Genome-Wide Identification and Bioinformatics Analysis of the FK506 Binding Protein Family in Rice
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of FKBP Genes
2.2. Phylogenetic Analysis of FKBP Genes
2.3. Chromosomal Localization and Collinearity Analysis of FKBP Genes
2.4. Analysis of FKBP Gene Structures
2.5. Analysis of Gene Promoter Cis-Acting Elements
2.6. Experimental Materials and Conditions
2.7. Total RNA Extraction, Reverse Transcription, and Quantitative RT-PCR
3. Results
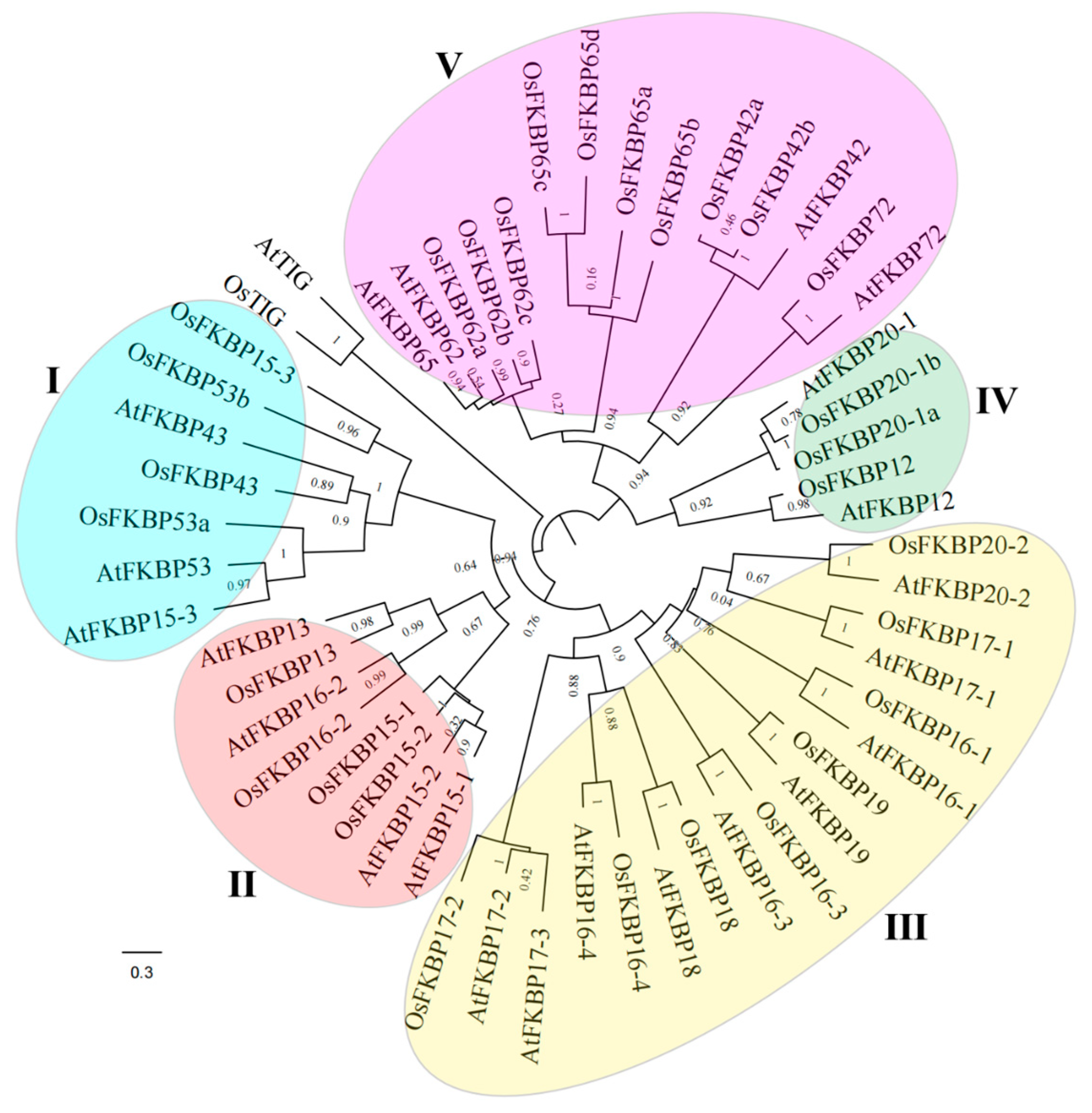
3.1. Identification and Evolutionary Analysis of FKBP Genes
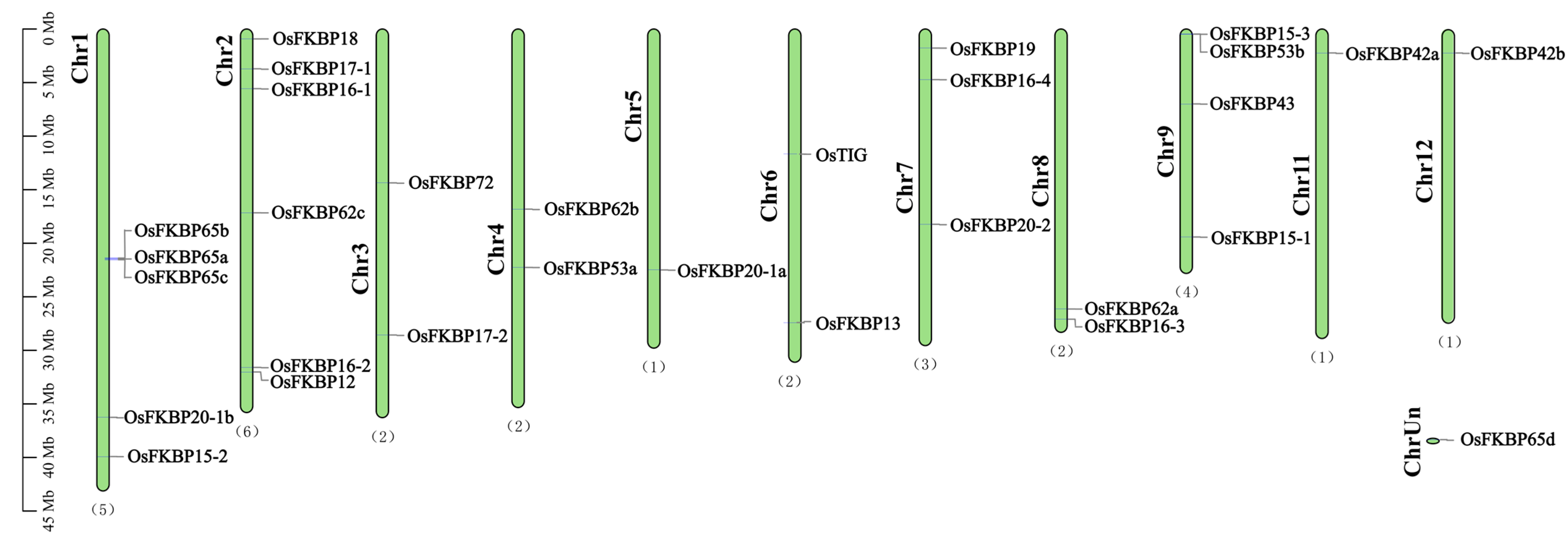
3.2. Chromosomal Localization Analysis of FKBP Genes
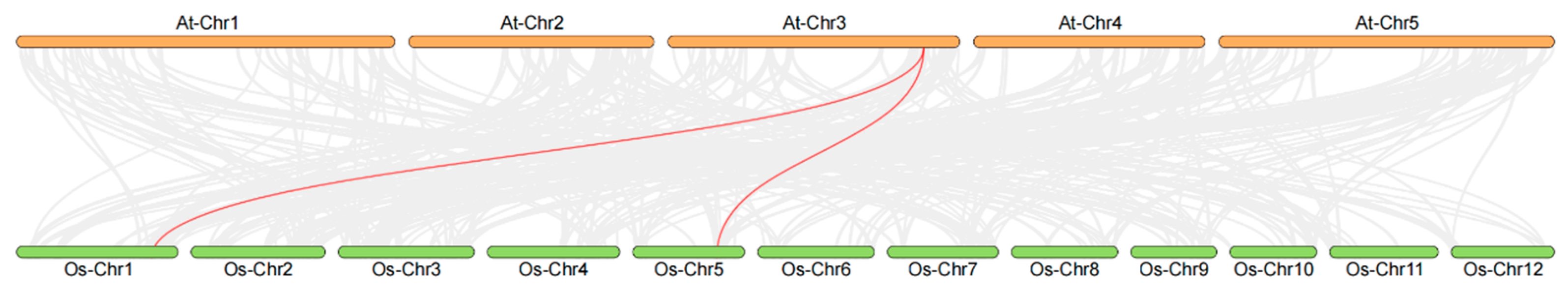
3.3. Collinearity Analysis of FKBP Genes
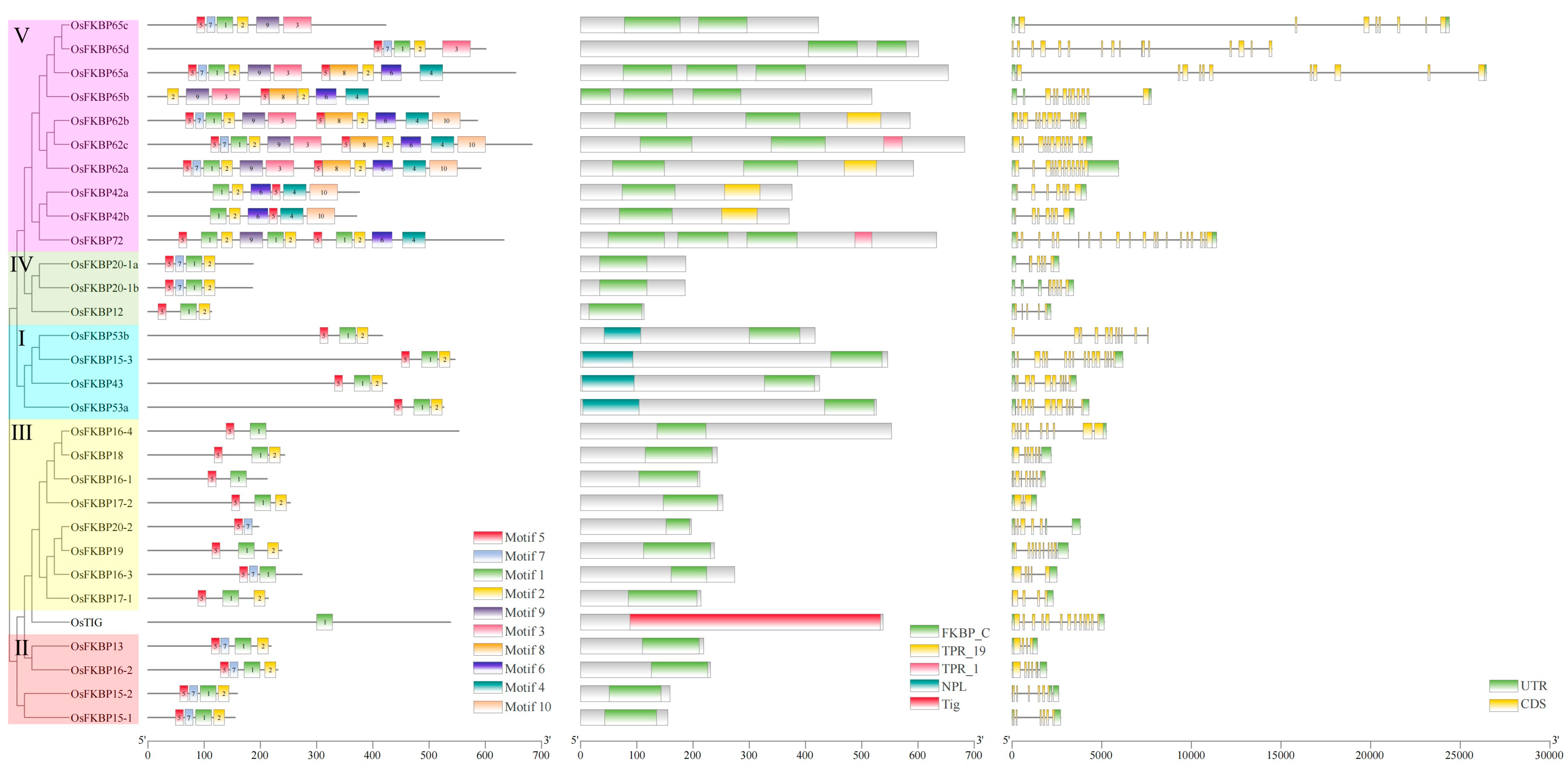
3.4. Analysis of Conserved Motifs, Domains, and the Gene Structures of FKBPs
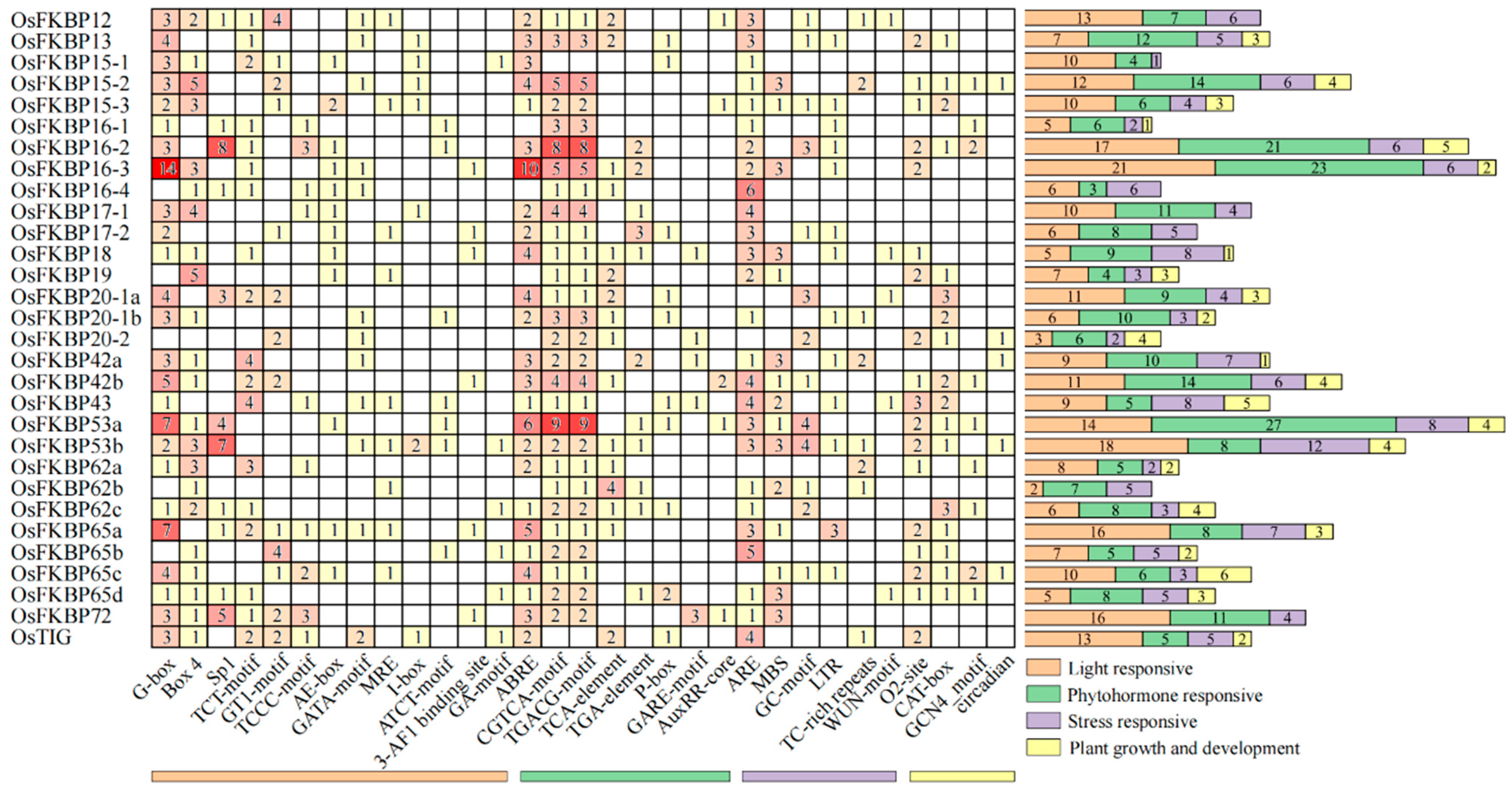
3.5. Analysis of Cis-Acting Elements in FKBP Gene Promoters
3.6. FKBP Gene Response to High-Temperature Stress
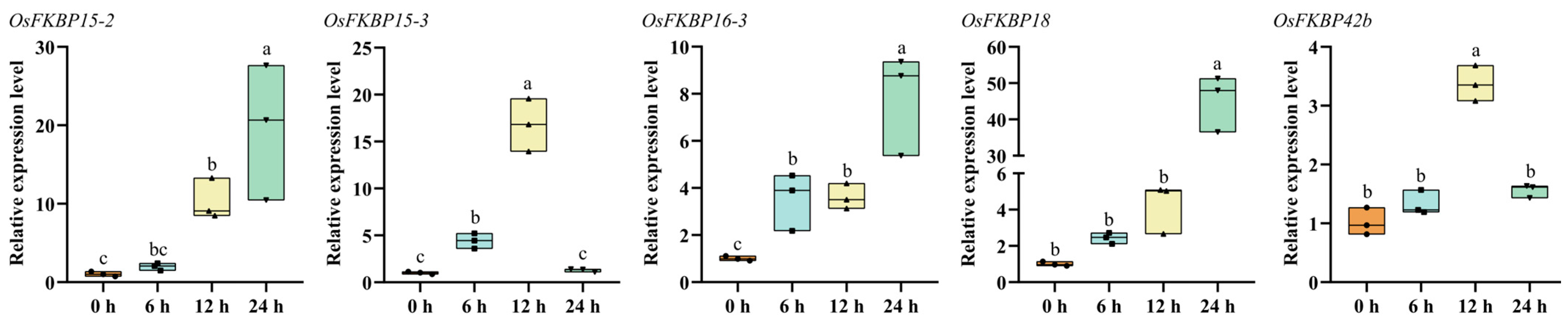
3.7. Analysis of FKBP Gene Responses to Drought Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shige, Z. Progress and clinical application evaluation of immunosuppressants. Eval. Anal. Drug-Use Hosp. China 2008, 8, 803–808. [Google Scholar]
- Schreiber, S.L. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 1991, 251, 283–287. [Google Scholar] [CrossRef]
- Fruman, D.A.; Burakoff, S.J.; Bierer, B.E. Immunophilins in protein folding and immunosuppression. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 1994, 8, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Van Duyne, G.D.; Standaert, R.F.; Karplus, P.A.; Schreiber, S.L.; Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993, 229, 105–124. [Google Scholar] [CrossRef]
- Patterson, C.E.; Gao, J.; Rooney, A.P.; Davis, E.C. Genomic organization of mouse and human 65 kDa FK506-binding protein genes and evolution of the FKBP multigene family. Genomics 2002, 79, 881–889. [Google Scholar] [CrossRef]
- Somarelli, J.A.; Lee, S.Y.; Skolnick, J.; Herrera, R.J. Structure-based classification of 45 FK506-binding proteins. Proteins 2008, 72, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Kurek, I.; Aviezer, K.; Erel, N.; Herman, E.; Breiman, A. The wheat peptidyl prolyl cis-trans-isomerase FKBP77 is heat induced and developmentally regulated. Plant Physiol. 1999, 119, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Aviezer-Hagai, K.; Skovorodnikova, J.; Galigniana, M.; Farchi-Pisanty, O.; Maayan, E.; Bocovza, S.; Efrat, Y.; von Koskull-Döring, P.; Ohad, N.; Breiman, A. Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol. Biol. 2007, 63, 237–255. [Google Scholar] [CrossRef]
- Hueros, G.; Rahfeld, J.; Salamini, F.; Thompson, R. A maize FK506-sensitive immunophilin, mzFKBP-66, is a peptidylproline cis-trans-isomerase that interacts with calmodulin and a 36-kDa cytoplasmic protein. Planta 1998, 205, 121–131. [Google Scholar] [CrossRef]
- Wang, J.; Sun, W.; Kong, X.; Zhao, C.; Li, J.; Chen, Y.; Gao, Z.; Zuo, K. The peptidyl-prolyl isomerases FKBP15-1 and FKBP15-2 negatively affect lateral root development by repressing the vacuolar invertase VIN2 in Arabidopsis. Planta 2002, 252, 52. [Google Scholar]
- Thirumalaikumar, V.P.; Gorka, M.; Schulz, K.; Masclaux-Daubresse, C.; Sampathkumar, A.; Skirycz, A.; Vierstra, R.D.; Balazadeh, S. Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90.1 and ROF1. Autophagy 2021, 17, 2184–2199. [Google Scholar] [CrossRef] [PubMed]
- Meiri, D.; Tazat, K.; Cohen-Peer, R.; Farchi-Pisanty, O.; Aviezer-Hagai, K.; Avni, A.; Breiman, A. Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol. Biol. 2010, 72, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Otegui, M.S.; Spalding, E.P. The ER-localized TWD1 immunophilin is necessary for localization of multidrug resistance-like proteins required for polar auxin transport in Arabidopsis roots. Plant Cell 2010, 22, 3295–3304. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.C.; Kim, D.W.; You, Y.N.; Seok, M.S.; Park, J.M.; Hwang, H.; Kim, B.G.; Luan, S.; Park, H.S.; Cho, H.S. Classification of rice (Oryza sativa L. Japonica nipponbare) immunophilins (FKBPs, CYPs) and expression patterns under water stress. BMC Plant Biol. 2010, 10, 253. [Google Scholar]
- Nigam, N.; Singh, A.; Sahi, C.; Chandramouli, A.; Grover, A. SUMO-conjugating enzyme (Sce) and FK506-binding protein (FKBP) encoding rice (Oryza sativa L.) genes: Genome-wide analysis, expression studies and evidence for their involvement in abiotic stress response. Mol. Genet. Genom. 2008, 279, 371–383. [Google Scholar]
- Suri, A.; Singh, H.; Kaur, K.; Kaachra, A.; Singh, P. Genome-wide characterization of FK506-binding proteins, parvulins and phospho-tyrosyl phosphatase activators in wheat and their regulation by heat stress. Front. Plant Sci. 2022, 13, 1053524. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Li, Y.; Zhu, M.; Cui, R.; Gao, J.; Shu, Y.; Lu, X.; Zhang, H.; Zhang, K. Genome-Wide Identification and Expression Analysis of the Cucumber FKBP Gene Family in Response to Abiotic and Biotic Stresses. Genes 2023, 14, 2006. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Feng, S.; Tian, Y.; Zhuang, L.; Cha, G.; Duan, S.; Li, H.; Nong, X.; Zhang, Z.; Tu, X.; et al. Identification, characterization and spatiotemporal expression analysis of the FKBP family genes in Locusta migratoria. Sci. Rep. 2023, 13, 4048. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, C.; Huang, J.; Liu, Q.; Wei, H.; Wang, H.; Liu, G.; Gu, L.; Yu, S. Genomic analyses reveal the genetic basis of early maturity and identification of loci and candidate genes in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 2021, 19, 109–123. [Google Scholar] [CrossRef]
- Zhan, Y.; Wu, T.; Zhao, X.; Wang, J.; Guo, S.; Chen, S.; Qu, S.; Zheng, Z. Genome-wide identification and expression of monoacylglycerol lipase (MAGL) gene family in peanut (Arachis hypogaea L.) and functional analysis of AhMGATs in neutral lipid metabolism. Int. J. Biol. Macromol. 2023, 243, 125300. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, 39–49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, Y.; Huan, Q.; Li, K.; Qian, W. Single-cell transcriptome atlas of the leaf and root of rice seedlings. J. Genet. Genom. 2021, 48, 881–898. [Google Scholar] [CrossRef]
- Zhao, J.; Meng, X.; Zhang, Z.; Wang, M.; Nie, F.; Liu, Q. OsLPR5 Encoding Ferroxidase Positively Regulates the Tolerance to Salt Stress in Rice. Int. J. Mol. Sci. 2023, 24, 8115. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, X.; Wang, M.; Xie, L.; Wu, Z.; Yu, J.; Wang, Y.; Zhang, Z.; Jia, Y.; Liu, Q. The miR528-D3 Module Regulates Plant Height in Rice by Modulating the Gibberellin and Abscisic Acid Metabolisms. Rice 2022, 15, 27. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Xiao, J.; Xu, K. Reduced photosynthetic dark reaction triggered by ABA application increases intercellular CO2 concentration, generates H2O2 and promotes closure of stomata in ginger leaves. Environ. Exp. Bot. 2015, 113, 11–17. [Google Scholar] [CrossRef]
- Sun, X.; Xiong, H.; Jiang, C.; Zhang, D.; Yang, Z.; Huang, Y.; Li, Z. Natural variation of DROT1 confers drought adaptation in upland rice. Nat. Commun. 2022, 13, 4265. [Google Scholar] [CrossRef]
- Gollan, P.J.; Bhave, M. Genome-wide analysis of genes encoding FK506-binding proteins in rice. Plant Mol. Biol. 2010, 72, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yanli, Y.; Yanjiao, L.; Kaiyuan, P.; Fajun, Z.; Sun, Q.; Wencai, L.; Zhaodong, M. The structure and biological function of the FKBP gene family in plants. Hereditas 2014, 536–546. [Google Scholar]
- Ge, Q.; Peng, P.; Cheng, M.; Meng, Y.; Cao, Y.; Zhang, S.; Long, Y.; Li, G.; Kang, G. Genome-Wide Identification and Analysis of FKBP Gene Family in Wheat (Triticum asetivum). Int. J. Mol. Sci. 2022, 23, 14501. [Google Scholar] [CrossRef] [PubMed]
- Luan, S.; Kudla, J.; Gruissem, W.; Schreiber, S.L. Molecular characterization of a FKBP-type immunophilin from higher plants. Proc. Natl. Acad. Sci. USA 1996, 93, 6964–6969. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Li, L.; Luan, S. Immunophilins and parvulins. Superfamily Pept. Prolyl Isomerases Arabidopsis. Plant Physiol. 2004, 134, 1248–1267. [Google Scholar] [CrossRef]
- Liu, J.; Ghelli, R.; Cardarelli, M.; Geisler, M.; Bartlett, M. Arabidopsis TWISTED DWARF1 regulates stamen elongation by differential activation of ABCB1,19-mediated auxin transport. J. Exp. Bot. 2022, 73, 4818–4831. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, F.; Wang, M.; Liu, L.; Ma, X.; Zhao, J. Genome-Wide Identification and Bioinformatics Analysis of the FK506 Binding Protein Family in Rice. Genes 2024, 15, 902. https://doi.org/10.3390/genes15070902
Nie F, Wang M, Liu L, Ma X, Zhao J. Genome-Wide Identification and Bioinformatics Analysis of the FK506 Binding Protein Family in Rice. Genes. 2024; 15(7):902. https://doi.org/10.3390/genes15070902
Chicago/Turabian StyleNie, Fanhao, Minghao Wang, Linlin Liu, Xuefei Ma, and Juan Zhao. 2024. "Genome-Wide Identification and Bioinformatics Analysis of the FK506 Binding Protein Family in Rice" Genes 15, no. 7: 902. https://doi.org/10.3390/genes15070902
APA StyleNie, F., Wang, M., Liu, L., Ma, X., & Zhao, J. (2024). Genome-Wide Identification and Bioinformatics Analysis of the FK506 Binding Protein Family in Rice. Genes, 15(7), 902. https://doi.org/10.3390/genes15070902




