Determinants of mer Promoter Activity from Pseudomonas aeruginosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Genes, Plasmids, and Primers
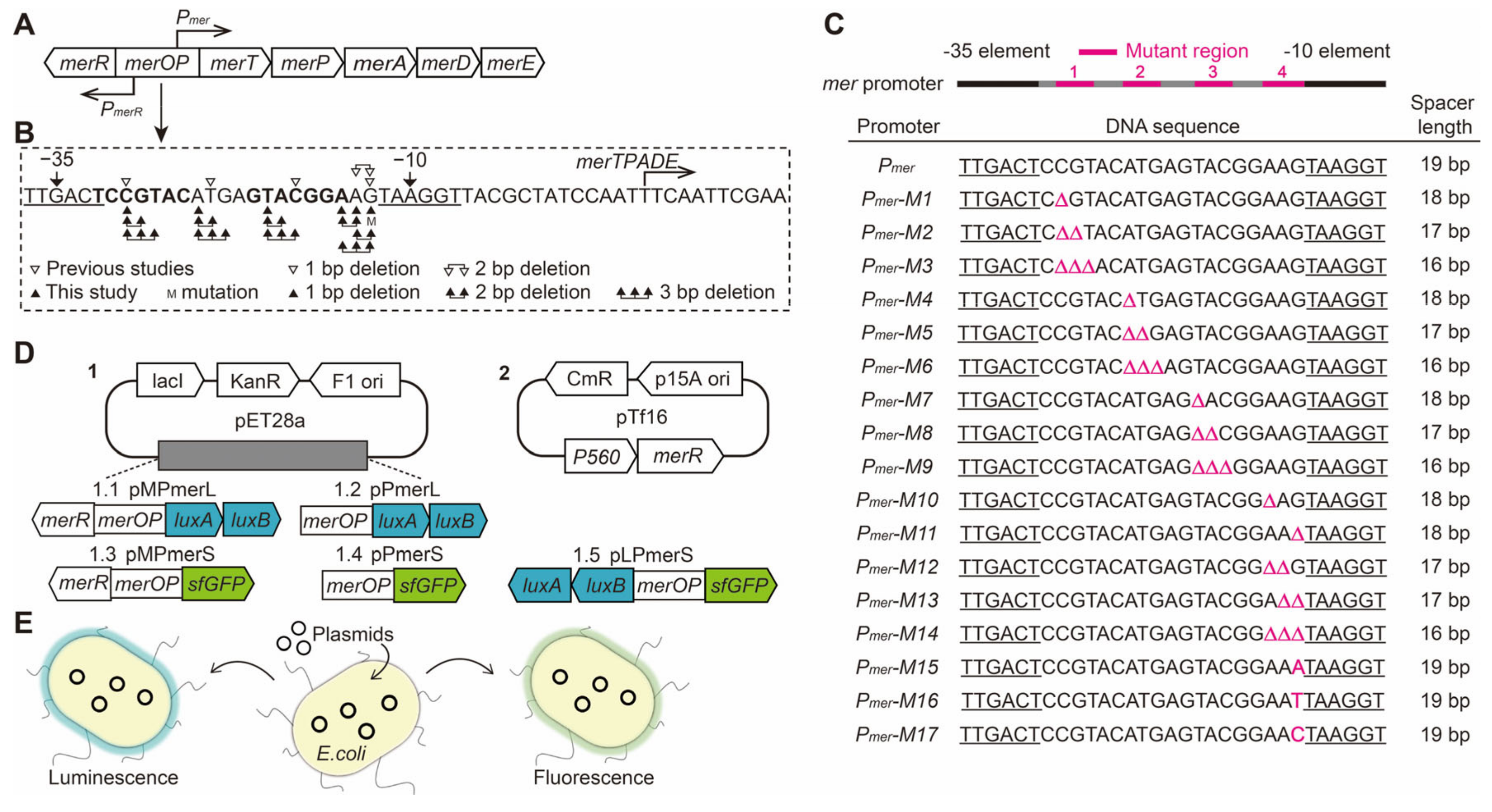
2.2. Designing Base Pair Deletion Derivatives of the mer Promoter
2.3. DNA Manipulations and Mutagenesis
2.4. Luminescence Assays
2.5. Fluorescence Assays
2.6. Data and Statistical Analysis
2.7. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis Assays (SDS-PAGE)
2.8. Quantitative Reverse Transcriptase PCR (qRT-PCR) Analysis
2.9. Analyzing the Expression Activity
2.10. Electrophoretic Mobility Shift Assays (EMSA)
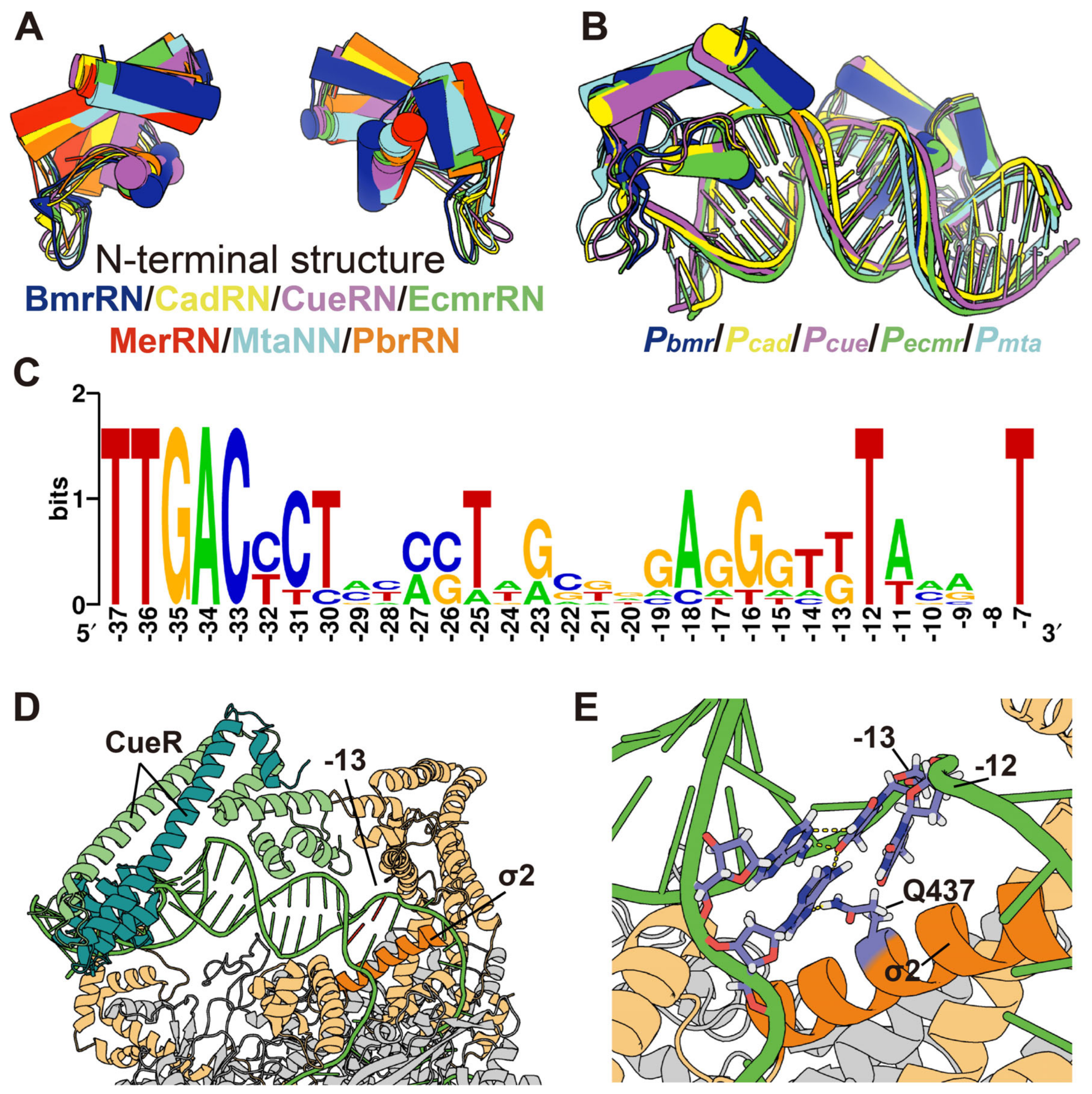
2.11. Alignment of MerR Family TFs and Regulated Promoters
3. Results and Discussion
3.1. Construction of mer Promoter Derivatives
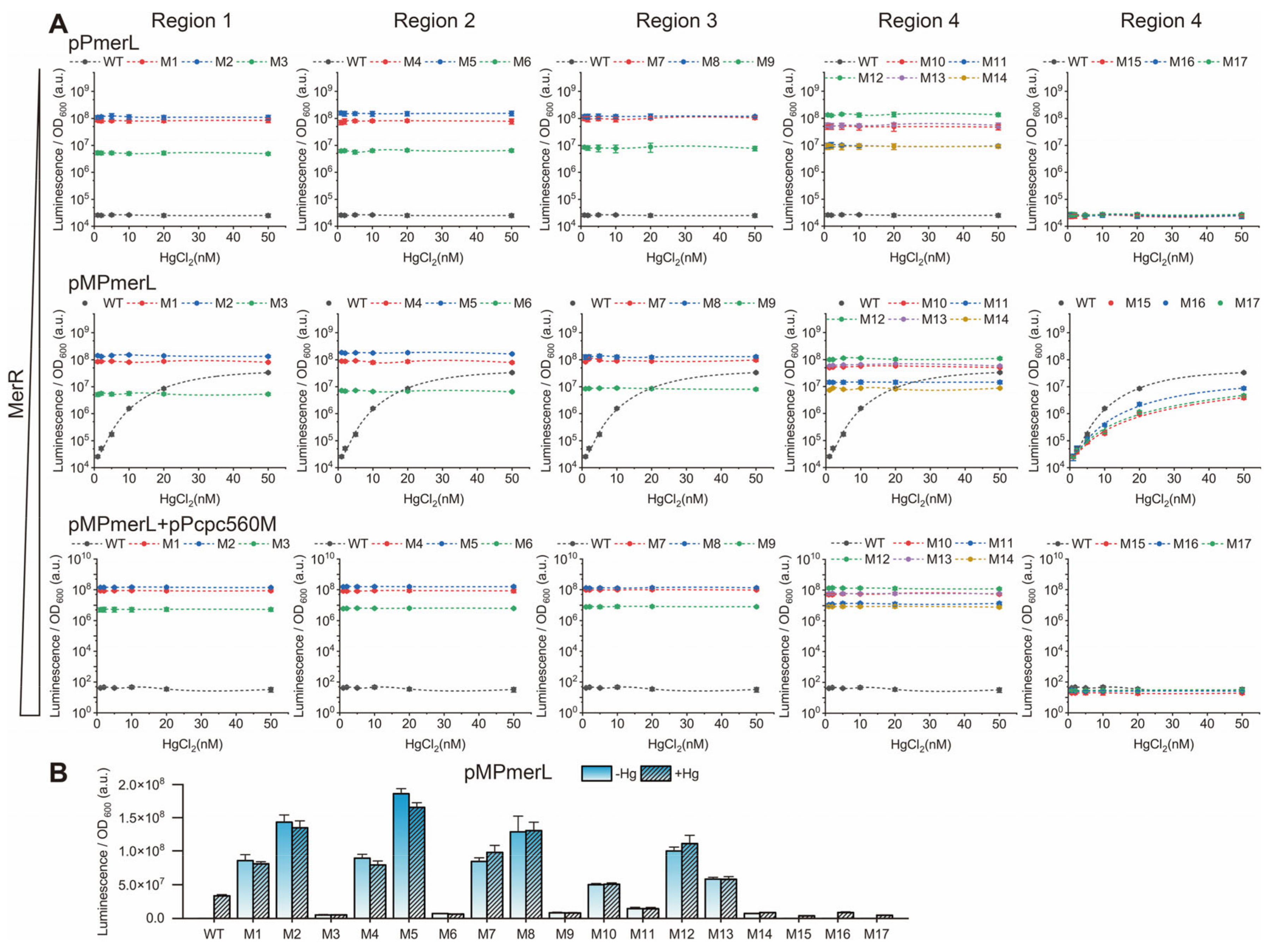
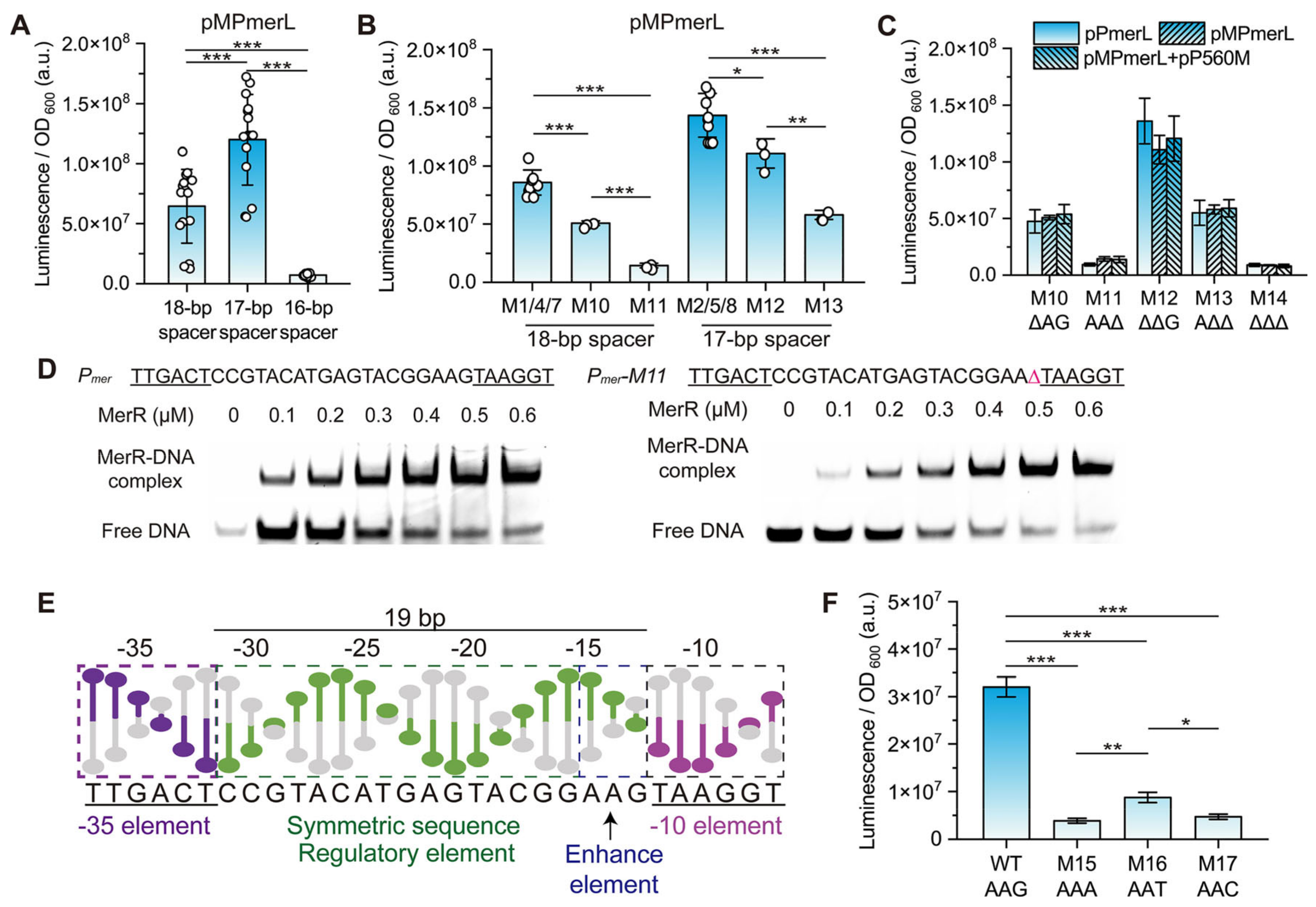
3.2. Effect of the Spacing between the −35 and −10 Elements on Pmer Activity
3.3. Role of Different Positions in the Spacer between the −35 and −10 Elements of Pmer
3.4. Role and Effects of the Regulatory Factor MerR and merR Gene
3.5. Conservation and Function of Guanine at Position −13 (−13G) in Pmer
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hou, D.Y.; O’Connor, D.; Igalavithana, A.D.; Alessi, D.S.; Luo, J.; Tsang, D.C.W.; Sparks, D.L.; Yamauchi, Y.; Rinklebe, J.; Ok, Y.S. Metal contamination and bioremediation of agricultural soils for food safety and sustainability. Nat. Rev. Earth Environ. 2020, 1, 366–381. [Google Scholar] [CrossRef]
- Malik, L.A.; Bashir, A.; Qureashi, A.; Pandith, A.H. Detection and removal of heavy metal ions: A review. Environ. Chem. Lett. 2019, 17, 1495–1521. [Google Scholar] [CrossRef]
- Bashir, A.; Malik, L.A.; Ahad, S.; Manzoor, T.; Bhat, M.A.; Dar, G.N.; Pandith, A.H. Removal of heavy metal ions from aqueous system by ion-exchange and biosorption methods. Environ. Chem. Lett. 2018, 17, 729–754. [Google Scholar] [CrossRef]
- Bolisetty, S.; Peydayesh, M.; Mezzenga, R. Sustainable technologies for water purification from heavy metals: Review and analysis. Chem. Soc. Rev. 2019, 48, 463–487. [Google Scholar] [CrossRef]
- Gao, Q.; Xu, J.; Bu, X.H. Recent advances about metal-organic frameworks in the removal of pollutants from wastewater. Coord. Chem. Rev. 2019, 378, 17–31. [Google Scholar] [CrossRef]
- Qasem, N.A.A.; Mohammed, R.H.; Lawal, D.U. Removal of heavy metal ions from wastewater: A comprehensive and critical review. NPJ Clean Water 2021, 4, 52. [Google Scholar] [CrossRef]
- Nolan, E.M.; Lippard, S.J. Tools and tactics for the optical detection of mercuric ion. Chem. Rev. 2008, 108, 3443–3480. [Google Scholar] [CrossRef]
- Gu, S.; Kang, X.; Wang, L.; Lichtfouse, E.; Wang, C. Clay mineral adsorbents for heavy metal removal from wastewater: A review. Environ. Chem. Lett. 2018, 17, 629–654. [Google Scholar] [CrossRef]
- Zhao, S.L.; Pudasainee, D.; Duan, Y.F.; Gupta, R.; Liu, M.; Lu, J.H. A review on mercury in coal combustion process: Content and occurrence forms in coal, transformation, sampling methods, emission and control technologies. Prog. Energy Combust. Sci. 2019, 73, 26–64. [Google Scholar] [CrossRef]
- Kim, K.H.; Kabir, E.; Jahan, S.A. A review on the distribution of Hg in the environment and its human health impacts. J. Hazard. Mater. 2016, 306, 376–385. [Google Scholar] [CrossRef]
- Walsh, C.T.; Distefano, M.D.; Moore, M.J.; Shewchuk, L.M.; Verdine, G.L. Molecular basis of bacterial resistance to organomercurial and inorganic mercuric salts. FASEB J. 1988, 2, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Stanisich, V.A.; Bennett, P.M.; Richmond, M.H. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J. Bacteriol. 1977, 129, 1227–1233. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.S.; Barkay, T. The mercury resistance operon: From an origin in a geothermal environment to an efficient detoxification machine. Front. Microbiol. 2012, 3, 349. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guo, Y.; Liu, Y.; Xiang, Y.; Liu, G.; Zhang, Q.; Yin, Y.; Cai, Y.; Jiang, G. Advances in bacterial whole-cell biosensors for the detection of bioavailable mercury: A review. Sci. Total Environ. 2023, 868, 161709. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Huang, S.; Liu, P.; Liu, X.; He, Y.; Chen, W.; Hu, Q.; Wei, T.; Gan, J.; Ma, J.; et al. Structural Analysis of the Hg(II)-Regulatory Protein Tn501 MerR from Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 33391. [Google Scholar] [CrossRef]
- Brown, N.L. Bacterial-Resistance to Mercury—Reductio Ad Absurdum. Trends Biochem. Sci. 1985, 10, 400–403. [Google Scholar] [CrossRef]
- Lund, P.A.; Ford, S.J.; Brown, N.L. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J. Gen. Microbiol. 1986, 132, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.L.; Ford, S.J.; Pridmore, R.D.; Fritzinger, D.C. Nucleotide sequence of a gene from the Pseudomonas transposon Tn501 encoding mercuric reductase. Biochemistry 1983, 22, 4089–4095. [Google Scholar] [CrossRef]
- Summers, A.O. Organization, expression, and evolution of genes for mercury resistance. Annu. Rev. Microbiol. 1986, 40, 607–634. [Google Scholar] [CrossRef]
- Ni’Bhriain, N.N.; Silver, S.; Foster, T.J. Tn5 insertion mutations in the mercuric ion resistance genes derived from plasmid R100. J. Bacteriol. 1983, 155, 690–703. [Google Scholar] [CrossRef]
- Park, S.J.; Wireman, J.; Summers, A.O. Genetic analysis of the Tn21 mer operator-promoter. J. Bacteriol. 1992, 174, 2160–2171. [Google Scholar] [CrossRef]
- Summers, A.O. Untwist and shout: A heavy metal-responsive transcriptional regulator. J. Bacteriol. 1992, 174, 3097–3101. [Google Scholar] [CrossRef]
- Lee, I.W.; Livrelli, V.; Park, S.J.; Totis, P.A.; Summers, A.O. Invivo DNA-Protein Interactions at the Divergent Mercury Resistance (Mer) Promoters. 2. Repressor Activator (Merr)-RNA Polymerase Interaction with Merop Mutants. J. Biol. Chem. 1993, 268, 2632–2639. [Google Scholar] [CrossRef]
- Lund, P.; Brown, N. Up-promoter mutations in the positively-regulated mer promoter of Tn501. Nucleic Acids Res. 1989, 17, 5517–5527. [Google Scholar] [CrossRef]
- Lund, P.A.; Brown, N.L. Regulation of transcription in Escherichia coli from the mer and merR promoters in the transposon Tn501. J. Mol. Biol. 1989, 205, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Parkhill, J.; Brown, N.L. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. 1990, 18, 5157–5162. [Google Scholar] [CrossRef] [PubMed]
- O’Halloran, T.V.; Frantz, B.; Shin, M.K.; Ralston, D.M.; Wright, J.G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell 1989, 56, 119–129. [Google Scholar] [CrossRef]
- Ansari, A.Z.; Chael, M.L.; O’Halloran, T.V. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature 1992, 355, 87–89. [Google Scholar] [CrossRef]
- Baksh, K.A.; Zamble, D.B. Allosteric control of metal-responsive transcriptional regulators in bacteria. J. Biol. Chem. 2020, 295, 1673–1684. [Google Scholar] [CrossRef]
- Priyadarshanee, M.; Chatterjee, S.; Rath, S.; Dash, H.R.; Das, S. Cellular and genetic mechanism of bacterial mercury resistance and their role in biogeochemistry and bioremediation. J. Hazard. Mater. 2022, 423, 126985. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Qiu, T.; Sun, Z.; Liu, F.; Yu, B. Mercury bioremediation by engineered Pseudomonas putida KT2440 with adaptationally optimized biosecurity circuit. Environ. Microbiol. 2022, 24, 3022–3036. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yu, H.; Zhang, B.; Liu, S.; Liu, C.G.; Li, F.; Song, H. Engineering whole-cell microbial biosensors: Design principles and applications in monitoring and treatment of heavy metals and organic pollutants. Biotechnol. Adv. 2022, 60, 108019. [Google Scholar] [CrossRef]
- Hui, C.Y.; Guo, Y.; Li, H.; Chen, Y.T.; Yi, J. Differential Detection of Bioavailable Mercury and Cadmium Based on a Robust Dual-Sensing Bacterial Biosensor. Front. Microbiol. 2022, 13, 846524. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Zhang, Y. Bacterial MerR family transcription regulators: Activationby distortion. Acta Biochim. Biophys. Sin. 2022, 54, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Heldwein, E.E.; Brennan, R.G. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 2001, 409, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Newberry, K.J.; Brennan, R.G. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J. Biol. Chem. 2004, 279, 20356–20362. [Google Scholar] [CrossRef] [PubMed]
- Bachas, S.; Eginton, C.; Gunio, D.; Wade, H. Structural contributions to multidrug recognition in the multidrug resistance (MDR) gene regulator, BmrR. Proc. Natl. Acad. Sci. USA 2011, 108, 11046–11051. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Q.; Yang, J.; Huang, S.; Wei, T.; Chen, W.; He, Y.; Wang, D.; Liu, Z.; Wang, K.; et al. Selective cadmium regulation mediated by a cooperative binding mechanism in CadR. Proc. Natl. Acad. Sci. USA 2019, 116, 20398–20403. [Google Scholar] [CrossRef]
- Fang, C.; Li, L.; Zhao, Y.; Wu, X.; Philips, S.J.; You, L.; Zhong, M.; Shi, X.; O’Halloran, T.V.; Li, Q.; et al. The bacterial multidrug resistance regulator BmrR distorts promoter DNA to activate transcription. Nat. Commun. 2020, 11, 6284. [Google Scholar] [CrossRef]
- Fang, C.; Philips, S.J.; Wu, X.; Chen, K.; Shi, J.; Shen, L.; Xu, J.; Feng, Y.; O’Halloran, T.V.; Zhang, Y. CueR activates transcription through a DNA distortion mechanism. Nat. Chem. Biol. 2021, 17, 57–64. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, B.; Jiang, Y.; Liu, C.; Zhou, W.; Chen, M.; Yang, Y.; Hu, Y.; Liu, B. Structural basis of copper-efflux-regulator-dependent transcription activation. iScience 2021, 24, 102449. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, C.; Zhou, W.; Shi, W.; Chen, M.; Zhang, B.; Schatz, D.G.; Hu, Y.; Liu, B. Structural visualization of transcription activated by a multidrug-sensing MerR family regulator. Nat. Commun. 2021, 12, 2702. [Google Scholar] [CrossRef]
- Tulin, G.; Figueroa, N.R.; Checa, S.K.; Soncini, F.C. The multifarious MerR family of transcriptional regulators. Mol. Microbiol. 2024, 121, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Philips, S.J.; Canalizo-Hernandez, M.; Yildirim, I.; Schatz, G.C.; Mondragon, A.; O’Halloran, T.V. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science 2015, 349, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Sameach, H.; Narunsky, A.; Azoulay-Ginsburg, S.; Gevorkyan-Aiapetov, L.; Zehavi, Y.; Moskovitz, Y.; Juven-Gershon, T.; Ben-Tal, N.; Ruthstein, S. Structural and Dynamics Characterization of the MerR Family Metalloregulator CueR in its Repression and Activation States. Structure 2017, 25, 988–996.e983. [Google Scholar] [CrossRef] [PubMed]
- Sameach, H.; Ghosh, S.; Gevorkyan-Airapetov, L.; Saxena, S.; Ruthstein, S. EPR Spectroscopy Detects Various Active State Conformations of the Transcriptional Regulator CueR. Angew. Chem. Int. Ed. Engl. 2019, 58, 3053–3056. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Kita, A.; Kobayashi, K.; Miki, K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc. Natl. Acad. Sci. USA 2008, 105, 4121–4126. [Google Scholar] [CrossRef] [PubMed]
- Counago, R.M.; Chen, N.H.; Chang, C.W.; Djoko, K.Y.; McEwan, A.G.; Kobe, B. Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region. Nucleic Acids Res. 2016, 44, 6981–6993. [Google Scholar] [CrossRef]
- Hubin, E.A.; Lilic, M.; Darst, S.A.; Campbell, E.A. Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures. Nat. Commun. 2017, 8, 16072. [Google Scholar] [CrossRef]
- Harley, C.B.; Reynolds, R.P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987, 15, 2343–2361. [Google Scholar] [CrossRef] [PubMed]
- Yeh, H.Y.; Chen, T.C.; Liou, K.M.; Hsu, H.T.; Chung, K.M.; Hsu, L.L.; Chang, B.Y. The core-independent promoter-specific interaction of primary sigma factor. Nucleic Acids Res. 2011, 39, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Lin, L.Y.; Zou, X.W.; Huang, C.C.; Chan, N.L. Structural basis of the mercury(II)-mediated conformational switching of the dual-function transcriptional regulator MerR. Nucleic Acids Res. 2015, 43, 7612–7623. [Google Scholar] [CrossRef] [PubMed]
- Yeh, H.Y.; Hsu, H.T.; Chen, T.C.; Chung, K.M.; Liou, K.M.; Chang, B.Y. The reduction in sigma-promoter recognition flexibility as induced by core RNAP is required for sigma to discern the optimal promoter spacing. Biochem. J. 2013, 455, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Jaurin, B.; Grundstrom, T.; Edlund, T.; Normark, S. The E. coli β-lactamase attenuator mediates growth rate-dependent regulation. Nature 1981, 290, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.R.; Bennett, G.N. Construction and analysis of in vivo activity of E. coli promoter hybrids and promoter mutants that alter the -35 to -10 spacing. Gene 1982, 20, 231–243. [Google Scholar] [CrossRef]
- Aoyama, T.; Takanami, M.; Ohtsuka, E.; Taniyama, Y.; Marumoto, R.; Sato, H.; Ikehara, M. Essential structure of E. coli promoter: Effect of spacer length between the two consensus sequences on promoter function. Nucleic Acids Res. 1983, 11, 5855–5864. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fang, C.; Zhuang, N.; Wang, T.; Zhang, Y. Structural basis for transcription initiation by bacterial ECF sigma factors. Nat. Commun. 2019, 10, 1153. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, M.; Peterfreund, G.L.; Tsou, A.M.; Selamoglu, N.; Daldal, F.; Zhong, Z.; Kan, B.; Zhu, J. Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proc. Natl. Acad. Sci. USA 2011, 108, 810–815. [Google Scholar] [CrossRef]
- Pedelacq, J.D.; Cabantous, S.; Tran, T.; Terwilliger, T.C.; Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006, 24, 79–88. [Google Scholar] [CrossRef]
- Yang, R.; Zhu, L.; Li, T.; Zhu, L.Y.; Ye, Z.; Zhang, D. Photosynthetic Conversion of CO(2) Into Pinene Using Engineered Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2021, 9, 779437. [Google Scholar] [CrossRef]
- Caicedo-Burbano, P.; Smit, T.; Pineda Hernandez, H.; Du, W.; Branco Dos Santos, F. Construction of Fully Segregated Genomic Libraries in Polyploid Organisms Such as Synechocystis sp. PCC 6803. ACS Synth. Biol. 2020, 9, 2632–2638. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, H.; Meng, H.; Zhu, Y.; Bao, G.; Zhang, Y.; Li, Y.; Ma, Y. Discovery of a super-strong promoter enables efficient production of heterologous proteins in cyanobacteria. Sci. Rep. 2014, 4, 4500. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, M.; Hihara, Y. Characterization of high-light-responsive promoters of the psaAB genes in Synechocystis sp. PCC 6803. Plant Cell Physiol. 2006, 47, 878–890. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.H.; Camsund, D.; Lindblad, P.; Heidorn, T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010, 38, 2577–2593. [Google Scholar] [CrossRef] [PubMed]
- Albers, S.C.; Gallegos, V.A.; Peebles, C.A. Engineering of genetic control tools in Synechocystis sp. PCC 6803 using rational design techniques. J. Biotechnol. 2015, 216, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.H.; Wang, X.; Ma, Y.X.; Wang, S. CMV enhancer/human PDGF-beta promoter for neuron-specific transgene expression. Gene Ther. 2004, 11, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.L.; Stoyanov, J.V.; Kidd, S.P.; Hobman, J.L. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003, 27, 145–163. [Google Scholar] [CrossRef]
- Delano, W.L. The PyMOL Molecular Graphics System; Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Changela, A.; Chen, K.; Xue, Y.; Holschen, J.; Outten, C.E.; O’Halloran, T.V.; Mondragon, A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 2003, 301, 1383–1387. [Google Scholar] [CrossRef]
- Huang, S.; Liu, X.; Wang, D.; Chen, W.; Hu, Q.; Wei, T.; Zhou, W.; Gan, J.; Chen, H. Structural Basis for the Selective Pb(II) Recognition of Metalloregulatory Protein PbrR691. Inorg. Chem. 2016, 55, 12516–12519. [Google Scholar] [CrossRef]
- Hubin, E.A.; Fay, A.; Xu, C.; Bean, J.M.; Saecker, R.M.; Glickman, M.S.; Darst, S.A.; Campbell, E.A. Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA. Elife 2017, 6, e22520. [Google Scholar] [CrossRef] [PubMed]
- Boyaci, H.; Chen, J.; Jansen, R.; Darst, S.A.; Campbell, E.A. Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding. Nature 2019, 565, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Qayyum, M.Z.; Pupov, D.; Esyunina, D.; Kulbachinskiy, A.; Murakami, K.S. Structural basis of ribosomal RNA transcription regulation. Nat. Commun. 2021, 12, 528. [Google Scholar] [CrossRef] [PubMed]
- Wosten, M.M. Eubacterial sigma-factors. FEMS Microbiol. Rev. 1998, 22, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Alvey, R.M.; Byrne, P.O.; Graham, J.E.; Shen, G.; Bryant, D.A. Expression of Genes in Cyanobacteria: Adaptation of Endogenous Plasmids as Platforms for High-Level Gene Expression in Synechococcus sp. PCC 7002. In Photosynthesis Research Protocols; Carpentier, R., Ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 273–293. [Google Scholar]
| Promoter | Expression Level (%) 1 | |||
|---|---|---|---|---|
| pPmerL (−MerR) | PMPmerL (+MerR) | |||
| Without Hg (II) | With Hg (II) | Without Hg (II) | With Hg (II) | |
| Pmer-WT | 0 | 0 | 0 | 100 |
| Pmer-M1 | 264 | 266 | 267 | 253 |
| Pmer-M2 | 345 | 344 | 447 | 421 |
| Pmer-M3 | 17 | 15 | 16 | 17 |
| Pmer-M4 | 222 | 246 | 279 | 247 |
| Pmer-M5 | 500 | 484 | 582 | 517 |
| Pmer-M6 | 19 | 20 | 22 | 20 |
| Pmer-M7 | 306 | 332 | 264 | 304 |
| Pmer-M8 | 376 | 373 | 401 | 407 |
| Pmer-M9 | 27 | 24 | 26 | 25 |
| Pmer-M10 | 142 | 142 | 156 | 158 |
| Pmer-M11 | 30 | 10 | 45 | 45 |
| Pmer-M12 | 126 | 128 | 311 | 346 |
| Pmer-M13 | 75 | 73 | 182 | 181 |
| Pmer-M14 | 28 | 28 | 23 | 27 |
| Pmer-M15 | 0 | 0 | 0 | 12 |
| Pmer-M16 | 0 | 0 | 0 | 27 |
| Pmer-M17 | 0 | 0 | 0 | 15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Q.; Wang, J.; Liu, C.; Feng, Y.; Chen, H. Determinants of mer Promoter Activity from Pseudomonas aeruginosa. Genes 2024, 15, 490. https://doi.org/10.3390/genes15040490
Hu Q, Wang J, Liu C, Feng Y, Chen H. Determinants of mer Promoter Activity from Pseudomonas aeruginosa. Genes. 2024; 15(4):490. https://doi.org/10.3390/genes15040490
Chicago/Turabian StyleHu, Qingyuan, Jue Wang, Chunhong Liu, Yu Feng, and Hao Chen. 2024. "Determinants of mer Promoter Activity from Pseudomonas aeruginosa" Genes 15, no. 4: 490. https://doi.org/10.3390/genes15040490





