Comparative Genome-Wide Alternative Splicing Analysis between Preadipocytes and Adipocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Isolation and Culture of Preadipocytes
2.3. Induced Differentiation of Preadipocytes
2.4. RNA Extraction, Quality Analysis, and Library Construction
2.5. Variable Splicing Analysis
2.6. Gene Ontology and Kyoto Encyclopedia of Genes and Genomics Enrichment Analysis
2.7. Identification of Differentially Alternative Splicing Transcripts
2.8. Quantitative Real-Time PCR
3. Results
3.1. RNA-Seq and Transcriptome Analysis of Preadipocytes and Adipocytes
3.2. Forms and Expression of Alternative Splicing
3.3. GO and KEGG Enrichment of Differentially Expressed Alternatively Spliced Genes
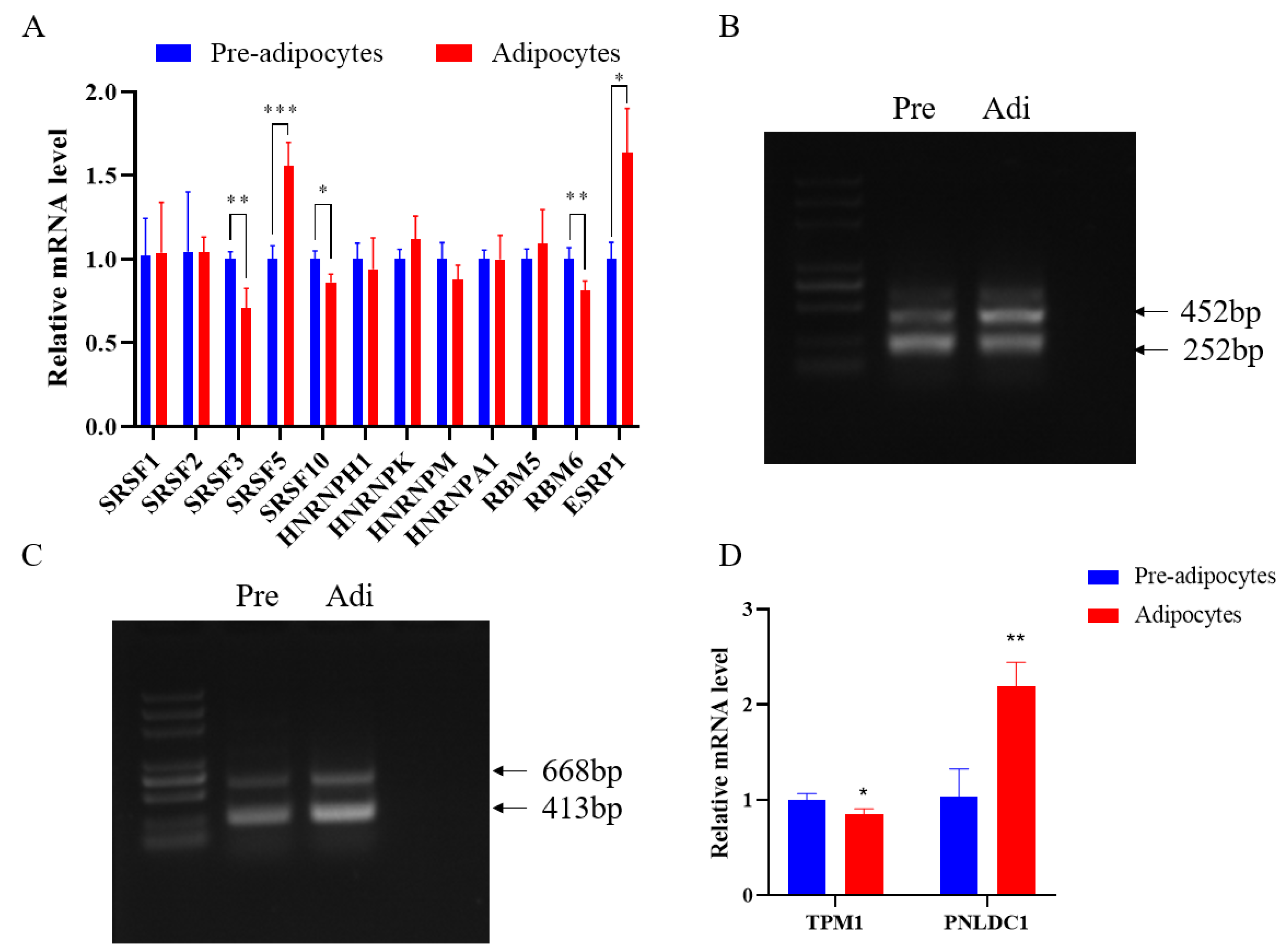
3.4. Identification of Alternative Splicing Types and Verification of Expression Levels of Transcripts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Silva, L.H.; Rodrigues, R.T.; Assis, D.E.; Benedeti, P.D.; Duarte, M.S.; Chizzotti, M.L. Explaining meat quality of bulls and steers by differential proteome and phosphoproteome analysis of skeletal muscle. J. Proteom. 2019, 199, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Brewer, M.S.; Zhu, L.G.; McKeith, F.K. Marbling effects on quality characteristics of pork loin chops: Consumer purchase intent, visual and sensory characteristics. Meat Sci. 2001, 59, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.X.; Lee, J.-Y. Epigenetic Regulation of Adipokines. Int. J. Mol. Sci. 2017, 18, 1740. [Google Scholar] [CrossRef] [PubMed]
- Musri, M.M.; Párrizas, M. Epigenetic regulation of adipogenesis. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Liu, X.; Zhou, Q.; Huang, T.; Zhang, L.; Gao, J.; Wang, Y.; Liu, Y.; Yan, T.; Zhang, S.; et al. DNA Methylation Modulates Aging Process in Adipocytes. Aging Dis. 2022, 13, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, R.; Liu, Y.; Zhao, Y.; Bi, Z.; Yao, Y.; Liu, Q.; Shi, H.; Wang, F.; Wang, Y. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 2020, 16, 1221–1235. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, J.; Chang, K.; Yang, F.; Jia, Z.; Sun, C.; Li, Q.; Xu, Y. Histone H3 methyltransferase Ezh2 promotes white adipocytes but inhibits brown and beige adipocyte differentiation in mice. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2021, 1866, 158901. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.-X.; Zou, Y.; Wang, G.-T.; Huang, S.-H.; Zhou, Y.-J.; Zhou, Y.-J. lnc TINCR induced by NOD1 mediates inflammatory response in 3T3-L1 adipocytes. Gene 2019, 698, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.J.; Smith, C.W.J.; Jiggins, C.D. Alternative splicing as a source of phenotypic diversity. Nat. Rev. Genet. 2022, 23, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Kalsotra, A.; Cooper, T.A. Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 2011, 12, 715–729. [Google Scholar] [CrossRef]
- Thomas, M.P.; Lieberman, J. Live or let die: Posttranscriptional gene regulation in cell stress and cell death. Immunol. Rev. 2013, 253, 237–252. [Google Scholar] [CrossRef]
- Kaida, D.; Motoyoshi, H.; Tashiro, E.; Nojima, T.; Hagiwara, M.; Ishigami, K.; Watanabe, H.; Kitahara, T.; Yoshida, T.; Nakajima, H.; et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 2007, 3, 576–583. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Ule, J.; Blencowe, B.J. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol. Cell 2019, 76, 329–345. [Google Scholar] [CrossRef]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Tan, B.; Zeng, J.; Meng, F.; Wang, S.; Xiao, L.; Zhao, X.; Hong, L.; Zheng, E.; Wu, Z.; Li, Z.; et al. Comprehensive analysis of pre-mRNA alternative splicing regulated by m6A methylation in pig oxidative and glycolytic skeletal muscles. BMC Genom. 2022, 23, 804. [Google Scholar] [CrossRef]
- Fang, X.; Xia, L.; Yu, H.; He, W.; Bai, Z.; Qin, L.; Jiang, P.; Zhao, Y.; Zhao, Z.; Yang, R. Comparative Genome-Wide Alternative Splicing Analysis of Longissimus Dorsi Muscles Between Japanese Black (Wagyu) and Chinese Red Steppes Cattle. Front. Veter- Sci. 2021, 8, 634577. [Google Scholar] [CrossRef]
- Lin, J.-C.; Lu, Y.-H.; Liu, Y.-R.; Lin, Y.-J. RBM4a-regulated splicing cascade modulates the differentiation and metabolic activities of brown adipocytes. Sci. Rep. 2016, 6, 20665. [Google Scholar] [CrossRef]
- Lin, J.-C. Impacts of Alternative Splicing Events on the Differentiation of Adipocytes. Int. J. Mol. Sci. 2015, 16, 22169–22189. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, M.; Manley, J.L. Phosphorylation switches the general splicing repressor SRp38 to a sequence-specific activator. Nat. Struct. Mol. Biol. 2008, 15, 1040–1048. [Google Scholar] [CrossRef]
- Reue, K.; Brindley, D.N. Thematic Review Series: Glycerolipids. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J. Lipid Res. 2008, 49, 2493–2503. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cheng, Y.; Wu, W.; Liu, Y.; Wei, N.; Feng, X.; Xie, Z.; Feng, Y. SRSF10 Regulates Alternative Splicing and Is Required for Adipocyte Differentiation. Mol. Cell. Biol. 2014, 34, 2198–2207. [Google Scholar] [CrossRef] [PubMed]
- Sun, C. The SF3b complex: Splicing and beyond. Cell. Mol. Life Sci. 2020, 77, 3583–3595. [Google Scholar] [CrossRef] [PubMed]
- Castellá, M.; Mestres-Arenas, A.; Gavaldà-Navarro, A.; Blasco-Roset, A.; Quesada-López, T.; Romero-Carramiñana, I.; Giralt, M.; Villarroya, F.; Cereijo, R. The splicing factor SF3B1 is involved in brown adipocyte thermogenic activation. Biochem. Pharmacol. 2024, 220, 116014. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.-Y.; Liang, Y.-C.; Tan, T.-H.; Chuang, H.-C.; Lin, Y.-J.; Lin, J.-C. RBM4a-SRSF3-MAP4K4 Splicing Cascade Constitutes a Molecular Mechanism for Regulating Brown Adipogenesis. Int. J. Mol. Sci. 2018, 19, 2646. [Google Scholar] [CrossRef] [PubMed]
- Brettle, M.; Patel, S.; Fath, T. Tropomyosins in the healthy and diseased nervous system. Brain Res. Bull. 2016, 126, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, J.; Wang, Q.; Cheng, M.; Lin, B. TPM1 mediates inflammation downstream of TREM2 via the PKA/CREB signaling pathway. J. Neuroinflamm. 2022, 19, 257. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; He, J.-X.; Zhu, M.; Dong, Y.-Q.; He, J.-X. Circ0001320 inhibits lung cancer cell growth and invasion by regulating TNFAIP1 and TPM1 expression through sponging miR-558. Hum. Cell 2021, 34, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Shen, J.; Yue, H.; Cao, Z. miRNA-183-5p.1 promotes the migration and invasion of gastric cancer AGS cells by targeting TPM1. Oncol. Rep. 2019, 42, 2371–2381. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, J.; Zhu, X.; Yang, B.; He, Z.; Yao, X. AZGP1P2/UBA1/RBM15 Cascade Mediates the Fate Determinations of Prostate Cancer Stem Cells and Promotes Therapeutic Effect of Docetaxel in Castration-Resistant Prostate Cancer via TPM1 m6A Modification. Research 2023, 6, 0252. [Google Scholar] [CrossRef]
- Qiao, Y.; Sun, Q.; Chen, X.; He, L.; Wang, D.; Su, R.; Xue, Y.; Sun, H.; Wang, H. Nuclear m6A reader YTHDC1 promotes muscle stem cell activation/proliferation by regulating mRNA splicing and nuclear export. eLife 2023, 12, e82703. [Google Scholar] [CrossRef] [PubMed]
- Anastasakis, D.; Skeparnias, I.; Shaukat, A.-N.; Grafanaki, K.; Kanellou, A.; Taraviras, S.; Papachristou, D.J.; Papakyriakou, A.; Stathopoulos, C. Mammalian PNLDC1 is a novel poly(A) specific exonuclease with discrete expression during early development. Nucleic Acids Res. 2016, 44, 8908–8920. [Google Scholar] [CrossRef] [PubMed]



| Samples | Total Reads | Clean Reads | Unique Mapped | Reads Filter % | Unique Mapped Rat % |
|---|---|---|---|---|---|
| Pre-1 | 82,899,462 | 81,014,722 | 74,318,042 | 97.73% | 91.73% |
| Pre-2 | 91,252,432 | 89,623,528 | 80,020,163 | 98.21% | 89.28% |
| Pre-3 | 83,152,664 | 80,554,612 | 72,714,018 | 96.88% | 90.27% |
| Adi-1 | 83,587,552 | 80,441,578 | 72,794,729 | 96.24% | 90.49% |
| Adi-2 | 79,558,574 | 77,525,064 | 69,168,973 | 97.44% | 89.22% |
| Adi-3 | 86,897,418 | 85,041,508 | 77,119,851 | 97.86% | 90.68% |
| Gene ID | Location | Adi Exp | Pre Exp | p-Value | FDR | Splicing Type |
|---|---|---|---|---|---|---|
| CD47 | chr1 | 11::2291 | 136::1943 | 0 | 0 | SE |
| HNRNPC | chr10 | 313::1 | 206::19 | 0 | 0 | SE |
| IRAK4 | chr5 | 391::32 | 403::0 | 0 | 0 | SE |
| CD200 | chr1 | 48::0 | 34::17 | 0 | 0 | SE |
| VCAN | chr7 | 525::668 | 8074::463 | 0 | 0 | SE |
| POLR2D | chr2 | 1172::37 | 1338::1 | 0 | 0 | SE |
| FBLN2 | chr22 | 34::457 | 234::864 | 0 | 0 | SE |
| CD200 | chr1 | 3::49 | 46::37 | 0 | 0 | MXE |
| CSNK1G1 | chr10 | 1::14 | 22::6 | 0 | 0 | MXE |
| HNRNPC | chr10 | 117::1 | 94::19 | 8.88 × 10−16 | 3.25 × 10−12 | SE |
| SNRPA1 | chr21 | 207::28 | 466::9 | 9.99 × 10−16 | 3.25 × 10−12 | SE |
| RTCA | chr3 | 5::15 | 11::0 | 1.89 × 10−15 | 6.38 × 10−12 | A3SS |
| POLQ | chr1 | 3::0 | 1::14 | 3.00 × 10−15 | 8.76 × 10−12 | SE |
| CCND3 | chr23 | 693::49 | 394::3 | 3.77 × 10−15 | 1.00 × 10−11 | SE |
| SGMS1 | chr26 | 2::7 | 5::0 | 4.88 × 10−15 | 1.19 × 10−11 | SE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Z.; Li, X.; Xu, M.; Meng, S.; Xu, H.; Li, M.; Cai, H. Comparative Genome-Wide Alternative Splicing Analysis between Preadipocytes and Adipocytes. Genes 2024, 15, 640. https://doi.org/10.3390/genes15050640
Hou Z, Li X, Xu M, Meng S, Xu H, Li M, Cai H. Comparative Genome-Wide Alternative Splicing Analysis between Preadipocytes and Adipocytes. Genes. 2024; 15(5):640. https://doi.org/10.3390/genes15050640
Chicago/Turabian StyleHou, Zhongyi, Xin Li, Maosheng Xu, Shengbo Meng, Huifen Xu, Ming Li, and Hanfang Cai. 2024. "Comparative Genome-Wide Alternative Splicing Analysis between Preadipocytes and Adipocytes" Genes 15, no. 5: 640. https://doi.org/10.3390/genes15050640
APA StyleHou, Z., Li, X., Xu, M., Meng, S., Xu, H., Li, M., & Cai, H. (2024). Comparative Genome-Wide Alternative Splicing Analysis between Preadipocytes and Adipocytes. Genes, 15(5), 640. https://doi.org/10.3390/genes15050640




