Association of Gene Polymorphisms with Normal Tension Glaucoma: A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.1.1. Inclusion Criteria
- (1)
- The diagnostic standard of NTG should be indicated clearly;
- (2)
- Cohort studies involving NTG patients and healthy controls which evaluate the potential association of specific gene mutations, SNPs, allele variations related to pathogenesis of the disease;
- (3)
- Some important information should be included: demographic features such as age and sex, allele or genotype frequencies of SNPs in both case and control groups, index of association strength such as odds ratio (OR) with 95% confidence interval (CI).
2.1.2. Exclusion Criteria
- (1)
- Studies published in the form of meta-analysis, review, case report, patent, guideline, conference abstract and book chapters;
- (2)
- Studied objects are animals;
- (3)
- Studies not written in English;
- (4)
- Studies which lack OR value, only refers to POAG but not NTG or did not indicate a clear definition of POAG.
2.2. Data Extraction
2.3. Quality Assessment
2.4. Meta-Analysis
3. Results
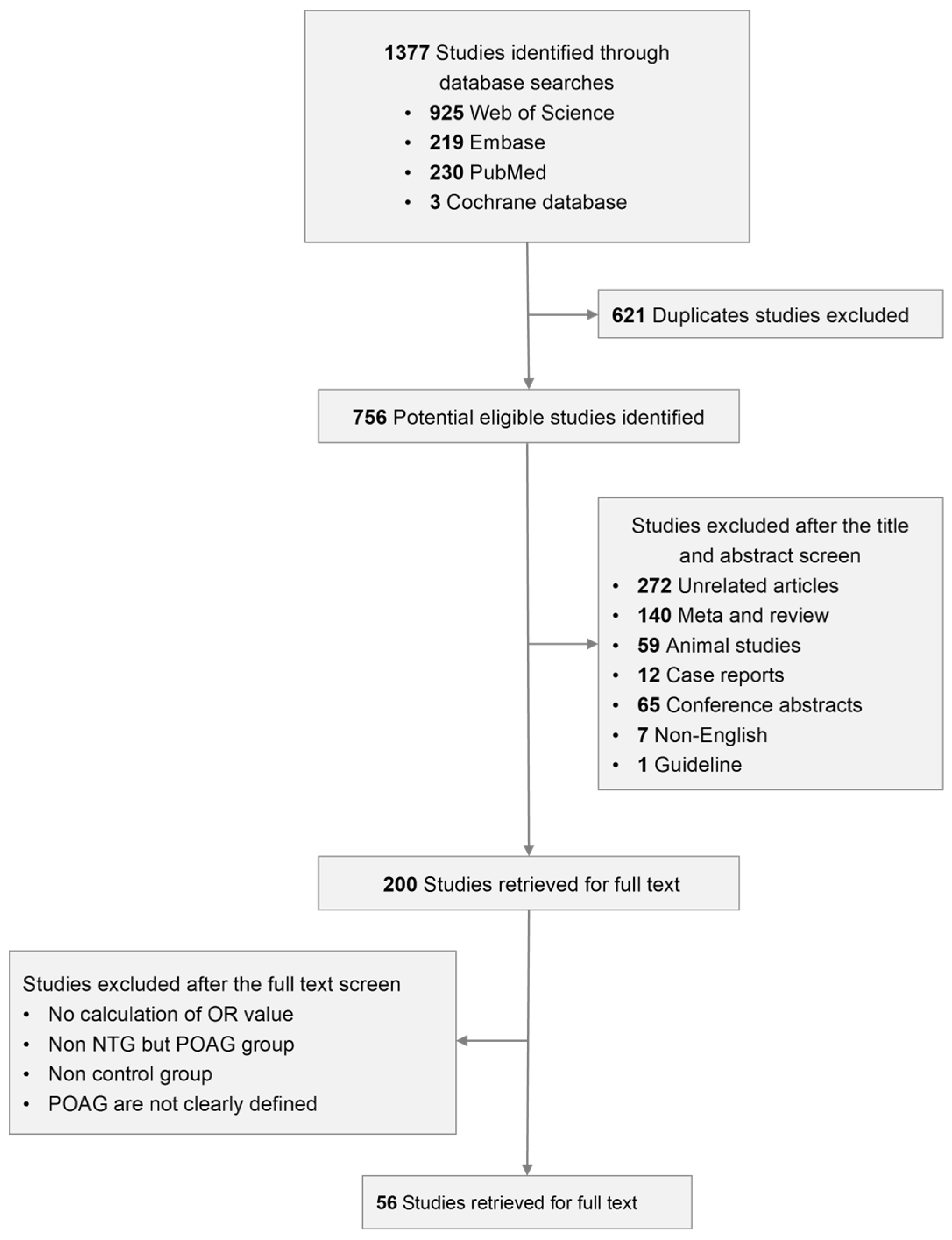
3.1. Selection of Qualified Literature
3.2. Characteristics of Qualified Studies
3.3. Meta-Analysis Results
3.3.1. Gene Polymorphisms Associated with NTG
EDNRA Polymorphisms
ELOVL5 Polymorphism
HK2 Polymorphism
NCK2 Polymorphism
OPA1 Polymorphisms
OPTN Polymorphisms
P53 Polymorphism
SRBD1 Polymorphism
TLR4 Polymorphisms
SIX1–SIX6 Polymorphism
3.3.2. Gene Polymorphisms Not Associated with NTG
3.3.3. Stratified Analysis in Different Ethnicities
3.4. Measurement of Publication Biases and Sensitivity Analysis
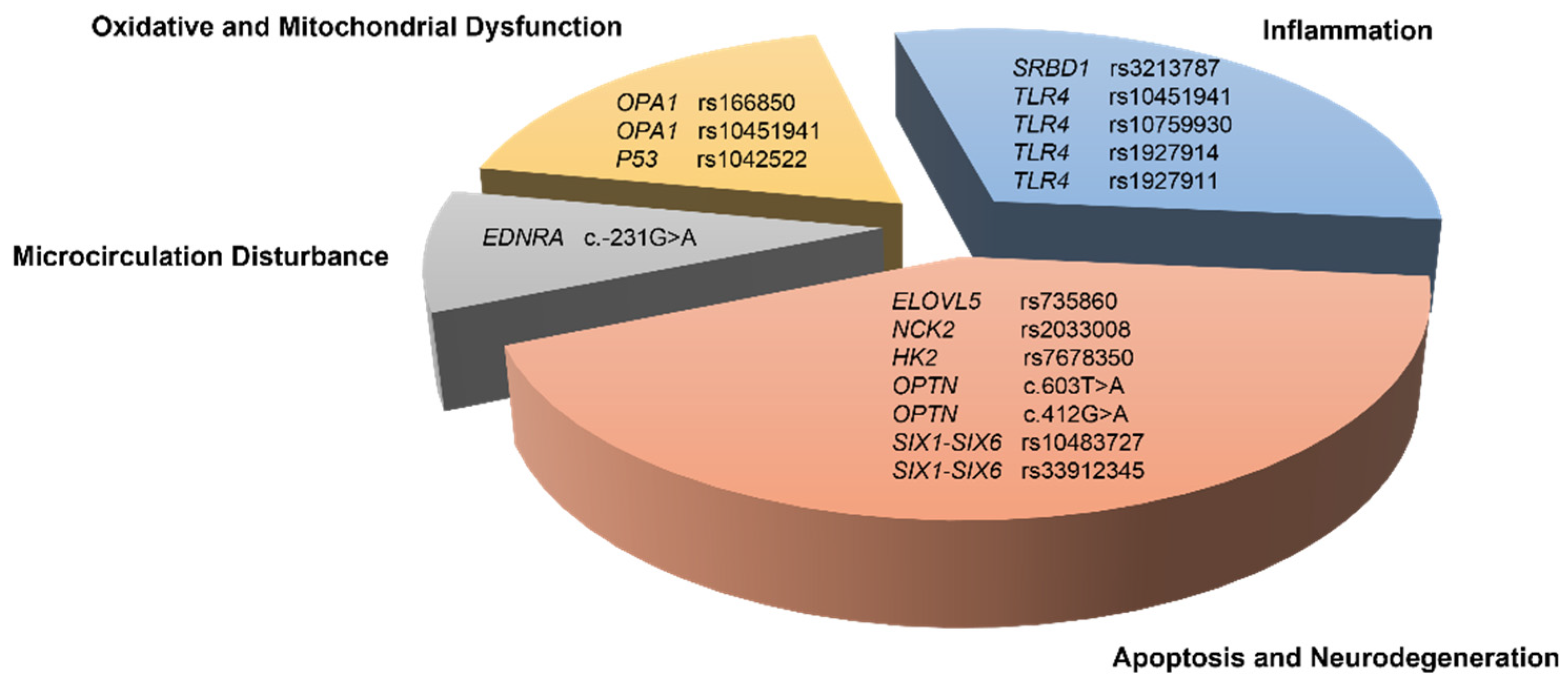
4. Discussion
4.1. Oxidative Stress-Related Genes
4.2. Neurodegeneration and Apoptosis-Related Genes
4.3. Inflammation-Related Genes
4.4. Microcirculation Disturbance-Related Gene
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stein, J.D.; Khawaja, A.P.; Weizer, J.S. Glaucoma in Adults-Screening, Diagnosis, and Management: A Review. JAMA 2021, 325, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Petrov, S.Y. [Modern view on normal-tension glaucoma]. Vestn. Oftalmol. 2020, 136, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Esporcatte, B.L.; Tavares, I.M. Normal-tension glaucoma: An update. Arq. Bras. Oftalmol. 2016, 79, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Killer, H.E.; Pircher, A. Normal tension glaucoma: Review of current understanding and mechanisms of the pathogenesis. Eye 2018, 32, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Shields, M.B. Normal-tension glaucoma: Is it different from primary open-angle glaucoma? Curr. Opin. Ophthalmol. 2008, 19, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Gosling, D.; Meyer, J.J. Normal Tension Glaucoma. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Chen, M.J. Normal tension glaucoma in Asia: Epidemiology, pathogenesis, diagnosis, and management. Taiwan. J. Ophthalmol. 2020, 10, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.Y.L.; Tham, C.C. Normal-tension glaucoma: Current concepts and approaches-A review. Clin. Exp. Ophthalmol. 2022, 50, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.F.; Gaier, E.D.; Wiggs, J.L. Genetics of Primary Inherited Disorders of the Optic Nerve: Clinical Applications. Cold Spring Harb. Perspect. Med. 2015, 5, a017277. [Google Scholar] [CrossRef]
- Wiggs, J.L.; Pasquale, L.R. Genetics of glaucoma. Hum. Mol. Genet. 2017, 26, R21–R27. [Google Scholar] [CrossRef]
- Alward, W.L.M.; van der Heide, C.; Khanna, C.L.; Roos, B.R.; Sivaprasad, S.; Kam, J.; Ritch, R.; Lotery, A.; Igo, R.P., Jr.; Cooke Bailey, J.N.; et al. Myocilin Mutations in Patients With Normal-Tension Glaucoma. JAMA Ophthalmol. 2019, 137, 559–563. [Google Scholar] [CrossRef]
- Lu, S.Y.; Rong, S.S.; Wu, Z.; Huang, C.; Matsushita, K.; Ng, T.K.; Leung, C.K.S.; Kawashima, R.; Usui, S.; Tam, P.O.S.; et al. Association of the CAV1-CAV2 locus with normal-tension glaucoma in Chinese and Japanese. Clin. Exp. Ophthalmol. 2020, 48, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Stang, A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010, 25, 603–605. [Google Scholar] [CrossRef]
- Higgins, J.P.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Egger, M.; Davey Smith, G.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Aung, T.; Ocaka, L.A.; Ebenezer, N.D.; Morris, A.G.; Brice, G.; Child, A.H.; Hitchings, R.A.; Lehmann, O.J.; Bhattacharya, S.S. Investigating the association between OPA1 polymorphisms and glaucoma: Comparison between normal tension and high tension primary open angle glaucoma. Hum. Genet. 2002, 110, 513–514. [Google Scholar] [CrossRef] [PubMed]
- Powell, B.L.; Toomes, C.; Scott, S.; Yeung, A.; Marchbank, N.J.; Spry, P.G.; Lumb, R.; Inglehearn, C.F.; Churchill, A.J. Polymorphisms in OPA1 are associated with normal tension glaucoma. Mol. Vis. 2003, 9, 460–464. [Google Scholar] [PubMed]
- Funayama, T.; Ishikawa, K.; Ohtake, Y.; Tanino, T.; Kurosaka, D.; Kimura, I.; Suzuki, K.; Ideta, H.; Nakamoto, K.; Yasuda, N.; et al. Variants in optineurin gene and their association with tumor necrosis factor-α polymorphisms in Japanese patients with glaucoma. Investig. Ophthalmol. Vis. Sci. 2004, 45, 4359–4367. [Google Scholar] [CrossRef] [PubMed]
- Fuse, N.; Takahashi, K.; Akiyama, H.; Nakazawa, T.; Seimiya, M.; Kuwahara, S.; Tamai, M. Molecular genetic analysis of optineurin gene for primary open-angle and normal tension glaucoma in the Japanese population. J. Glaucoma 2004, 13, 299–303. [Google Scholar] [CrossRef]
- Woo, S.J.; Kim, D.M.; Kim, J.Y.; Park, S.S.; Ko, H.S.; Yoo, T. Investigation of the association between OPA1 polymorphisms and normal-tension glaucoma in Korea. J. Glaucoma 2004, 13, 492–495. [Google Scholar] [CrossRef]
- Dimasi, D.P.; Hewitt, A.W.; Green, C.M.; Mackey, D.A.; Craig, J.E. Lack of association of p53 polymorphisms and haplotypes in high and normal tension open angle glaucoma. J. Med. Genet. 2005, 42, e55. [Google Scholar] [CrossRef]
- Fan, B.J.; Wang, D.Y.; Fan, D.S.; Tam, P.O.; Lam, D.S.; Tham, C.C.; Lam, C.Y.; Lau, T.C.; Pang, C.P. SNPs and interaction analyses of myocilin, optineurin, and apolipoprotein E in primary open angle glaucoma patients. Mol. Vis. 2005, 11, 625–631. [Google Scholar]
- Hashizume, K.; Mashima, Y.; Fumayama, T.; Ohtake, Y.; Kimura, I.; Yoshida, K.; Ishikawa, K.; Yasuda, N.; Fujimaki, T.; Asaoka, R.; et al. Genetic polymorphisms in the angiotensin II receptor gene and their association with open-angle glaucoma in a Japanese population. Investig. Ophthalmol. Vis. Sci. 2005, 46, 1993–2001. [Google Scholar] [CrossRef]
- Inagaki, Y.; Mashima, Y.; Fuse, N.; Funayama, T.; Ohtake, Y.; Yasuda, N.; Murakami, A.; Hotta, Y.; Fukuchi, T.; Tsubota, K. Polymorphism of β-adrenergic receptors and susceptibility to open-angle glaucoma. Mol. Vis. 2006, 12, 673–680. [Google Scholar]
- Kim, S.H.; Kim, J.Y.; Kim, D.M.; Ko, H.S.; Kim, S.Y.; Yoo, T.; Hwang, S.; Park, S.S. Investigations on the association between normal tension glaucoma and single nucleotide polymorphisms of the endothelin-1 and endothelin receptor genes. Mol. Vis. 2006, 12, 1016–1021. [Google Scholar]
- Lam, C.Y.; Fan, B.J.; Wang, D.Y.; Tam, P.O.; Yung Tham, C.C.; Leung, D.Y.; Ping Fan, D.S.; Chiu Lam, D.S.; Pang, C.P. Association of apolipoprotein E polymorphisms with normal tension glaucoma in a Chinese population. J. Glaucoma 2006, 15, 218–222. [Google Scholar] [CrossRef]
- Mabuchi, F.; Tang, S.; Kashiwagi, K.; Yamagata, Z.; Iijima, H.; Tsukahara, S. Methylenetetrahydrofolate reductase gene polymorphisms c.677C/T and c.1298A/C are not associated with open angle glaucoma. Mol. Vis. 2006, 12, 735–739. [Google Scholar]
- Yao, W.; Jiao, X.; Hejtmancik, J.F.; Leske, M.C.; Hennis, A.; Nemesure, B.; He, Q.; Wu, S.Y.; Mendell, N.; Jiang, L.; et al. Evaluation of the association between OPA1 polymorphisms and primary open-angle glaucoma in Barbados families. Mol. Vis. 2006, 12, 649–654. [Google Scholar]
- How, A.C.; Aung, T.; Chew, X.; Yong, V.H.; Lim, M.C.; Lee, K.Y.; Toh, J.Y.; Li, Y.; Liu, J.; Vithana, E.N. Lack of association between interleukin-1 gene cluster polymorphisms and glaucoma in Chinese subjects. Investig. Ophthalmol. Vis. Sci. 2007, 48, 2123–2126. [Google Scholar] [CrossRef]
- Jeoung, J.W.; Kim, D.M.; Ko, H.S.; Park, S.S.; Kim, J.Y.; Kim, S.Y.; Yoo, T.W. Investigation of the association between normal-tension glaucoma and single nucleotide polymorphisms in natriuretic peptide gene. Korean J. Ophthalmol. 2007, 21, 33–38. [Google Scholar] [CrossRef]
- Mabuchi, F.; Tang, S.; Kashiwagi, K.; Yamagata, Z.; Iijima, H.; Tsukahara, S. The OPA1 gene polymorphism is associated with normal tension and high tension glaucoma. Am. J. Ophthalmol. 2007, 143, 125–130. [Google Scholar] [CrossRef]
- Miyazawa, A.; Fuse, N.; Mengkegale, M.; Ryu, M.; Seimiya, M.; Wada, Y.; Nishida, K. Association between primary open-angle glaucoma and WDR36 DNA sequence variants in Japanese. Mol. Vis. 2007, 13, 1912–1919. [Google Scholar]
- Tosaka, K.; Mashima, Y.; Funayama, T.; Ohtake, Y.; Kimura, I.; Glaucoma Gene Research, G. Association between open-angle glaucoma and gene polymorphism for heat-shock protein 70-1. Jpn. J. Ophthalmol. 2007, 51, 417–423. [Google Scholar] [CrossRef]
- Shibuya, E.; Meguro, A.; Ota, M.; Kashiwagi, K.; Mabuchi, F.; Iijima, H.; Kawase, K.; Yamamoto, T.; Nakamura, M.; Negi, A.; et al. Association of Toll-like receptor 4 gene polymorphisms with normal tension glaucoma. Investig. Ophthalmol. Vis. Sci. 2008, 49, 4453–4457. [Google Scholar] [CrossRef]
- Clement, C.I.; Goldberg, I.; Healey, P.R.; Graham, S.L. Plasma homocysteine, MTHFR gene mutation, and open-angle glaucoma. J. Glaucoma 2009, 18, 73–78. [Google Scholar] [CrossRef]
- Daugherty, C.L.; Curtis, H.; Realini, T.; Charlton, J.F.; Zareparsi, S. Primary open angle glaucoma in a Caucasian population is associated with the p53 codon 72 polymorphism. Mol. Vis. 2009, 15, 1939–1944. [Google Scholar]
- Fan, B.J.; Wang, D.Y.; Cheng, C.Y.; Ko, W.C.; Lam, S.C.; Pang, C.P. Different WDR36 mutation pattern in Chinese patients with primary open-angle glaucoma. Mol. Vis. 2009, 15, 646–653. [Google Scholar]
- Mabuchi, F.; Sakurada, Y.; Kashiwagi, K.; Yamagata, Z.; Iijima, H.; Tsukahara, S. Lack of association between p53 gene polymorphisms and primary open angle glaucoma in the Japanese population. Mol. Vis. 2009, 15, 1045–1049. [Google Scholar]
- Woo, S.J.; Kim, J.Y.; Kim, D.M.; Park, S.S.; Ko, H.S.; Yoo, T. Investigation of the association between 677C>T and 1298A>C 5,10-methylenetetra- hydrofolate reductase gene polymorphisms and normal-tension glaucoma. Eye 2009, 23, 17–24. [Google Scholar] [CrossRef]
- Fan, B.J.; Liu, K.; Wang, D.Y.; Tham, C.C.; Tam, P.O.; Lam, D.S.; Pang, C.P. Association of polymorphisms of tumor necrosis factor and tumor protein p53 with primary open-angle glaucoma. Investig. Ophthalmol. Vis. Sci. 2010, 51, 4110–4116. [Google Scholar] [CrossRef]
- Mabuchi, F.; Sakurada, Y.; Kashiwagi, K.; Yamagata, Z.; Iijima, H.; Tsukahara, S. Estrogen receptor β gene polymorphism and intraocular pressure elevation in female patients with primary open-angle glaucoma. Am. J. Ophthalmol. 2010, 149, 826–830.e821–e822. [Google Scholar] [CrossRef]
- Mabuchi, F.; Sakurada, Y.; Kashiwagi, K.; Yamagata, Z.; Iijima, H.; Tsukahara, S. Lack of association of common variants on chromosome 2p with primary open-angle glaucoma in the Japanese population. Proc. Natl. Acad. Sci. USA 2010, 107, E90–E91. [Google Scholar] [CrossRef]
- Wolf, C.; Gramer, E.; Muller-Myhsok, B.; Pasutto, F.; Gramer, G.; Wissinger, B.; Weisschuh, N. Lysyl oxidase-like 1 gene polymorphisms in German patients with normal tension glaucoma, pigmentary glaucoma and exfoliation glaucoma. J. Glaucoma 2010, 19, 136–141. [Google Scholar] [CrossRef]
- Writing Committee for the Normal Tension Glaucoma Genetic Study Group of Japan Glaucoma Society; Meguro, A.; Inoko, H.; Ota, M.; Mizuki, N.; Bahram, S. Genome-wide association study of normal tension glaucoma: Common variants in SRBD1 and ELOVL5 contribute to disease susceptibility. Ophthalmology 2010, 117, 1331–1338.e5. [Google Scholar] [CrossRef]
- Yu-Wai-Man, P.; Stewart, J.D.; Hudson, G.; Andrews, R.M.; Griffiths, P.G.; Birch, M.K.; Chinnery, P.F. OPA1 increases the risk of normal but not high tension glaucoma. J. Med. Genet. 2010, 47, 120–125. [Google Scholar] [CrossRef]
- Mabuchi, F.; Sakurada, Y.; Kashiwagi, K.; Yamagata, Z.; Iijima, H.; Tsukahara, S. Association between SRBD1 and ELOVL5 gene polymorphisms and primary open-angle glaucoma. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4626–4629. [Google Scholar] [CrossRef]
- Suh, W.; Kim, S.; Ki, C.S.; Kee, C. Toll-like receptor 4 gene polymorphisms do not associate with normal tension glaucoma in a Korean population. Mol. Vis. 2011, 17, 2343–2348. [Google Scholar]
- Yasumura, R.; Meguro, A.; Ota, M.; Nomura, E.; Uemoto, R.; Kashiwagi, K.; Mabuchi, F.; Iijima, H.; Kawase, K.; Yamamoto, T.; et al. Investigation of the association between SLC1A3 gene polymorphisms and normal tension glaucoma. Mol. Vis. 2011, 17, 792–796. [Google Scholar]
- Takano, Y.; Shi, D.; Shimizu, A.; Funayama, T.; Mashima, Y.; Yasuda, N.; Fukuchi, T.; Abe, H.; Ideta, H.; Zheng, X.; et al. Association of Toll-like receptor 4 gene polymorphisms in Japanese subjects with primary open-angle, normal-tension, and exfoliation glaucoma. Am. J. Ophthalmol. 2012, 154, 825–832. [Google Scholar] [CrossRef]
- Wiggs, J.L.; Hewitt, A.W.; Fan, B.J.; Wang, D.Y.; Figueiredo Sena, D.R.; O’Brien, C.; Realini, A.; Craig, J.E.; Dimasi, D.P.; Mackey, D.A.; et al. The p53 codon 72 PRO/PRO genotype may be associated with initial central visual field defects in caucasians with primary open angle glaucoma. PLoS ONE 2012, 7, e45613. [Google Scholar] [CrossRef]
- Shi, D.; Funayama, T.; Mashima, Y.; Takano, Y.; Shimizu, A.; Yamamoto, K.; Mengkegale, M.G.; Miyazawa, A.; Yasuda, N.; Fukuchi, T.; et al. Association of HK2 and NCK2 with normal tension glaucoma in the Japanese population. PLoS ONE 2013, 8, e54115. [Google Scholar] [CrossRef]
- Shi, D.; Takano, Y.; Nakazawa, T.; Mengkegale, M.; Yokokura, S.; Nishida, K.; Fuse, N. Molecular genetic analysis of primary open-angle glaucoma, normal tension glaucoma, and developmental glaucoma for the VAV2 and VAV3 gene variants in Japanese subjects. Biochem. Biophys. Res. Commun. 2013, 432, 509–512. [Google Scholar] [CrossRef]
- Lin, K.H.; Feng, S.C.; Shen, Y.C.; Wei, L.C.; Liang, C.Y.; Chang, C.J.; Yang, Y.Y.; Chiu, C.H.; Wang, C.Y. Interleukin-6(-174) locus polymorphism and serum IL-6 levels in normal tension glaucoma. Ophthalmic Genet. 2014, 35, 255–257. [Google Scholar] [CrossRef]
- Gao, Y.; Li, W.; Yin, Z.; Ma, Y.; Cai, H.; Tang, X. Association between Genetic Polymorphisms of the β Adrenergic Receptor and Diurnal Intraocular Pressure in Chinese Volunteers and Glaucoma Patients. Curr. Eye Res. 2016, 41, 1553–1560. [Google Scholar] [CrossRef]
- Kosior-Jarecka, E.; Wróbel-Dudzińska, D.; Łukasik, U.; Aung, T.; Khor, C.C.; Kocki, J.; Żarnowski, T. Plasma endothelin-1 and single nucleotide polymorphisms of endothelin-1 and endothelin type A receptor genes as risk factors for normal tension glaucoma. Mol. Vis. 2016, 22, 1256–1266. [Google Scholar]
- Nishisako, M.; Meguro, A.; Nomura, E.; Yamane, T.; Takeuchi, M.; Ota, M.; Kashiwagi, K.; Mabuchi, F.; Iijima, H.; Kawase, K.; et al. SLC1A1 Gene Variants and Normal Tension Glaucoma: An Association Study. Ophthalmic Genet. 2016, 37, 194–200. [Google Scholar] [CrossRef]
- Sang, J.; Jia, L.; Zhao, B.; Wang, H.; Zhang, N.; Wang, N. Association of three single nucleotide polymorphisms at the SIX1-SIX6 locus with primary open angle glaucoma in the Chinese population. Sci. China Life Sci. 2016, 59, 694–699. [Google Scholar] [CrossRef]
- Jeoung, J.W.; Kim, D.M.; Oh, S.; Lee, J.S.; Park, S.S.; Kim, J.Y. The Relation Between Endothelial Nitric Oxide Synthase Polymorphisms and Normal Tension Glaucoma. J. Glaucoma 2017, 26, 1030–1035. [Google Scholar] [CrossRef]
- Suh, W.; Won, H.H.; Kee, C. The Association of Single-Nucleotide Polymorphisms in the MMP-9 Gene with Normal Tension Glaucoma and Primary Open-Angle Glaucoma. Curr. Eye Res. 2018, 43, 534–538. [Google Scholar] [CrossRef]
- Jung, S.H.; Lee, Y.C.; Lee, M.Y.; Shin, H.Y. Association of HK2 and NCK2 with normal-tension glaucoma in a population from the Republic of Korea. Graefes Arch. Clin. Exp. Ophthalmol. 2019, 257, 2717–2721. [Google Scholar] [CrossRef]
- Kosior-Jarecka, E.; Sagan, M.; Wrobel-Dudzinska, D.; Lukasik, U.; Aung, T.; Khor, C.C.; Kocki, J.; Zarnowski, T. Estrogen receptor gene polymorphisms and their influence on clinical status of Caucasian patients with primary open angle glaucoma. Ophthalmic Genet. 2019, 40, 323–328. [Google Scholar] [CrossRef]
- Jung, S.H.; Lee, Y.C.; Lee, M.Y.; Shin, H.Y. Lack of Correlation between ASB10 and Normal-tension Glaucoma in a Population from the Republic of Korea. Curr. Eye Res. 2020, 45, 521–525. [Google Scholar] [CrossRef]
- Jung, S.H.; Lee, Y.C.; Lee, M.Y.; Shin, H.Y. Lack of correlation between S1 RNA binding domain 1 SNP rs3213787/rs11884064 and normal-tension glaucoma in a population from the Republic of Korea. Medicine 2020, 99, e20066. [Google Scholar] [CrossRef]
- Lee, J.S.; Jeoung, J.W.; Oh, S.; Kim, D.M.; Ahn, J.H.; Kim, M.J.; Seong, M.W.; Park, S.S.; Kim, J.Y. No association between POU4F1, POU4F2, ISL1 polymorphisms and normal-tension glaucoma. Ophthalmic Genet. 2020, 41, 427–431. [Google Scholar] [CrossRef]
- Milanowski, P.; Kosior-Jarecka, E.; Lukasik, U.; Wrobel-Dudzinska, D.; Milanowska, J.; Khor, C.C.; Aung, T.; Kocki, J.; Żarnowski, T. Associations between OPA1, MFN1, and MFN2 polymorphisms and primary open angle glaucoma in Polish participants of European ancestry. Ophthalmic Genet. 2022, 43, 42–47. [Google Scholar] [CrossRef]
- Yue, J.L.; Zheng, S.F. Analysis of association between MALAT1 haplotype and the severity of normal-tension glaucoma (NTG). J. Cell Mol. Med. 2021, 25, 9918–9926. [Google Scholar] [CrossRef]
- Kim, Y.W.; Bak, E.; Wy, S.; Lee, S.C.; Kim, Y.J.; Kim, Y.K.; Park, K.H.; Jeoung, J.W. Genetic Risk and Phenotype Correlation of Primary Open-Angle Glaucoma Based on Rho-Kinase Gene Polymorphisms. J. Clin. Med. 2021, 10, 1953. [Google Scholar] [CrossRef]
- Liuska, P.J.; Lemmela, S.; Havulinna, A.S.; Kaarniranta, K.; Uusitalo, H.; Laivuori, H.; Kiiskinen, T.; Daly, M.J.; Palotie, A.; Turunen, J.A.; et al. Association of the MYOC p.(Gln368Ter) Variant With Glaucoma in a Finnish Population. JAMA Ophthalmol. 2021, 139, 762–768. [Google Scholar] [CrossRef]
- He, J.N.; Ng, T.K.; Lu, S.Y.; Tam, P.O.S.; Chan, P.P.; Tham, C.C.; Pang, C.P.; Chen, L.J.; Chu, W.K. Genetic association of ANGPT2 with primary open-angle glaucoma. Eye Vis. 2022, 9, 37. [Google Scholar] [CrossRef]
- Lee, C.C.; Wang, T.C.; Wang, H.Y.; Hsu, B.; Shih, R.J.; Lo, N.W.; Wang, C.Y. Association between HSPA5 Promoter Polymorphisms and a Reduced Risk of Normal Tension Glaucoma. Ophthalmic Res. 2022, 65, 474–480. [Google Scholar] [CrossRef]
- Shin, H.Y.; Lee, Y.C.; Lee, M.Y. Association of Polymorphisms at the SIX1/SIX6 Locus With Normal Tension Glaucoma in a Korean Population. J. Glaucoma 2022, 31, 763–766. [Google Scholar] [CrossRef]
- Olichon, A.; Emorine, L.J.; Descoins, E.; Pelloquin, L.; Brichese, L.; Gas, N.; Guillou, E.; Delettre, C.; Valette, A.; Hamel, C.P.; et al. The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett. 2002, 523, 171–176. [Google Scholar] [CrossRef]
- Frezza, C.; Cipolat, S.; Martins de Brito, O.; Micaroni, M.; Beznoussenko, G.V.; Rudka, T.; Bartoli, D.; Polishuck, R.S.; Danial, N.N.; De Strooper, B.; et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 2006, 126, 177–189. [Google Scholar] [CrossRef]
- Varanita, T.; Soriano, M.E.; Romanello, V.; Zaglia, T.; Quintana-Cabrera, R.; Semenzato, M.; Menabò, R.; Costa, V.; Civiletto, G.; Pesce, P.; et al. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab. 2015, 21, 834–844. [Google Scholar] [CrossRef]
- Liu, R.; Chan, D.C. OPA1 and cardiolipin team up for mitochondrial fusion. Nat. Cell Biol. 2017, 19, 760–762. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, X.; Zhang, H.; Li, N.; Yang, X.; Cheng, W.; Zhao, K. Association of OPA1 polymorphisms with NTG and HTG: A meta-analysis. PLoS ONE 2012, 7, e42387. [Google Scholar] [CrossRef]
- Toledo, F.; Wahl, G.M. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 2006, 6, 909–923. [Google Scholar] [CrossRef]
- Levine, A.J. p53, the cellular gatekeeper for growth and division. Cell 1997, 88, 323–331. [Google Scholar] [CrossRef]
- Vousden, K.H.; Lane, D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 275–283. [Google Scholar] [CrossRef]
- Artandi, S.E.; Attardi, L.D. Pathways connecting telomeres and p53 in senescence, apoptosis, and cancer. Biochem. Biophys. Res. Commun. 2005, 331, 881–890. [Google Scholar] [CrossRef]
- Jeong, B.S.; Hu, W.; Belyi, V.; Rabadan, R.; Levine, A.J. Differential levels of transcription of p53-regulated genes by the arginine/proline polymorphism: p53 with arginine at codon 72 favors apoptosis. FASEB J. 2010, 24, 1347–1353. [Google Scholar] [CrossRef]
- Kihara, A. Very long-chain fatty acids: Elongation, physiology and related disorders. J. Biochem. 2012, 152, 387–395. [Google Scholar] [CrossRef]
- Bazinet, R.P.; Laye, S. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 2014, 15, 771–785. [Google Scholar] [CrossRef]
- Serhan, C.N.; Chiang, N.; Dalli, J.; Levy, B.D. Lipid mediators in the resolution of inflammation. Cold Spring Harb. Perspect. Biol. 2014, 7, a016311. [Google Scholar] [CrossRef]
- Guillou, H.; Zadravec, D.; Martin, P.G.; Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010, 49, 186–199. [Google Scholar] [CrossRef]
- SanGiovanni, J.P.; Chew, E.Y. The role of omega-3 long-chain polyunsaturated fatty acids in health and disease of the retina. Prog. Retin. Eye Res. 2005, 24, 87–138. [Google Scholar] [CrossRef]
- Ren, H.; Magulike, N.; Ghebremeskel, K.; Crawford, M. Primary open-angle glaucoma patients have reduced levels of blood docosahexaenoic and eicosapentaenoic acids. Prostaglandins Leukot. Essent. Fatty Acids 2006, 74, 157–163. [Google Scholar] [CrossRef]
- Centenera, M.M.; Scott, J.S.; Machiels, J.; Nassar, Z.D.; Miller, D.C.; Zinonos, I.; Dehairs, J.; Burvenich, I.J.G.; Zadra, G.; Chetta, P.M.; et al. ELOVL5 Is a Critical and Targetable Fatty Acid Elongase in Prostate Cancer. Cancer Res. 2021, 81, 1704–1718. [Google Scholar] [CrossRef]
- Lee, J.Y.; Nam, M.; Son, H.Y.; Hyun, K.; Jang, S.Y.; Kim, J.W.; Kim, M.W.; Jung, Y.; Jang, E.; Yoon, S.J.; et al. Polyunsaturated fatty acid biosynthesis pathway determines ferroptosis sensitivity in gastric cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 32433–32442. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Z.; Zheng, Y.; Zhu, Y.; Wei, Z.; Xu, H.; Tang, Q.; Kong, X.; Hu, L. SKAP2, a novel target of HSF4b, associates with NCK2/F-actin at membrane ruffles and regulates actin reorganization in lens cell. J. Cell Mol. Med. 2011, 15, 783–795. [Google Scholar] [CrossRef]
- Dubrac, A.; Genet, G.; Ola, R.; Zhang, F.; Pibouin-Fragner, L.; Han, J.; Zhang, J.; Thomas, J.L.; Chedotal, A.; Schwartz, M.A.; et al. Targeting NCK-Mediated Endothelial Cell Front-Rear Polarity Inhibits Neovascularization. Circulation 2016, 133, 409–421. [Google Scholar] [CrossRef]
- Stoilova, D.; Child, A.; Trifan, O.C.; Crick, R.P.; Coakes, R.L.; Sarfarazi, M. Localization of a locus (GLC1B) for adult-onset primary open angle glaucoma to the 2cen-q13 region. Genomics 1996, 36, 142–150. [Google Scholar] [CrossRef]
- Akiyama, M.; Yatsu, K.; Ota, M.; Katsuyama, Y.; Kashiwagi, K.; Mabuchi, F.; Iijima, H.; Kawase, K.; Yamamoto, T.; Nakamura, M.; et al. Microsatellite analysis of the GLC1B locus on chromosome 2 points to NCK2 as a new candidate gene for normal tension glaucoma. Br. J. Ophthalmol. 2008, 92, 1293–1296. [Google Scholar] [CrossRef]
- Roberts, D.J.; Tan-Sah, V.P.; Ding, E.Y.; Smith, J.M.; Miyamoto, S. Hexokinase-II positively regulates glucose starvation-induced autophagy through TORC1 inhibition. Mol. Cell 2014, 53, 521–533. [Google Scholar] [CrossRef]
- Majewski, N.; Nogueira, V.; Bhaskar, P.; Coy, P.E.; Skeen, J.E.; Gottlob, K.; Chandel, N.S.; Thompson, C.B.; Robey, R.B.; Hay, N. Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak. Mol. Cell 2004, 16, 819–830. [Google Scholar] [CrossRef]
- Zhou, Z.; Doggett, T.A.; Sene, A.; Apte, R.S.; Ferguson, T.A. Autophagy supports survival and phototransduction protein levels in rod photoreceptors. Cell Death Differ. 2015, 22, 488–498. [Google Scholar] [CrossRef]
- Medchalmi, S.; Tare, P.; Sayyad, Z.; Swarup, G. A glaucoma- and ALS-associated mutant of OPTN induces neuronal cell death dependent on Tbk1 activity, autophagy and ER stress. FEBS J. 2021, 288, 4576–4595. [Google Scholar] [CrossRef]
- Liu, Y.; Allingham, R.R. Molecular genetics in glaucoma. Exp. Eye Res. 2011, 93, 331–339. [Google Scholar] [CrossRef]
- Ayala-Lugo, R.M.; Pawar, H.; Reed, D.M.; Lichter, P.R.; Moroi, S.E.; Page, M.; Eadie, J.A.; Azócar, V.; Maul, E.J.; Ntim-Amponsah, C.T.; et al. Variation in optineurin (OPTN) allele frequencies between and within populations. Mol. Vis. 2007, 13, 151–163. [Google Scholar]
- Alward, W.L.; Kwon, Y.H.; Kawase, K.; Craig, J.E.; Hayreh, S.S.; Johnson, A.T.; Khanna, C.L.; Yamamoto, T.; Mackey, D.A.; Roos, B.R.; et al. Evaluation of optineurin sequence variations in 1048 patients with open-angle glaucoma. Am. J. Ophthalmol. 2003, 136, 904–910. [Google Scholar] [CrossRef]
- Chen, M.; Yu, X.; Xu, J.; Ma, J.; Chen, X.; Chen, B.; Gu, Y.; Wang, K. Association of Gene Polymorphisms With Primary Open Angle Glaucoma: A Systematic Review and Meta-Analysis. Investig. Ophthalmol. Vis. Sci. 2019, 60, 1105–1121. [Google Scholar] [CrossRef]
- Kawakami, K.; Sato, S.; Ozaki, H.; Ikeda, K. Six family genes--structure and function as transcription factors and their roles in development. Bioessays 2000, 22, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Carnes, M.U.; Liu, Y.P.; Allingham, R.R.; Whigham, B.T.; Havens, S.; Garrett, M.E.; Qiao, C.; Neighborhood Consortium Investigators; Katsanis, N.; Wiggs, J.L.; et al. Discovery and functional annotation of SIX6 variants in primary open-angle glaucoma. PLoS Genet. 2014, 10, e1004372. [Google Scholar] [CrossRef]
- Kuo, J.Z.; Zangwill, L.M.; Medeiros, F.A.; Liebmann, J.M.; Girkin, C.A.; Hammel, N.; Rotter, J.I.; Weinreb, R.N. Quantitative Trait Locus Analysis of SIX1-SIX6 With Retinal Nerve Fiber Layer Thickness in Individuals of European Descent. Am. J. Ophthalmol. 2015, 160, 123–130.e121. [Google Scholar] [CrossRef] [PubMed]
- Skowronska-Krawczyk, D.; Zhao, L.; Zhu, J.; Weinreb, R.N.; Cao, G.; Luo, J.; Flagg, K.; Patel, S.; Wen, C.; Krupa, M.; et al. P16INK4a Upregulation Mediated by SIX6 Defines Retinal Ganglion Cell Pathogenesis in Glaucoma. Mol. Cell 2015, 59, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.N.; Loomis, S.J.; Kang, J.H.; Allingham, R.R.; Gharahkhani, P.; Khor, C.C.; Burdon, K.P.; Aschard, H.; Chasman, D.I.; Igo, R.P., Jr.; et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat. Genet. 2016, 48, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Allingham, R.R.; Aung, T.; Tham, Y.C.; Hauser, M.A.; Vithana, E.N.; Khor, C.C.; Wong, T.Y. Association of common SIX6 polymorphisms with peripapillary retinal nerve fiber layer thickness: The Singapore Chinese Eye Study. Investig. Ophthalmol. Vis. Sci. 2014, 56, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Lebedeva, I.V.; Emdad, L.; Kang, D.C.; Baldwin, A.S., Jr.; Fisher, P.B. Human polynucleotide phosphorylase (hPNPaseold-35): A potential link between aging and inflammation. Cancer Res. 2004, 64, 7473–7478. [Google Scholar] [CrossRef] [PubMed]
- Leszczyniecka, M.; Kang, D.C.; Sarkar, D.; Su, Z.Z.; Holmes, M.; Valerie, K.; Fisher, P.B. Identification and cloning of human polynucleotide phosphorylase, hPNPase old-35, in the context of terminal differentiation and cellular senescence. Proc. Natl. Acad. Sci. USA 2002, 99, 16636–16641. [Google Scholar] [CrossRef]
- Sarkar, D.; Leszczyniecka, M.; Kang, D.C.; Lebedeva, I.V.; Valerie, K.; Dhar, S.; Pandita, T.K.; Fisher, P.B. Down-regulation of Myc as a potential target for growth arrest induced by human polynucleotide phosphorylase (hPNPaseold-35) in human melanoma cells. J. Biol. Chem. 2003, 278, 24542–24551. [Google Scholar] [CrossRef]
- Kanemaki, N.; Tchedre, K.T.; Imayasu, M.; Kawarai, S.; Sakaguchi, M.; Yoshino, A.; Itoh, N.; Meguro, A.; Mizuki, N. Dogs and humans share a common susceptibility gene SRBD1 for glaucoma risk. PLoS ONE 2013, 8, e74372. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Coyne, C.B.; Zeh, H.J.; Lotze, M.T. PAMPs and DAMPs: Signal 0s that spur autophagy and immunity. Immunol. Rev. 2012, 249, 158–175. [Google Scholar] [CrossRef] [PubMed]
- Tezel, G.; Hernandez, R.; Wax, M.B. Immunostaining of heat shock proteins in the retina and optic nerve head of normal and glaucomatous eyes. Arch. Ophthalmol. 2000, 118, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, H.; Medina-Ortiz, W.E.; Luan, T.; Clark, A.F.; McDowell, C.M. Crosstalk Between Transforming Growth Factor β-2 and Toll-Like Receptor 4 in the Trabecular Meshwork. Investig. Ophthalmol. Vis. Sci. 2017, 58, 1811–1823. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, J.; Meguro, A.; Ota, M.; Nomura, E.; Nishide, T.; Kashiwagi, K.; Mabuchi, F.; Iijima, H.; Kawase, K.; Yamamoto, T.; et al. Association of toll-like receptor 2 gene polymorphisms with normal tension glaucoma. Mol. Vis. 2009, 15, 2905–2910. [Google Scholar]
- Inoue, A.; Yanagisawa, M.; Kimura, S.; Kasuya, Y.; Miyauchi, T.; Goto, K.; Masaki, T. The human endothelin family: Three structurally and pharmacologically distinct isopeptides predicted by three separate genes. Proc. Natl. Acad. Sci. USA 1989, 86, 2863–2867. [Google Scholar] [CrossRef] [PubMed]
- Ripodas, A.; de Juan, J.A.; Roldan-Pallares, M.; Bernal, R.; Moya, J.; Chao, M.; Lopez, A.; Fernandez-Cruz, A.; Fernandez-Durango, R. Localisation of endothelin-1 mRNA expression and immunoreactivity in the retina and optic nerve from human and porcine eye. Evidence for endothelin-1 expression in astrocytes. Brain Res. 2001, 912, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Durango, R.; Rollin, R.; Mediero, A.; Roldan-Pallares, M.; Garcia Feijo, J.; Garcia Sanchez, J.; Fernandez-Cruz, A.; Ripodas, A. Localization of endothelin-1 mRNA expression and immunoreactivity in the anterior segment of human eye: Expression of ETA and ETB receptors. Mol. Vis. 2003, 9, 103–109. [Google Scholar] [PubMed]
- Sugiyama, T.; Moriya, S.; Oku, H.; Azuma, I. Association of endothelin-1 with normal tension glaucoma: Clinical and fundamental studies. Surv. Ophthalmol. 1995, 39 (Suppl. S1), S49–S56. [Google Scholar] [CrossRef] [PubMed]
- Cellini, M.; Possati, G.L.; Profazio, V.; Sbrocca, M.; Caramazza, N.; Caramazza, R. Color Doppler imaging and plasma levels of endothelin-1 in low-tension glaucoma. Acta Ophthalmol. Scand. Suppl. 1997, 75, 11–13. [Google Scholar] [CrossRef]
- Cioffi, G.A.; Sullivan, P. The effect of chronic ischemia on the primate optic nerve. Eur. J. Ophthalmol. 1999, 9 (Suppl. S1), S34–S36. [Google Scholar] [CrossRef]
- Oku, H.; Sugiyama, T.; Kojima, S.; Watanabe, T.; Azuma, I. Experimental optic cup enlargement caused by endothelin-1-induced chronic optic nerve head ischemia. Surv. Ophthalmol. 1999, 44 (Suppl. S1), S74–S84. [Google Scholar] [CrossRef] [PubMed]
- Stokely, M.E.; Brady, S.T.; Yorio, T. Effects of endothelin-1 on components of anterograde axonal transport in optic nerve. Investig. Ophthalmol. Vis. Sci. 2002, 43, 3223–3230. [Google Scholar]
- Liang, Y.J.; Wang, Y.Y.; Rong, S.S.; Chen, Z.J.; Chen, S.Y.; Tham, J.A.; Chan, P.P.; Yam, J.C.; Wiggs, J.L.; Pang, C.P.; et al. Genetic Associations of Primary Angle-Closure Disease: A Systematic Review and Meta-Analysis. JAMA Ophthalmol. 2024, 123, e240363. [Google Scholar] [CrossRef] [PubMed]
- Rong, S.S.; Tang, F.Y.; Chu, W.K.; Ma, L.; Yam, J.C.; Tang, S.M.; Li, J.; Gu, H.; Young, A.L.; Tham, C.C.; et al. Genetic Associations of Primary Angle-Closure Disease: A Systematic Review and Meta-analysis. Ophthalmology 2016, 123, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
| No. | Reference | Country/City (Ethnicity) | Sample Size | Male/Female | Age, y | Quality Assessment | Genotyping Methods | Genotype Frequency | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||||||||||
| Cases | Controls | Cases | Controls | Cases | Controls | AA | AB | BB | AA | AB | BB | |||||
| 1 | Lee et al., 2022 [70] | China Taiwan (Chinese) | 222 | 236 | 122/100 | 127/109 | 69 ± 9 | 68 ± 10 | 8☆ | allelic | 126 | 80 | 16 | 108 | 101 | 27 |
| 2 | Shin et al., 2022 [71] | Korea (Korean) | 210 | 117 | NA | NA | NA | NA | 8☆ | allelic | 130 | 76 | 4 | 64 | 45 | 8 |
| 3 | He et al., 2022 [69] | China Hongkong (Chinese) | 537 | 496 | 278/259 | 184/312 | 63.2 ± 12.8 | 70.2 ± 10.8 | 9☆ | allelic | NA | NA | NA | NA | NA | NA |
| China Shantou (Chinese) | 135 | 543 | 79/56 | 283/260 | 61.6 ± 14.6 7 | 74.4 ± 6.9 | ||||||||||
| 4 | Liuska et al., 2021 [68] | Finland (Finnish) | 892 | 205,435 | NA | NA | NA | NA | 9☆ | allelic | 884 | 8 | 0 | 204,378 | 1053 | 4 |
| 5 | Kim et al., 2021 [67] | Korea (Korean) | 282 | 213 | 127/155 | 120/93 | 54.3 ± 13.3 | 54.6 ± 9.7 | 9☆ | allelic | NA | NA | NA | NA | NA | NA |
| 6 | Milanowski et al., 2021 [65] | Poland (Caucasian) | 204 | 258 | 48/156 | 80/178 | 71.6 ± 11.1 | 70.9 ± 11.6 | 8☆ | allelic | 121 | 70 | 0 | 168 | 79 | 6 |
| 7 | Yue et al., 2021 [66] | China (Chinese) | 402 | 425 | 226/176 | 254/171 | 63.8 ± 6.5 | 64.5 ± 5.1 | 7☆ | allelic | 311 | 79 | 12 | 338 | 79 | 8 |
| 8 | Jung et al., 2020 [63] | Korea (Korean) | 159 | 103 | 60/99 | 44/59 | 61.14 ± 11.94 | 68.78 ± 9.82 | 7☆ | allelic | 260 | 44 | 1 | 241 | 96 | 18 |
| 9 | Lee et al., 2020 [64] | Korea (Korean) | 435 | 419 | 206/229 | 231/188 | 58.8 ± 13.6 | 56.2 ± 10.3 | 7☆ | allelic | 288 | 127 | 20 | 290 | 116 | 13 |
| 10 | Jung et al., 2019 [60] | Korea (Korean) | 154 | 101 | 58/96 | 42/59 | 61.23 ± 11.95 | 67.29 ±11.37 | 7☆ | allelic | 70 | 68 | 16 | 62 | 31 | 8 |
| 11 | Jung et al., 2019 [62] | Korea (Korean) | 157 | 106 | 57/100 | 43/63 | 61.06 ± 12.16 | 67.19 ± 10.53 | 7☆ | allelic | 148 | 9 | 0 | 98 | 8 | 0 |
| 12 | Kosior-Jarecka et al., 2019 [61] | Poland (Caucasian) | 143 | 165 | 43/100 | NA | 74 | NA | 8☆ | allelic | 77 | 57 | 6 | 90 | 68 | 6 |
| 13 | Jeoung et al., 2017 [58] | Korea (Korean) | 245 | 231 | 117/128 | 115/116 | 60.2 ± 12.7 | 58.6 ± 12.4 | 8☆ | allelic | 211 | 39 | 1 | 212 | 33 | 0 |
| 14 | Suh et al., 2017 [59] | Korea (Korean) | 140 | 352 | NA | NA | NA | NA | 7☆ | allelic | 62 | 61 | 16 | 158 | 151 | 33 |
| 15 | Nishisako et al., 2016 [56] | Japan (Japanese) | 292 | 500 | 140/152 | 246/254 | 46.7 ± 8.4 | 50.2 ± 10.6 | 7☆ | allelic | 93 | 135 | 64 | 147 | 248 | 105 |
| 16 | Gao et al., 2016 [54] | China (Chinese) | 55 | 50 | 29/26 | 31/19 | 52.5 ± 14.0 | 49.1 ± 13.6 | 7☆ | allelic | 39 | 15 | 1 | 38 | 12 | 0 |
| 17 | Sang et al., 2016 [57] | China (Chinese) | 181 | 266 | 104/77 | 114/152 | 53.5 ± 16.8 | 67.6 ± 11.3 | 7☆ | allelic | 131 | 45 | 5 | 140 | 103 | 23 |
| 18 | Kosior-Jarecka et al., 2016 [55] | Poland (Caucasian) | 160 | 165 | 50/110 | 50/115 | 72.01 ± 11.61 | 72.52 ± 11.06 | 6☆ | allelic | 83 | 6 | 71 | 81 | 15 | 69 |
| 19 | Lin et al., 2014 [53] | China (Chinese) | 249 | 262 | 123/117 | 140/122 | 63.2 ± 10.2 | 61.3 ± 11.4 | 6☆ | allelic | 231 | 18 | 0 | 241 | 21 | 0 |
| 20 | Shi et al., 2013 [52] | Japan (Japanese) | 163 | 180 | 86/77 | 95/85 | 61.8 ± 13.7 | 68.0 ± 7.7 | 6☆ | allelic | 147 | 16 | 0 | 168 | 11 | 1 |
| 21 | Shi et al., 2013 [51] | Japan (Japanese) | stage 1 120 | 121 | 61/60 | 61/59 | 54.0 ± 12.2 | 70.3 ± 10.2 | 6☆ | allelic | 159 | 111 | 16 | 130 | 105 | 36 |
| stage 2 286 | 271 | 139/147 | 145/126 | 56.4 ± 13.3 | 69.7 ± 9.3 | allelic | ||||||||||
| 22 | Wiggs et al., 2012 [50] | U.S. (Caucasian) | 64 | 400 | 23/41 | 179/221 | 61.06 ± 11.6 | 66.06 ± 11.3 | 7☆ | allelic | 36 | 15 | 1 | 82 | 72 | 13 |
| 23 | TAKANO et al., 2012 [49] | Japan (Japanese) | 365 | 216 | 171/194 | 116/100 | 58.6 ± 13.1 | 69.7 ± 11.3 | 8☆ | allelic | 141 | 159 | 65 | 103 | 85 | 28 |
| 24 | Suh et al., 2011 [47] | Korea (Korean) | 147 | 380 | NA | NA | NA | NA | 9☆ | allelic | 52 | 72 | 23 | 126 | 191 | 63 |
| 25 | Mabuchi et al., 2011 [46] | Japan (Japanese) | 158 | 191 | 65/93 | 70/121 | 68.6 ± 11.8 | 65.7 ± 11.4 | 7☆ | allelic | 51 | 84 | 23 | 71 | 89 | 31 |
| 26 | Yasumura et al., 2011 [48] | Japan (Japanese) | 295 | 518 | 142/153 | NA | 46.4 ± 8.1 | NA | 8☆ | allelic | 241 | 52 | 2 | 404 | 110 | 4 |
| 27 | Wolf et al., 2010 [43] | Germany (German) | 273 | 280 | 96/177 | 115/165 | 63.9 ± 14.2 | 66 ± 13 | 7☆ | allelic | 74 | 131 | 68 | 75 | 135 | 10 |
| 28 | Meguro et al., 2010 [44] | Japan (Japanese) | 305 | 355 | 145/160 | 174/181 | 46.6 ± 8.5 | 61.7 ± 8.9 | 8☆ | genomic | 51 | 138 | 116 | 100 | 162 | 93 |
| 29 | Mabuchi et al., 2010 [42] | Japan (Japanese) | 213 | 191 | 91/122 | 70/121 | NA | NA | 7☆ | allelic | 79 | 100 | 34 | 77 | 84 | 30 |
| 30 | Mabuchi et al., 2010 [41] | Japan (Japanese) | 213 | 191 | 91/122 | 70/121 | NA | NA | 7☆ | allelic | 59 | 107 | 47 | 49 | 84 | 58 |
| 31 | Fan et al., 2010 [40] | China (Chinese) | 100 | 201 | 54/46 | 120/81 | 63.2 ± 11.5 | 69.8 ± 8.7 | 8☆ | allelic | 89 | 9 | 1 | 173 | 27 | 1 |
| 32 | Yu-Wai-Man et al., 2010 [45] | England (Caucasian) | 70 | 75 | NA | NA | NA | 79.3 | 7☆ | allelic | 41 | 26 | 3 | 59 | 13 | 3 |
| 33 | Fan et al., 2009 [37] | China (Chinese) | 42 | 77 | 33/9 | 58/19 | 66.7 ± 10.1 | 72.0 ± 8.5 | 8☆ | allelic | 27 | 13 | 2 | 47 | 27 | 3 |
| 34 | Clement et al., 2009 [35] | Australia (75 Caucasian, 1 Asian) | 34 | 42 | 9/25 | 16/26 | 72.5 ± 9.4 | 70.4 ± 7.8 | 9☆ | allelic | 21 | 11 | 2 | 25 | 14 | 3 |
| 35 | Daugherty et al., 2009 [36] | U.S. (Caucasian) | 52 | 167 | 18/34 | 62/105 | 69.8 ± 12.0 | 60.3 ± 12.0 | 7☆ | allelic | 29 | 28 | 5 | 109 | 57 | 12 |
| 36 | Mabuchi et al., 2009 [38] | Japan (Japanese) | 213 | 189 | 91/122 | 70/119 | 63.9 ± 13.7 | 65.5 ± 11.4 | 7☆ | allelic | 92 | 95 | 26 | 83 | 83 | 23 |
| 37 | Woo et al., 2009 [39] | Korea (Korean) | 78 | 100 | 32/46 | 47/53 | 46.2 ± 11.7 | 49.3 ± 9.2 | 8☆ | allelic | 25 | 34 | 19 | 31 | 50 | 19 |
| 38 | Shibuya et al., 2008 [34] | Japan (Japanese) | 250 | 318 | 119/131 | 157/161 | 46.1 ± 7.7 | 61.2 ± 8.3 | 8☆ | allelic | 81 | 127 | 42 | 137 | 141 | 40 |
| 39 | Tosaka et al., 2007 [33] | Japan (Japanese) | 290 | 241 | 142/148 | 114/127 | 55.8 ± 13.0 | 69.7 ± 11.3 | 7☆ | allelic | 106 | 130 | 54 | 67 | 130 | 44 |
| 40 | Mabuchi et al., 2007 [31] | Japan (Japanese) | 194 | 185 | NA | NA | 63.6 ± 13.3 | 65.3 ± 11.5 | 8☆ | allelic | 190 | 4 | 0 | 182 | 3 | 0 |
| 41 | Miyazawa et al., 2007 [32] | Japan (Japanese) | 103 | 118 | 53/50 | 62/56 | 61.8 ± 11.7 | 68.0 ± 7.7 | 7☆ | allelic | 76 | 25 | 2 | 72 | 41 | 5 |
| 42 | Jeoung et al., 2007 [30] | Korea (Korean) | 67 | 100 | 28/39 | 47/53 | 48.8 ± 10.2 | 49.3 ± 9.2 | 8☆ | allelic | 53 | 13 | 1 | 83 | 16 | 1 |
| 43 | How et al., 2007 [29] | Singapore (Chinese) | 94 | 79 | 64/30 | 32/47 | 72.9 | 67.7 | 7☆ | allelic | 71 | 17 | 1 | 64 | 13 | 2 |
| 44 | Kim et al., 2006 [25] | Korea (Korean) | 67 | 100 | 28/39 | 47/53 | 48.8 ± 10.2 | 49.3 ± 9.2 | 8☆ | allelic | 29 | 32 | 6 | 44 | 39 | 17 |
| 45 | Lam et al., 2006 [26] | China (Chinese) | 106 | 300 | NA | 191/109 | NA | 70.4 ± 9.3 | 7☆ | allelic | 102 | 3 | 1 | 286 | 13 | 1 |
| 46 | Inagaki et al., 2006 [24] | Japan (Japanese) | 294 | 240 | 144/150 | 114/126 | 58.8 ± 13.2 | 69.7 ± 11.2 | 7☆ | allelic | 219 | 72 | 3 | 176 | 63 | 1 |
| 47 | Mabuchi et al., 2006 [27] | Japan (Japanese) | 131 | 106 | NA | NA | 62.8 ± 13.3 | 65.0 ± 10.5 | 7☆ | allelic | 54 | 58 | 19 | 48 | 39 | 19 |
| 48 | Yao et al., 2006 [28] | Africa (African-Caribbean) | 61 | 48 | NA | NA | 52.1 | 61.3 | 9☆ | allelic | 58 | 3 | 0 | 46 | 2 | 0 |
| 49 | Hashizume et al., 2005 [23] | Japan (Japanese) | 268 | 240 | 129/139 | 113/127 | 58.8 ± 13.4 | 69.7 ± 11.2 | 7☆ | allelic | 164 | 90 | 14 | 163 | 66 | 11 |
| 50 | Dimasi et al., 2005 [21] | Australia (Caucasian) | 62 | 178 | NA | NA | NA | NA | 8☆ | allelic | 34 | 43 | 22 | 38 | 108 | 55 |
| 51 | Fan et al., 2005 [22] | China (Chinese) | 106 | 281 | NA | 180/101 | NA | 69.8 ± 9.8 | 6☆ | allelic | 67 | 36 | 3 | 200 | 74 | 7 |
| 52 | Funayama et al., 2004 [18] | Japan (Japanese) | 217 | 218 | 97/120 | 92/126 | 60.3 ± 12.4 | 70.6 ± 10.9 | 7☆ | allelic | 169 | 43 | 5 | 182 | 35 | 1 |
| 53 | Fuse et al., 2004 [19] | Japan (Japanese) | 65 | 100 | 27/38 | 62/38 | 61.8 ± 13.7 | 68 ± 7.7 | 6☆ | allelic | 55 | 9 | 1 | 95 | 5 | 0 |
| 54 | Woo et al., 2004 [20] | Korea (Korean) | 65 | 101 | 26/39 | 48/53 | 47.0 ± 10.3 | 49.0 ± 9.2 | 8☆ | allelic | 62 | 3 | 0 | 101 | 0 | 0 |
| 55 | Powell et al., 2003 [17] | England (Caucasian) | 61 | 168 | 26/35 | 109/59 | NA | NA | 6☆ | allelic | 41 | 16 | 4 | 111 | 53 | 4 |
| 56 | Aung et al., 2002 [16] | England (Caucasian) | 163 | 186 | NA | NA | NA | NA | 7☆ | allelic | 57 | 26 | 0 | 86 | 14 | 0 |
| No. | Gene Symbol | SNP | Minor Allele | No. of Cohorts | Ethnicity | Pooled Sample Size | Genetic Model | Heterogeneity Test | Fixed or Random Effect Model | OR | 95%CI | p | Begg’s Test | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | p (Q) | I² (%) | z | p | |||||||||||
| 1 | APOE | -491A>T | T | 2 | Chinese | 312 | 581 | B vs. A | 0.928 | 0.0 | fixed | 0.91 | 0.44–1.88 | 0.800 | 0.000 | 1.000 |
| BB + AB vs. AA | 0.933 | 0.0 | fixed | 0.77 | 0.35–1.73 | 0.531 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.974 | 0.0 | fixed | 2.76 | 0.39–19.68 | 0.312 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.940 | 0.0 | fixed | 0.63 | 0.25–1.54 | 0.307 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.973 | 0.0 | fixed | 2.71 | 0.38–19.36 | 0.321 | 0.000 | 1.000 | ||||||||
| -427T>C | C | 2 | 312 | 582 | B vs. A | 0.941 | 0.0 | fixed | 0.50 | 0.11–2.25 | 0.365 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.940 | 0.0 | fixed | 0.50 | 0.11–2.25 | 0.363 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | excluded | excluded | NA | NA | NA | NA | NA | NA | ||||||||
| AB vs. AA | 0.940 | 0.0 | fixed | 0.50 | 0.11–2.25 | 0.363 | 0.000 | 1.000 | ||||||||
| BB vs. AA | excluded | excluded | NA | NA | NA | NA | NA | NA | ||||||||
| -219T>G | G | 2 | 312 | 581 | B vs. A | 0.885 | 0.0 | fixed | 0.98 | 0.78–1.25 | 0.899 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.865 | 0.0 | fixed | 0.80 | 0.59–1.10 | 0.172 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.957 | 0.0 | fixed | 1.65 | 1.01–2.71 | 0.046 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.874 | 0.0 | fixed | 0.70 | 0.50–0.98 | 0.039 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.916 | 0.0 | fixed | 1.39 | 0.83–2.33 | 0.215 | 0.000 | 1.000 | ||||||||
| 2 | EDNRA | c.-231G>A | A | 2 | Caucasian Korean | 227 | 265 | B vs. A | 0.578 | 0.0 | fixed | 0.95 | 0.73–1.22 | 0.683 | 0.000 | 1.000 |
| BB + AB vs. AA | 0.731 | 0.0 | fixed | 0.86 | 0.60–1.22 | 0.384 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.128 | 56.7 | random | 0.95 | 0.64–1.42 | 0.816 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.057 | 72.4 | random | 0.85 | 0.50–1.45 | 0.554 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.118 | 59.0 | random | 0.61 | 0.39–0.97 | 0.035 | 0.000 | 1.000 | ||||||||
| c.*70C>G | G | 2 | 227 | 265 | B vs. A | 0.005 | 87.4 | random | 1.29 | 0.60–2.78 | 0.513 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.168 | 47.4 | fixed | 1.67 | 1.08–2.56 | 0.020 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.008 | 86.0 | random | 1.14 | 0.37–3.56 | 0.817 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.769 | 0.0 | fixed | 1.45 | 0.89–2.35 | 0.135 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.026 | 79.7 | random | 1.44 | 0.44–4.72 | 0.543 | 0.000 | 1.000 | ||||||||
| 3 | ELOVL5 | rs735860 | C | 2 | Japanese | 463 | 546 | B vs. A | 0.001 | 91.3 | random | 1.49 | 0.79–2.80 | 0.216 | 0.000 | 1.000 |
| BB + AB vs. AA | 0.015 | 83.2 | random | 1.81 | 0.88–3.70 | 0.106 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.049 | 74.2 | random | 1.29 | 0.67–2.49 | 0.447 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.448 | 0.0 | fixed | 1.51 | 1.11–2.05 | 0.009 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.030 | 78.7 | random | 1.65 | 0.71–3.82 | 0.246 | 0.000 | 1.000 | ||||||||
| 4 | HK2 | rs678350 | G | 2 | Korean Japanese | 440 | 372 | B vs. A | 0.580 | 0.0 | fixed | 1.54 | 1.23–1.91 | 0.000 | 0.000 | 1.000 |
| BB + AB vs. AA | 0.689 | 0.0 | fixed | 1.75 | 1.32–2.31 | 0.000 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.499 | 0.0 | fixed | 1.75 | 1.09–2.80 | 0.020 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.482 | 0.0 | fixed | 1.65 | 1.22–2.23 | 0.001 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.633 | 0.0 | fixed | 2.14 | 1.31–3.48 | 0.002 | 0.000 | 1.000 | ||||||||
| 5 | NCK2 | rs2033008 | A | 2 | Korean Japanese | 440 | 372 | B vs. A | 0.799 | 0.0 | fixed | 0.70 | 0.57–0.87 | 0.001 | 0.000 | 1.000 |
| BB + AB vs. AA | 0.636 | 0.0 | fixed | 0.77 | 0.58–1.02 | 0.065 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.526 | 0.0 | fixed | 0.44 | 0.27–0.70 | 0.001 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.968 | 0.0 | fixed | 0.86 | 0.64–1.16 | 0.321 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.558 | 0.0 | fixed | 0.41 | 0.25–0.67 | 0.000 | 0.000 | 1.000 | ||||||||
| 6 | MTHFR | rs397507444, c.677 C/T | T | 3 | Caucasian Asian Korean Japanese | 243 | 248 | B vs. A | 0.923 | 0.0 | fixed | 1.02 | 0.79–1.33 | 0.855 | 0.000 | 1.000 |
| BB + AB vs. AA | 0.828 | 0.0 | fixed | 1.05 | 0.73–1.52 | 0.778 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.517 | 0.0 | fixed | 1.00 | 0.62–1.62 | 0.986 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.582 | 0.0 | fixed | 1.07 | 0.72–1.60 | 0.725 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.813 | 0.0 | fixed | 1.01 | 0.59–1.72 | 0.969 | 0.000 | 1.000 | ||||||||
| rs1217691063c.1298 A/C | C | 2 | Korean Japanese | 209 | 206 | B vs. A | 0.604 | 0.0 | fixed | 0.94 | 0.65–1.34 | 0.720 | 0.000 | 1.000 | ||
| BB + AB vs. AA | 0.572 | 0.0 | fixed | 0.95 | 0.63–1.43 | 0.797 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.712 | 0.0 | fixed | 0.55 | 0.12–2.65 | 0.459 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.574 | 0.0 | fixed | 0.97 | 0.64–1.47 | 0.873 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.511 | 0.0 | fixed | 0.63 | 0.13–2.97 | 0.556 | 0.000 | 1.000 | ||||||||
| 7 | NOS3 | rs1799983, 894 G>T | T | 2 | Korean Chinese | 350 | 446 | B vs. A | 0.248 | 25.0 | fixed | 1.03 | 0.71–1.47 | 0.888 | 0.000 | 1.000 |
| BB + AB vs. AA | 0.219 | 33.7 | fixed | 1.00 | 0.68–1.46 | 0.989 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.865 | 0.0 | fixed | 2.43 | 0.30–19.58 | 0.404 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.215 | 35.0 | fixed | 0.97 | 0.66–1.43 | 0.879 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.833 | 0.0 | fixed | 2.38 | 0.30–19.03 | 0.414 | 0.000 | 1.000 | ||||||||
| rs2070744, -786T>C | C | 2 | 350 | 446 | B vs. A | 0.315 | 0.8 | fixed | 1.04 | 0.75–1.43 | 0.816 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.363 | 0.0 | fixed | 1.00 | 0.70–1.42 | 0.987 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.261 | 20.7 | fixed | 1.97 | 0.52–7.39 | 0.315 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.427 | 0.0 | fixed | 0.96 | 0.67–1.37 | 0.814 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.248 | 25.1 | fixed | 1.92 | 0.51–7.17 | 0.334 | 0.000 | 1.000 | ||||||||
| 8 | OPA1 | rs166850, IVS8+4C⬎T | T | 9 | Caucasian Chinese Japanese Korean African-Caribbean | 904 | 1217 | B vs. A | 0.034 | 52.1 | random | 1.49 | 1.03–2.15 | 0.034 | 0.940 | 0.348 |
| BB + AB vs. AA | 0.000 | 76.1 | random | 1.93 | 1.09–3.45 | 0.025 | 0.520 | 0.602 | ||||||||
| BB vs. AA + AB | 0.174 | 39.6 | fixed | 0.96 | 0.41–2.24 | 0.931 | 1.020 | 0.308 | ||||||||
| AB vs. AA | 0.000 | 73.0 | random | 1.82 | 1.04–3.19 | 0.038 | 0.310 | 0.754 | ||||||||
| BB vs. AA | 0.216 | 32.6 | fixed | 1.04 | 0.44–2.43 | 0.930 | 1.020 | 0.308 | ||||||||
| rs10451941, IVS8+32T⬎C | C | 9 | 944 | 1220 | B vs. A | 0.405 | 3.5 | fixed | 1.49 | 1.30–1.71 | 0.000 | 0.100 | 0.917 | |||
| BB + AB vs. AA | 0.243 | 22.5 | fixed | 1.55 | 1.29–1.87 | 0.000 | 1.150 | 0.251 | ||||||||
| BB vs. AA + AB | 0.603 | 0.0 | fixed | 1.87 | 1.43–2.45 | 0.000 | 0.300 | 0.764 | ||||||||
| AB vs. AA | 0.130 | 36.0 | fixed | 1.41 | 1.16–1.71 | 0.001 | 0.520 | 0.602 | ||||||||
| BB vs. AA | 0.564 | 0.0 | fixed | 2.16 | 1.59–2.95 | 0.000 | 0.000 | 1.000 | ||||||||
| 9 | OPTN | c.603T>A, Met98Lys | A | 3 | Chinese Japanese | 388 | 599 | B vs. A | 0.239 | 30.1 | fixed | 1.51 | 1.14–2.02 | 0.005 | 1.040 | 0.296 |
| BB + AB vs. AA | 0.341 | 7.0 | fixed | 1.55 | 1.12–2.14 | 0.007 | 1.040 | 0.296 | ||||||||
| BB vs. AA + AB | 0.417 | 0.0 | fixed | 2.20 | 0.82–5.95 | 0.119 | 1.040 | 0.296 | ||||||||
| AB vs. AA | 0.401 | 0.0 | fixed | 1.49 | 1.07–2.07 | 0.018 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.459 | 0.0 | fixed | 2.41 | 0.88–6.58 | 0.087 | 0.000 | 1.000 | ||||||||
| c.412G>A, Thr34Thr | A | 3 | 388 | 599 | B vs. A | 0.185 | 40.8 | fixed | 1.66 | 1.29–2.13 | 0.000 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.346 | 5.7 | fixed | 1.69 | 1.27–2.25 | 0.000 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.272 | 23.3 | fixed | 3.72 | 1.41–9.79 | 0.008 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.529 | 0.0 | fixed | 1.58 | 1.17–2.12 | 0.002 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.258 | 26.3 | fixed | 4.22 | 1.59–11.18 | 0.004 | 0.000 | 1.000 | ||||||||
| IVS6-5T>C | C | 2 | 171 | 381 | B vs. A | 0.048 | 74.4 | random | 1.26 | 0.64–2.50 | 0.507 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.141 | 53.8 | random | 1.07 | 0.59–1.97 | 0.817 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.135 | 55.3 | random | 2.08 | 0.53–8.18 | 0.296 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.058 | 72.2 | random | 2.03 | 0.98–4.17 | 0.055 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.387 | 0.0 | fixed | 1.01 | 0.41–2.50 | 0.976 | 0.000 | 1.000 | ||||||||
| IVS6-10G>A | A | 2 | 171 | 381 | B vs. A | 0.532 | 0.0 | fixed | 1.31 | 0.79–2.18 | 0.296 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.499 | 0.0 | fixed | 1.33 | 0.78–2.27 | 0.299 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | . | . | fixed | 1.55 | 0.10–25.17 | 0.759 | NA | NA | ||||||||
| AB vs. AA | 0.474 | 0.0 | fixed | 1.32 | 0.77–2.28 | 0.316 | 0.000 | 1.000 | ||||||||
| BB vs. AA | . | . | fixed | 1.56 | 0.10–25.40 | 0.757 | NA | NA | ||||||||
| IVS7+24G>A | A | 2 | 171 | 381 | B vs. A | 0.760 | 0.0 | fixed | 1.58 | 0.99–2.51 | 0.053 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.148 | 52.3 | random | 1.29 | 0.60–2.75 | 0.517 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.398 | 0.0 | fixed | 2.75 | 0.51–14.87 | 0.241 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.061 | 71.6 | random | 1.17 | 0.42–3.32 | 0.761 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.856 | 0.0 | fixed | 2.71 | 0.49–14.92 | 0.253 | 0.000 | 1.000 | ||||||||
| 10 | p53 | rs1042522, -Arg72Pro | C | 5 | Caucasian Chinese Japanese | 490 | 1135 | B vs. A | 0.000 | 81.3 | random | 0.97 | 0.64–1.45 | 0.868 | 0.240 | 0.806 |
| BB + AB vs. AA | 0.000 | 93.3 | random | 2.32 | 1.02–5.28 | 0.045 | 0.240 | 0.806 | ||||||||
| BB vs. AA + AB | 0.013 | 68.5 | random | 1.140 | 0.58–2.25 | 0.704 | 0.240 | 0.806 | ||||||||
| AB vs. AA | 0.003 | 75.1 | random | 0.880 | 0.53–1.46 | 0.630 | -0.240 | 1.000 | ||||||||
| BB vs. AA | 0.001 | 79.3 | random | 1.020 | 0.41–2.51 | 0.973 | -0.240 | 1.000 | ||||||||
| 11 | SRBD1 | rs3213787 | G | 3 | Korean Japanese | 622 | 649 | B vs. A | 0.050 | 66.6 | random | 0.40 | 0.30–0.52 | 0.001 | 0.000 | 1.000 |
| BB + AB vs. AA | 0.080 | 60.4 | random | 0.38 | 0.26–0.51 | 0.001 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.082 | 60.1 | random | 0.23 | 0.09–0.59 | 0.252 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.067 | 63.0 | random | 0.41 | 0.30–0.56 | 0.002 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.075 | 61.4 | random | 0.20 | 0.08–0.50 | 0.201 | 0.000 | 1.000 | ||||||||
| 12 | TLR4 | rs10759930 | C | 3 | Korean Japanese | 762 | 914 | B vs. A | 0.087 | 59.0 | random | 1.21 | 0.97–1.53 | 0.097 | 1.040 | 0.296 |
| BB + AB vs. AA | 0.095 | 57.6 | random | 1.29 | 0.94–1.78 | 0.114 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.186 | 40.5 | fixed | 1.20 | 0.91–1.58 | 0.192 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.178 | 42.1 | fixed | 1.27 | 1.02–1.59 | 0.031 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.151 | 47.1 | fixed | 1.43 | 1.06–1.94 | 0.001 | 1.040 | 0.296 | ||||||||
| rs1927914 | G | 3 | 762 | 914 | B vs. A | 0.002 | 84.2 | random | 1.29 | 0.89–1.87 | 0.180 | 1.040 | 0.296 | |||
| BB + AB vs. AA | 0.074 | 61.7 | random | 1.32 | 0.94–1.85 | 0.104 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.489 | 0.0 | fixed | 1.24 | 0.94–1.64 | 0.125 | 1.040 | 0.296 | ||||||||
| AB vs. AA | 0.129 | 51.1 | random | 1.30 | 0.94–1.78 | 0.108 | 1.040 | 0.296 | ||||||||
| BB vs. AA | 0.172 | 43.1 | fixed | 1.43 | 1.06–1.94 | 0.020 | 0.000 | 1.000 | ||||||||
| rs1927911 | A | 3 | 762 | 914 | B vs. A | 0.001 | 85.5 | random | 1.33 | 0.91–1.96 | 0.141 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.091 | 58.2 | random | 1.31 | 0.95–1.80 | 0.102 | 1.040 | 0.296 | ||||||||
| BB vs. AA + AB | 0.486 | 0.0 | fixed | 1.25 | 0.94–1.66 | 0.125 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.155 | 46.3 | fixed | 1.29 | 1.04–1.61 | 0.021 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.013 | 76.9 | random | 1.48 | 0.78–2.79 | 0.227 | 1.040 | 0.296 | ||||||||
| rs12377632 | T | 3 | 762 | 914 | B vs. A | 0.114 | 54.0 | random | 1.16 | 0.93–1.44 | 0.181 | 1.040 | 0.296 | |||
| BB + AB vs. AA | 0.060 | 64.4 | random | 1.27 | 0.90–1.80 | 0.171 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.379 | 0.0 | random | 1.08 | 0.81–1.45 | 0.589 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.057 | 65.0 | random | 1.29 | 0.89–1.87 | 0.179 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.016 | 75.8 | random | 1.46 | 0.79–2.72 | 0.231 | 0.000 | 1.000 | ||||||||
| rs2149356 | T | 3 | 762 | 914 | B vs. A | 0.020 | 74.5 | random | 1.25 | 0.93–1.68 | 0.133 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.057 | 65.0 | random | 1.29 | 0.91–1.83 | 0.155 | 1.040 | 0.296 | ||||||||
| BB vs. AA + AB | 0.359 | 2.3 | fixed | 1.31 | 0.99–1.74 | 0.062 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.120 | 52.8 | random | 1.24 | 0.90–1.71 | 0.181 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.019 | 74.9 | random | 1.47 | 0.80–2.72 | 0.219 | 1.040 | 0.296 | ||||||||
| rs11536889 | C | 3 | 762 | 914 | B vs. A | 0.315 | 13.5 | fixed | 1.05 | 0.89–1.24 | 0.527 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.000 | 93.8 | random | 0.86 | 0.37–1.98 | 0.720 | 1.040 | 0.296 | ||||||||
| BB vs. AA + AB | 0.508 | 0.0 | fixed | 0.88 | 0.57–1.35 | 0.559 | 1.040 | 0.296 | ||||||||
| AB vs. AA | 0.448 | 0.0 | fixed | 1.13 | 0.92–1.40 | 0.247 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.017 | 75.5 | random | 1.34 | 0.59–3.02 | 0.867 | 0.000 | 1.000 | ||||||||
| rs7037117 | G | 3 | 762 | 914 | B vs. A | 0.000 | 89.1 | random | 1.12 | 0.67–1.89 | 0.665 | 1.040 | 0.296 | |||
| BB + AB vs. AA | 0.051 | 66.4 | random | 1.34 | 0.93–1.92 | 0.112 | 1.040 | 0.296 | ||||||||
| BB vs. AA + AB | 0.746 | 0.0 | fixed | 1.28 | 0.82–2.01 | 0.280 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.055 | 65.6 | random | 1.33 | 0.91–1.93 | 0.138 | 1.040 | 0.296 | ||||||||
| BB vs. AA | 0.079 | 60.6 | random | 1.48 | 0.71–3.08 | 0.290 | 1.040 | 0.296 | ||||||||
| rs7045953 | G | 3 | 762 | 914 | B vs. A | 0.307 | 15.4 | fixed | 1.12 | 0.86–1.45 | 0.414 | 1.040 | 0.296 | |||
| BB + AB vs. AA | 0.339 | 7.5 | fixed | 1.11 | 0.84–1.47 | 0.467 | 1.040 | 0.296 | ||||||||
| BB vs. AA + AB | 0.506 | 0.0 | fixed | 1.56 | 0.51–4.75 | 0.436 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.393 | 0.0 | fixed | 1.09 | 0.82–1.46 | 0.534 | 1.040 | 0.296 | ||||||||
| BB vs. AA | 0.491 | 0.0 | fixed | 1.42 | 0.46–4.35 | 0.542 | 0.000 | 1.000 | ||||||||
| 13 | WDR36 | rs17553936, IVS16–30A>G | G | 2 | Chinese Japanese | 145 | 195 | B vs. A | 0.287 | 11.6 | fixed | 0.70 | 0.47–1.04 | 0.078 | 0.000 | 1.000 |
| BB + AB vs. AA | 0.365 | 0.0 | fixed | 0.65 | 0.41–1.03 | 0.068 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.440 | 0.0 | fixed | 0.68 | 0.20–2.28 | 0.528 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.469 | 0.0 | fixed | 0.66 | 0.41–1.06 | 0.086 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.378 | 0.0 | fixed | 0.61 | 0.18–2.06 | 0.426 | 0.000 | 1.000 | ||||||||
| 14 | SIX1-SIX6 | rs10483727 | C | 2 | Korean Japanese | 391 | 383 | B vs. A | 0.141 | 53.8 | random | 0.55 | 0.38–0.80 | 0.002 | 0.000 | 1.000 |
| BB + AB vs. AA | 0.062 | 71.2 | random | 0.56 | 0.32–0.99 | 0.047 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.543 | 0.0 | fixed | 0.25 | 0.12–0.54 | 0.000 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.041 | 76.0 | random | 0.65 | 0.34–1.24 | 0.188 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.748 | 0.0 | fixed | 0.21 | 0.10–0.46 | 0.000 | 0.000 | 1.000 | ||||||||
| rs33912345 | A | 2 | 391 | 383 | B vs. A | 0.069 | 69.7 | random | 0.56 | 0.35–0.89 | 0.013 | 0.000 | 1.000 | |||
| BB + AB vs. AA | 0.067 | 70.3 | random | 0.55 | 0.32–0.77 | 0.038 | 0.000 | 1.000 | ||||||||
| BB vs. AA + AB | 0.856 | 0.0 | fixed | 0.24 | 0.11–0.54 | 0.001 | 0.000 | 1.000 | ||||||||
| AB vs. AA | 0.086 | 66.1 | random | 0.62 | 0.36–1.08 | 0.089 | 0.000 | 1.000 | ||||||||
| BB vs. AA | 0.696 | 0.0 | fixed | 0.20 | 0.08–0.45 | 0.000 | 0.000 | 1.000 | ||||||||
| Gene | SNP | OR (95%CI) | p Value | Involved Mechanisms | Possible Function in NTG | |
|---|---|---|---|---|---|---|
| Name | Symbol | |||||
| optic atrophy 1 | OPA1 | rs166850, IVS8+4C⬎T | 1.49 (1.03–2.15) | 0.034 | Encoding proteins crucial for normal mitochondrial function | Downregulation of OPA1 gene is associated with increased mitochondrial fission in optic nerve, increasing cell death of RGC-5 cells |
| rs10451941, IVS8+32T⬎C | 1.49 (1.30–1.71) | 0.000 | ||||
| elongation of long-chain fatty acids family member 5 | ELOVL5 | rs735860 | 1.51 (1.11–2.05) | 0.009 | Encoding elongases of polyunsaturated fatty acids | Enhanced ELOVL5 expression may cause apoptosis and cell growth inhibition in retinal ganglion cells |
| non-catalytic region of tyrosine kinase adaptor | NCK2 | rs2033008 | 0.70 (0.57–0.87) | 0.001 | Regulating the cellular actin dynamics and polarity | Participating in neural regeneration and protection, especially for the transition of glia cells into photoreceptors |
| hexokinase 2 | HK2 | rs678350 | 1.54 (1.23–1.91) | 0.000 | Catalyzing the first step of glycolysis | Important for photoreceptors’ function and preventing cell apoptosis |
| optineurin | OPTN | c.603T>A, Met98Lys | 1.51 (1.14–2.02) | 0.005 | An adaptor protein involved in many cellular functions | Inhibition of autophagy and induced cell death of RGCs |
| c.412G>A, Thr34Thr | 1.66 (1.29–2.13) | 0.000 | ||||
| S1 RNA binding domain 1 | SRBD1 | rs3213787 | 0.40 (0.30–0.52) | 0.001 | Modulating signal transduction | Prevent cell proliferation, promote proinflammatory cytokines accumulation and accelerate cell apoptosis of RGCs |
| toll-like receptor 4 | TLR4 | rs10759930 | 1.27 (1.02–1.59) | 0.031 | Participating in innate immunity and initiating inflammatory response | Inflammation and immunity lead to RGC apoptosis and optic nerve damage |
| rs1927914 | 1.43 (1.06–1.94) | 0.020 | ||||
| rs1927911 | 1.29 (1.04–1.61) | 0.021 | ||||
| endothelin receptor type A | EDNRA | c.-231G>A | 0.61 (0.39–0.97) | 0.035 | Bind with ET-1 to activate vasoconstriction | Damaging optic nerve resulted from vascular dysfunction and promoting astrocytes proliferation |
| c.*70C>G | 1.67 (1.08–2.56) | 0.020 | ||||
| tumor protein p53 | p53 | rs1042522, -Arg72Pro | 2.32 (1.02–5.28) | 0.045 | Regulating cell circle, cell metabolism, senescence and DNA repair | Producing ROS causing mitochondria damage and inducing cell apoptosis |
| sine oculis homeobox homolog 1- sine oculis homeobox homolog 6 | SIX1-SIX6 | rs10483727 | 0.55 (0.38–0.80) | 0.002 | Regulating the development of the visual system | Reducing the number of retinal ganglion cells, especially during the aging process |
| rs33912345 | 0.56 (0.35–0.89) | 0.013 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, L.; Wu, J.; Wang, N. Association of Gene Polymorphisms with Normal Tension Glaucoma: A Systematic Review and Meta-Analysis. Genes 2024, 15, 491. https://doi.org/10.3390/genes15040491
Pan L, Wu J, Wang N. Association of Gene Polymorphisms with Normal Tension Glaucoma: A Systematic Review and Meta-Analysis. Genes. 2024; 15(4):491. https://doi.org/10.3390/genes15040491
Chicago/Turabian StylePan, Lijie, Jian Wu, and Ningli Wang. 2024. "Association of Gene Polymorphisms with Normal Tension Glaucoma: A Systematic Review and Meta-Analysis" Genes 15, no. 4: 491. https://doi.org/10.3390/genes15040491







