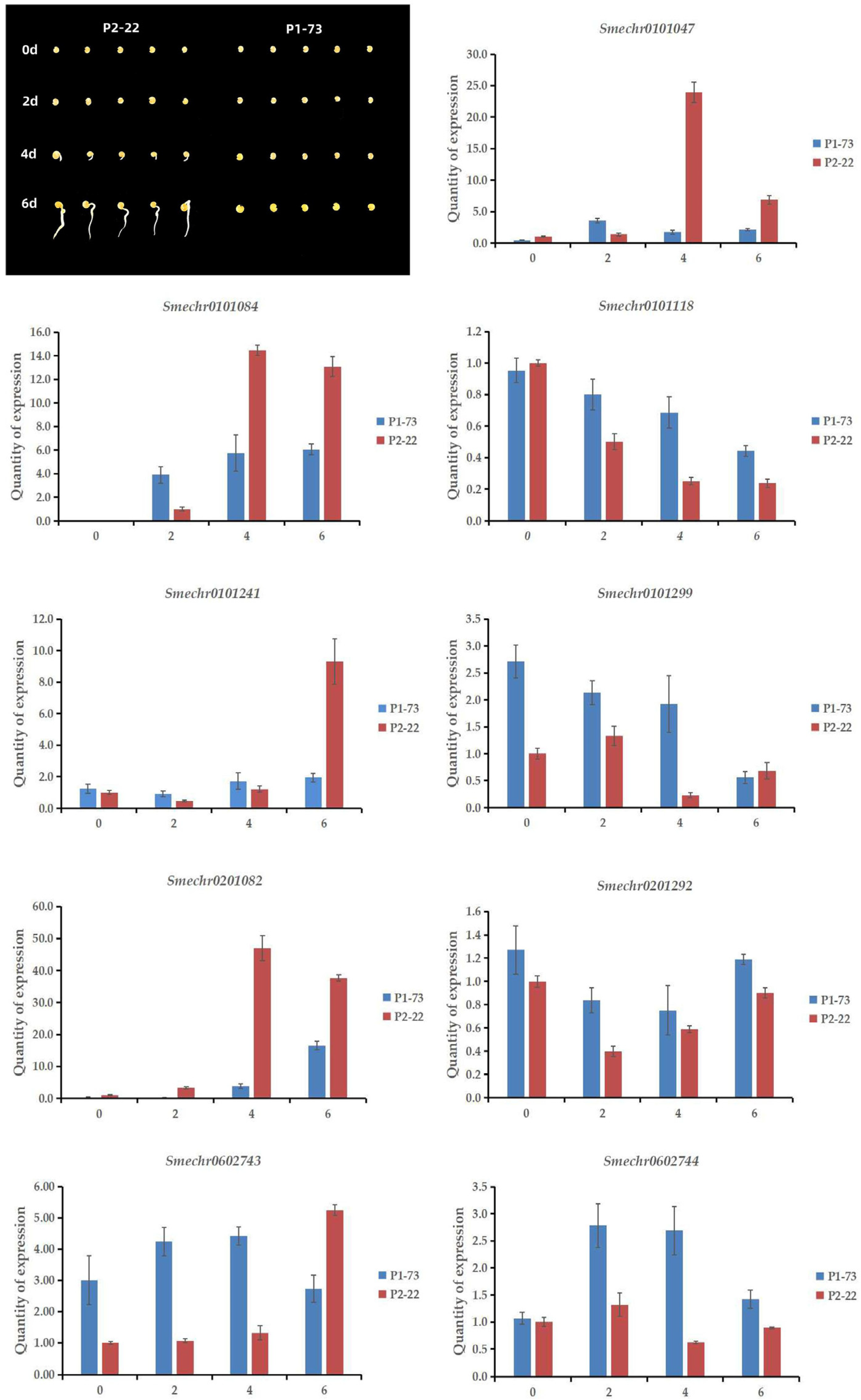
| Test Material | 1 d | 2 d | 3 d | 4 d | 5 d | 6 d | 7 d | 8 d | 9 d | 10 d | 11 d | 12 d | 13 d | 14 d |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1-73 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 5.3% | 14.0% | 33.0% | 52.0% | 63.3% | 67.3% | 72.3% | 76.3% |
| P2-22 | 0.0% | 0.0% | 0.0% | 2.3% | 15.7% | 38.0% | 75.7% | 94.3% | 98.3% | 98.7% | 100.0% | 100.0% | 100.0% | 100.0% |
| F1 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 2.0% | 3.3% | 14.3% | 32.0% | 44.7% | 56.0% | 69.7% | 80.7% | 84.3% |
| F2 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 1.0% | 5.7% | 42.9% | 77.2% | 94.1% | 98.2% | 99.4% | 99.6% | 99.6% |
| Sample ID | Raw Reads | Raw Bases | Q30 (%) | GC (%) | Mapped (%) | Genome Coverage 1× (%) | Average Depth | SNP Number | Indel Number |
|---|---|---|---|---|---|---|---|---|---|
| P1-73 | 83,794,760 | 12,653,008,760 | 91.48 | 37.9758 | 93.99 | 89.13 | 10.09 | 685,891 | 117,150 |
| P2-22 | 77,173,102 | 11,653,138,402 | 91.46 | 37.3624 | 95.53 | 89.16 | 9.45 | 521,259 | 96,169 |
| E-pool | 252,479,026 | 38,124,332,926 | 91.55 | 38.1669 | 93.46 | 90.45 | 30.22 | 912,593 | 196,108 |
| L-pool | 232,529,434 | 35,111,944,534 | 91.58 | 37.9924 | 94.40 | 90.40 | 28.05 | 909,687 | 191,146 |
| QTL | Chromosome | Start/Mb | End/Mb | Interval/Mb | No. of Genes Associated with Effective Mutations |
|---|---|---|---|---|---|
| dr1.1 | chr1 | 10.295 | 12.763 | 2.468 | 38 |
| dr2.1 | chr2 | 52.221 | 55.671 | 3.450 | 13 |
| dr6.1 | chr6 | 85.328 | 85.987 | 0.659 | 17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ai, J.; Wang, W.; Hu, T.; Hu, H.; Wang, J.; Yan, Y.; Pang, H.; Wang, Y.; Bao, C.; Wei, Q. Identification of Quantitative Trait Loci and Candidate Genes Controlling Seed Dormancy in Eggplant (Solanum melongena L.). Genes 2024, 15, 415. https://doi.org/10.3390/genes15040415
Ai J, Wang W, Hu T, Hu H, Wang J, Yan Y, Pang H, Wang Y, Bao C, Wei Q. Identification of Quantitative Trait Loci and Candidate Genes Controlling Seed Dormancy in Eggplant (Solanum melongena L.). Genes. 2024; 15(4):415. https://doi.org/10.3390/genes15040415
Chicago/Turabian StyleAi, Jiaqi, Wuhong Wang, Tianhua Hu, Haijiao Hu, Jinglei Wang, Yaqin Yan, Hongtao Pang, Yong Wang, Chonglai Bao, and Qingzhen Wei. 2024. "Identification of Quantitative Trait Loci and Candidate Genes Controlling Seed Dormancy in Eggplant (Solanum melongena L.)" Genes 15, no. 4: 415. https://doi.org/10.3390/genes15040415
APA StyleAi, J., Wang, W., Hu, T., Hu, H., Wang, J., Yan, Y., Pang, H., Wang, Y., Bao, C., & Wei, Q. (2024). Identification of Quantitative Trait Loci and Candidate Genes Controlling Seed Dormancy in Eggplant (Solanum melongena L.). Genes, 15(4), 415. https://doi.org/10.3390/genes15040415





