MicroRNA166: Old Players and New Insights into Crop Agronomic Traits Improvement
Abstract
:1. Introduction
2. Conservation and Diversification of MIR165/166 in Model Plants and Main Crops
3. Functions of miR166 in Crop Development and Stress Response
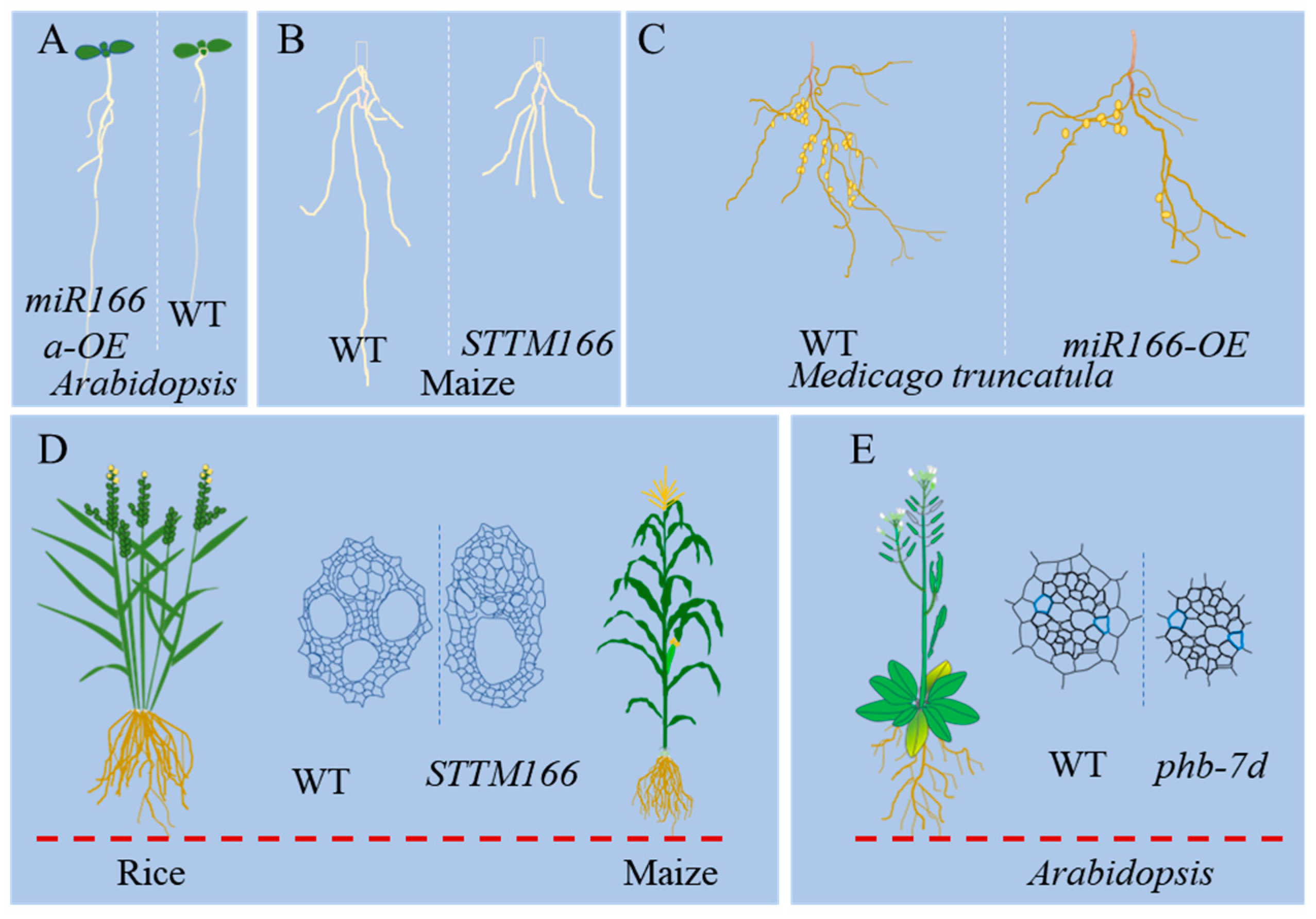
3.1. miR166 as a Determinant in Plant Morphogenesis
3.2. miR166 Regulates Root and Vascular Development
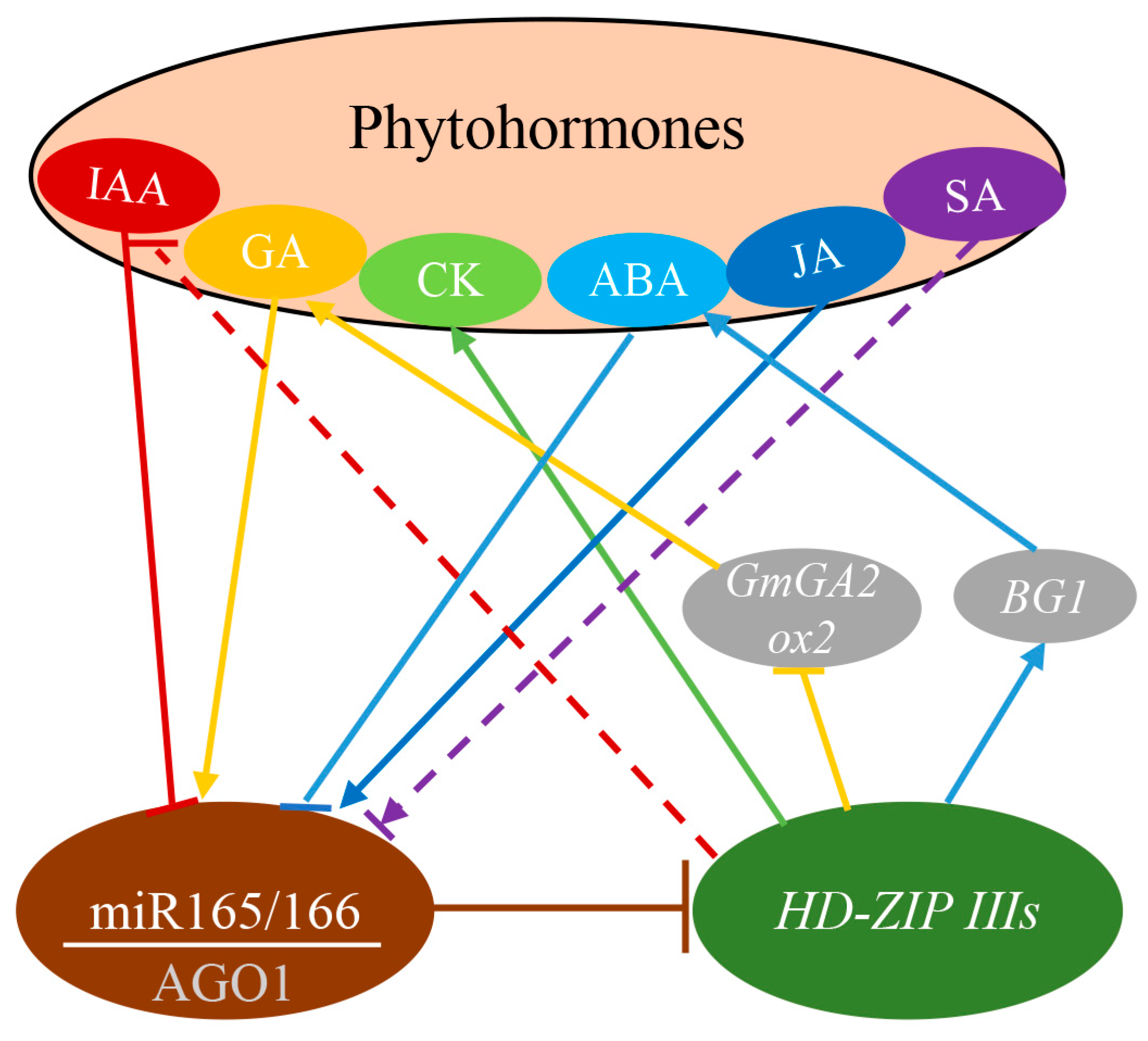
3.3. The Regulatory Role of miR165/166 in Phytohormones Signaling
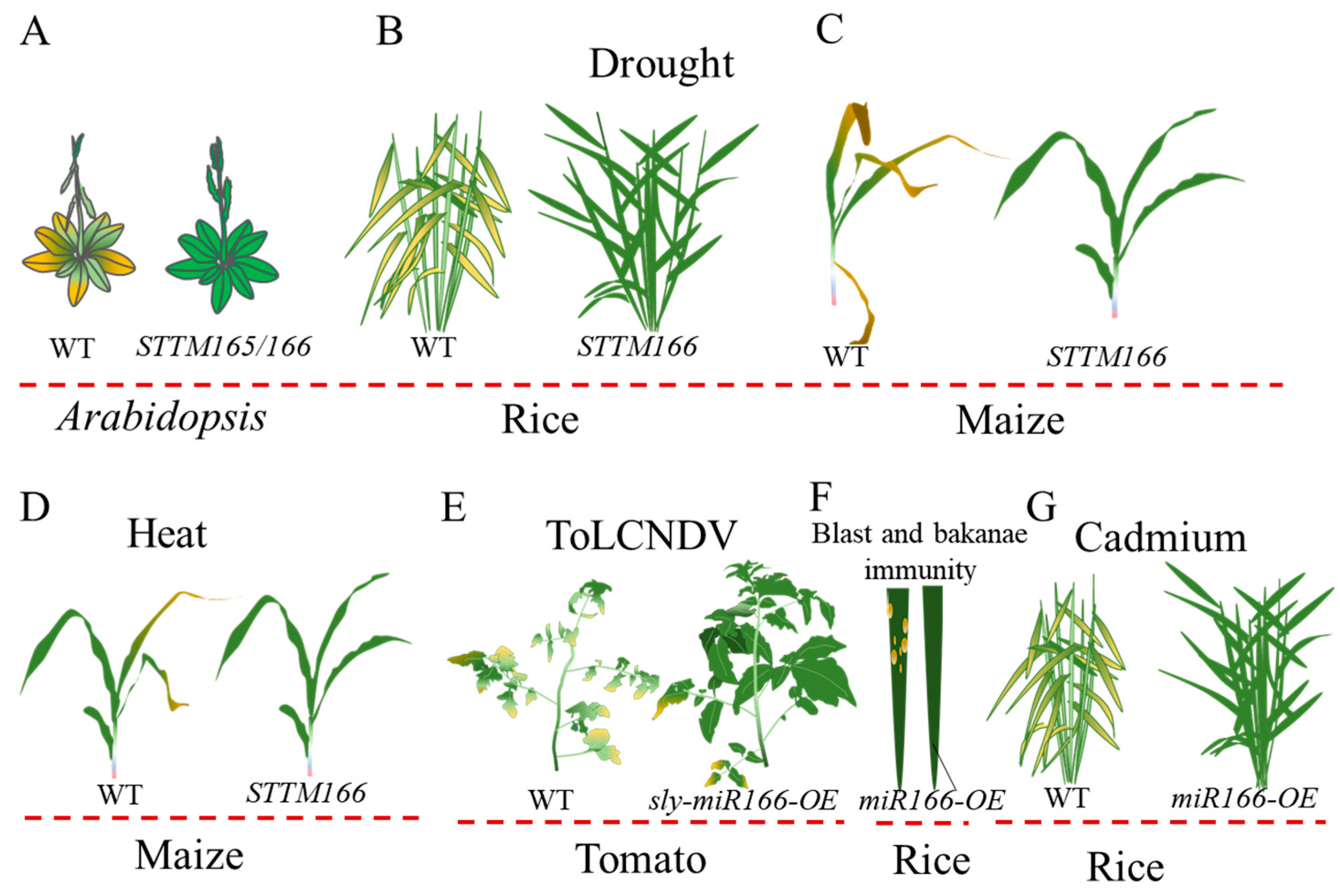
3.4. miR166 in Response to Abiotic Stress and Pathogenic Infection
3.5. Other Functions of miR166 in Crops
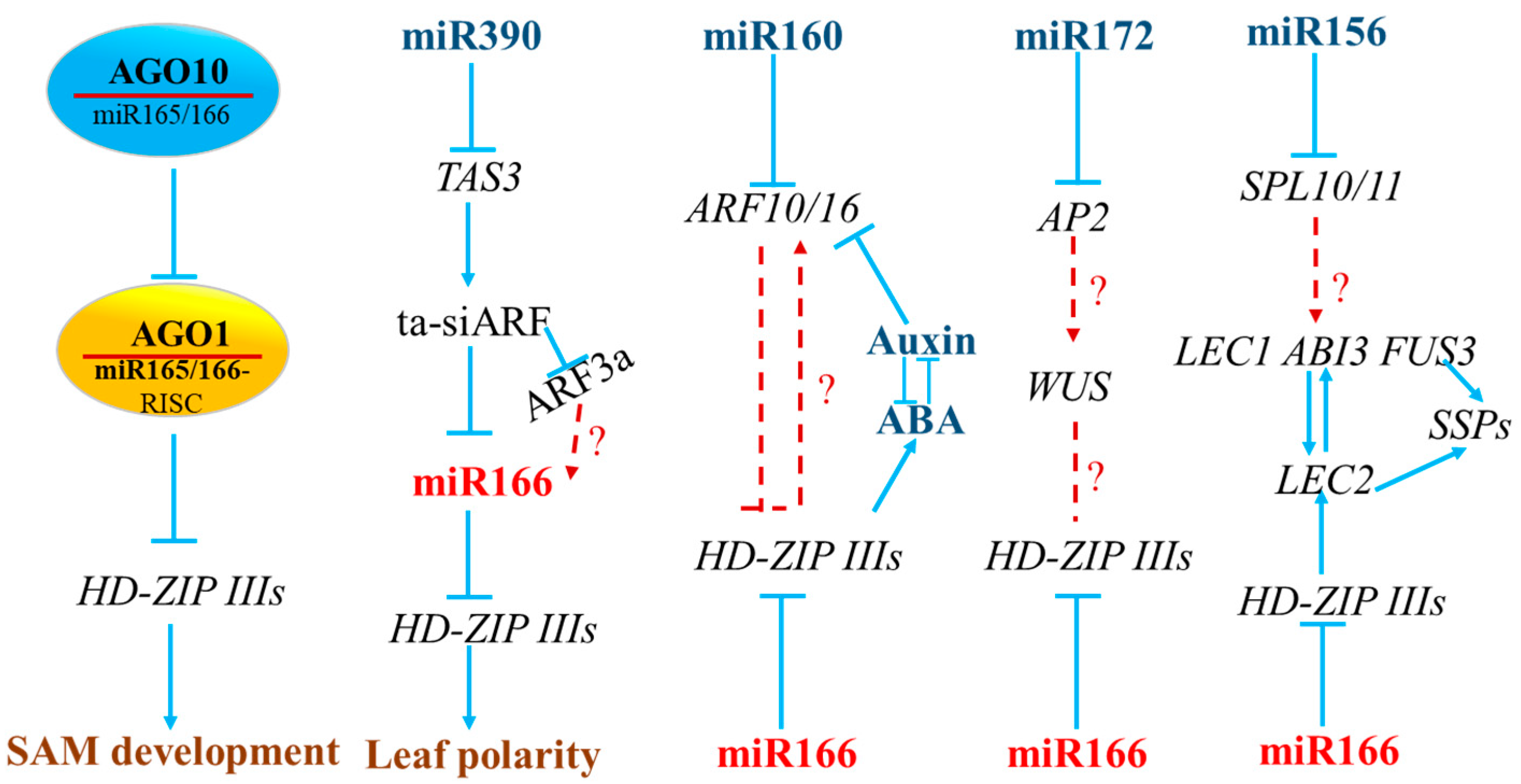
4. The Interactions between miR166 and Other miRNAs in Model and Crop Plants
5. Exploring the miR166-HD-ZIP IIIs Module to Improve Complex Agronomic Traits
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ray, D.K.; Gerber, J.S.; MacDonald, G.K.; West, P.C. Climate variation explains a third of global crop yield variability. Nat. Commun. 2015, 6, 5989. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef]
- Tang, J.; Chu, C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nature Plants 2017, 3, 17077. [Google Scholar] [CrossRef]
- Zhang, Z.; Teotia, S.; Tang, J.; Tang, G. Perspectives on microRNAs and phased small interfering RNAs in maize (Zea mays L.): Functions and big Impact on agronomic traits enhancement. Plants 2019, 8, 170. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. Small RNAs in development—Insights from plants. Curr. Opin. Genet. Dev. 2012, 22, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, C.; Yu, T.; Zhang, R.; Zheng, H.; Yan, W. MicroRNAs control mRNA fate by compartmentalization based on 3′ UTR length in male germ cells. Genome Biol. 2017, 18, 105. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef]
- Gandikota, M.; Birkenbihl, R.P.; Höhmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. Cell Mol. Biol. 2007, 49, 683–693. [Google Scholar] [CrossRef]
- Yan, J.; Gu, Y.; Jia, X.; Kang, W.; Pan, S.; Tang, X.; Chen, X.; Tang, G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 2012, 24, 415–427. [Google Scholar] [CrossRef]
- Peng, T.; Qiao, M.; Liu, H.; Teotia, S.; Zhang, Z.; Zhao, Y.; Wang, B.; Zhao, D.; Shi, L.; Zhang, C.; et al. A resource for inactivation of microRNAs using short tandem target mimic technology in model and crop plants. Mol. Plant 2018, 11, 1400–1417. [Google Scholar] [CrossRef]
- Li, N.; Yang, T.; Guo, Z.; Wang, Q.; Chai, M.; Wu, M.; Li, X.; Li, W.; Li, G.; Tang, J.; et al. Maize microRNA166 Inactivation Confers Plant Development and Abiotic Stress Resistance. Int. J. Mol. Sci. 2020, 21, 9506. [Google Scholar] [CrossRef]
- Sakaguchi, J.; Watanabe, Y. miR165/166 and the development of land plants. Dev. Growth Differ. 2012, 54, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Gong, S.; Wang, Y.; Wang, F.; Bao, H.; Sun, J.; Cai, C.; Yi, K.; Chen, Z.; Zhu, C. MicroRNA166 modulates Cadmium tolerance and accumulation in rice. Plant Physiol. 2018, 177, 1691–1703. [Google Scholar] [CrossRef]
- Maher, C.G.; Stein, L.; Ware, D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006, 16, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.S.; Tarver, J.E.; Hiscock, S.J.; Donoghue, P.C.J. Evolutionary history of plant microRNAs. Trends Plant Sci. 2014, 19, 175–182. [Google Scholar] [CrossRef]
- Hashimoto, K.; Miyashima, S.; Sato-Nara, K.; Yamada, T.; Nakajima, K. Functionally diversified members of the MIR165/6 gene family regulate ovule morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2018, 47, D155–D162. [Google Scholar] [CrossRef]
- Cui, J.; You, C.; Chen, X. The evolution of microRNAs in plants. Curr. Opin. Plant Biol. 2017, 35, 61–67. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Baldrich, P.; Hsing, Y.-I.C.; San Segundo, B. Genome-Wide Analysis of Polycistronic MicroRNAs in Cultivated and Wild Rice. Genome Biol. Evol. 2016, 8, 1104–1114. [Google Scholar] [CrossRef] [PubMed]
- Barik, S.; SarkarDas, S.; Singh, A.; Gautam, V.; Kumar, P.; Majee, M.; Sarkar, A.K. Phylogenetic analysis reveals conservation and diversification of micro RNA166 genes among diverse plant species. Genomics 2014, 103, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Todesco, M.; Rubiosomoza, I.; Pazares, J.; Weigel, D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001031. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Honda, M.; Zhu, H.; Zhang, Z.; Guo, X.; Li, T.; Li, Z.; Peng, X.; Nakajima, K.; Duan, L. Spatiotemporal sequestration of miR165/166 by Arabidopsis Argonaute10 promotes shoot apical meristem maintenance. Cell Rep. 2015, 10, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Merelo, P.; Ram, H.; Caggiano, M.P.; Ohno, C.; Ott, F.; Straub, D.; Graeff, M.; Cho, S.K.; Yang, S.W.; Wenkel, S.; et al. Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc. Natl. Acad. Sci. USA 2016, 113, 11973–11978. [Google Scholar] [CrossRef]
- Jia, X.; Ding, N.; Fan, W.; Yan, J.; Gu, Y.; Tang, X.; Li, R.; Tang, G. Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Sci. Int. J. Exp. Plant Biol. 2015, 233, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhao, C.; Zhou, J.; Yang, Y.; Wang, P.; Zhu, X.; Tang, G.; Bressan, R.A.; Zhu, J.K. The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006416. [Google Scholar] [CrossRef]
- Kidner, C.A.; Martienssen, R.A. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 2004, 428, 81–84. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, F.; Wang, R.; Zhou, X.; Sze, S.H.; Liou, L.W.; Barefoot, A.; Dickman, M.; Zhang, X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 2011, 145, 242–256. [Google Scholar] [CrossRef]
- Singh, A.; Roy, S.; Singh, S.; Das, S.S.; Gautam, V.; Yadav, S.; Kumar, A.; Singh, A.; Samantha, S.; Sarkar, A.K. Phytohormonal crosstalk modulates the expression of miR166/165s, target Class III HD-ZIPs, and KANADI genes during root growth in Arabidopsis thaliana. Sci. Rep. 2017, 7, 3408. [Google Scholar] [CrossRef]
- Donner, T.J.; Sherr, I.; Scarpella, E. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 2009, 136, 3235–3246. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jung, J.H.; Reyes, J.L.; Kim, Y.S.; Kim, S.Y.; Chung, K.S.; Kim, J.A.; Lee, M.; Lee, Y.; Narry Kim, V.; et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. Cell Mol. Biol. 2005, 42, 84–94. [Google Scholar] [CrossRef]
- Tatematsu, K.; Toyokura, K.; Miyashima, S.; Nakajima, K.; Okada, K. A molecular mechanism that confines the activity pattern of miR165 in Arabidopsis leaf primordia. Plant J. Cell Mol. Biol. 2015, 82, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Wang, H.; Li, H.; Yuan, Z.; Li, F.; Yang, L.; Huang, H. Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIR165a and MIR166a genes. FEBS Lett. 2009, 583, 3711–3717. [Google Scholar] [CrossRef]
- Gu, Z.; Huang, C.; Li, F.; Zhou, X. A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 2014, 12, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Sha, A.; Zhao, J.; Yin, K.; Tang, Y.; Wang, Y.; Wei, X.; Hong, Y.; Liu, Y. Virus-based microRNA silencing in plants. Plant Physiol. 2014, 164, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, H.; Srivastava, A.K.; Pan, Y.; Bai, J.; Fang, J.; Shi, H.; Zhu, J.K. Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 2018, 176, 2082–2094. [Google Scholar] [CrossRef]
- Juarez, M.T.; Kui, J.S.; Thomas, J.; Heller, B.A.; Timmermans, M.C. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 2004, 428, 84–88. [Google Scholar] [CrossRef]
- Juarez, M.T.; Twigg, R.W.; Timmermans, M.C.P. Specification of adaxial cell fate during maize leaf development. Development 2004, 131, 4533–4544. [Google Scholar] [CrossRef]
- Jiang, D.; Hua, L.; Zhang, C.; Li, H.; Wang, Z.; Li, J.; Wang, G.; Song, R.; Shen, T.; Li, H.; et al. Mutations in the miRNA165/166 binding site of the HB2 gene result in pleiotropic effects on morphological traits in wheat. Crop J. 2023, 11, 9–20. [Google Scholar] [CrossRef]
- Nogueira, F.T.S.; Chitwood, D.H.; Madi, S.; Ohtsu, K.; Schnable, P.S.; Scanlon, M.J.; Timmermans, M.C.P. Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet. 2009, 5, e1000320. [Google Scholar] [CrossRef] [PubMed]
- Itoh, J.; Hibara, K.; Sato, Y.; Nagato, Y. Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 2008, 147, 1960–1975. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, F.T.; Madi, S.; Chitwood, D.H.; Juarez, M.T.; Timmermans, M.C. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 2007, 21, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Eshed Williams, L. Genetics of Shoot Meristem and Shoot Regeneration. Annu. Rev. Genet. 2021, 55, 661–681. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Meng, S.; Weng, J.; Wu, Q. Fine-tuning shoot meristem size to feed the world. Trends Plant Sci. 2022, 27, 355–363. [Google Scholar] [CrossRef]
- Grigg, S.P.; Canales, C.; Hay, A.; Tsiantis, M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 2005, 437, 1022–1026. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.; Grigg, S.P.; Xie, M.; Christensen, S.; Fletcher, J.C. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 2005, 132, 3657–3668. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Fan, L.; Le, B.H.; Ye, P.; Mo, B.; Chen, X. Regulation of ARGONAUTE10 Expression Enables Temporal and Spatial Precision in Axillary Meristem Initiation in Arabidopsis. Dev. Cell 2020, 55, 603–616.e5. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, X. Argonautes compete for miR165/166 to regulate shoot apical meristem development. Curr. Opin. Plant Biol. 2012, 15, 652–658. [Google Scholar] [CrossRef]
- Sun, W.; Xiang, X.; Zhai, L.; Zhang, D.; Cao, Z.; Liu, L.; Zhang, Z. AGO18b negatively regulates determinacy of spikelet meristems on the tassel central spike in maize. J. Integr. Plant Biol. 2018, 60, 65–78. [Google Scholar] [CrossRef]
- Iwamoto, M.; Tagiri, A. MicroRNA-targeted transcription factor gene RDD1 promotes nutrient ion uptake and accumulation in rice. Plant J. Cell Mol. Biol. 2016, 85, 466–477. [Google Scholar] [CrossRef]
- Iwamoto, M. The transcription factor gene RDD1 promotes carbon and nitrogen transport and photosynthesis in rice. Plant Physiol. Biochem. 2020, 155, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Clepet, C.; Devani, R.S.; Boumlik, R.; Hao, Y.; Morin, H.; Marcel, F.; Verdenaud, M.; Mania, B.; Brisou, G.; Citerne, S.; et al. The miR166- SlHB15A Regulatory Module Controls Ovule Development and Parthenocarpic Fruit Set under Adverse Temperatures in Tomato. Mol. Plant 2021, 14, 1185–1198. [Google Scholar] [CrossRef]
- Li, X.; Lian, H.; Zhao, Q.; He, Y. MicroRNA166 Monitors SPOROCYTELESS/NOZZLE for Building of the Anther Internal Boundary. Plant Physiol. 2019, 181, 208–220. [Google Scholar] [CrossRef]
- Barrera-Rojas, C.H.; Otoni, W.C.; Nogueira, F.T.S. Shaping the root system: The interplay between miRNA regulatory hubs and phytohormones. J. Exp. Bot. 2021, 72, 6822–6835. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; Wang, G.; Augstein, F.; de Vries, J.; Carlsbecker, A. Continuous root xylem formation and vascular acclimation to water deficit involves endodermal ABA signalling via miR165. Development 2018, 145, dev159202. [Google Scholar] [CrossRef] [PubMed]
- Carlsbecker, A.; Lee, J.Y.; Roberts, C.J.; Dettmer, J.; Lehesranta, S.; Zhou, J.; Lindgren, O.; Moreno-Risueno, M.A.; Vaten, A.; Thitamadee, S.; et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 2010, 465, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Miyashima, S.; Koi, S.; Hashimoto, T.; Nakajima, K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 2011, 138, 2303–2313. [Google Scholar] [CrossRef]
- Singh, A.; Singh, S.; Panigrahi, K.C.; Reski, R.; Sarkar, A.K. Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain-leucine zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 945–953. [Google Scholar] [CrossRef]
- Wei, H.; Song, Z.; Xie, Y.; Cheng, H.; Yan, H.; Sun, F.; Liu, H.; Shen, J.; Li, L.; He, X.; et al. High temperature inhibits vascular development via the PIF4-miR166-HB15 module in Arabidopsis. Curr. Biol. 2023, 33, 3203–3214.e4. [Google Scholar] [CrossRef]
- Boualem, A.; Laporte, P.; Jovanovic, M.; Laffont, C.; Plet, J.; Combier, J.P.; Niebel, A.; Crespi, M.; Frugier, F. MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. Cell Mol. Biol. 2008, 54, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Gautam, V.; Singh, A.; Yadav, S.; Singh, S.; Kumar, P.; Sarkar Das, S.; Sarkar, A.K. Conserved LBL1-ta-siRNA and miR165/166-RLD1/2 modules regulate root development in maize. Development 2021, 148, dev190033. [Google Scholar] [CrossRef]
- Zhou, G.K.; Kubo, M.; Zhong, R.; Demura, T.; Ye, Z.H. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007, 48, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fang, R.; Deng, R.; Li, J. The OsmiRNA166b-OsHox32 pair regulates mechanical strength of rice plants by modulating cell wall biosynthesis. Plant Biotechnol. J. 2021, 19, 1468–1480. [Google Scholar] [CrossRef]
- Curaba, J.; Singh, M.B.; Bhalla, P.L. miRNAs in the crosstalk between phytohormone signalling pathways. J. Exp. Bot. 2014, 65, 1425–1438. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Ma, J.; Zhang, Y.; Yang, S.; Feng, X.; Yan, J. The miR166 mediated regulatory module controls plant height by regulating gibberellic acid biosynthesis and catabolism in soybean. J. Integr. Plant Biol. 2022, 64, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Jain, D.; Pandey, J.; Yadav, M.; Bansal, K.C.; Singh, I.K. Deciphering the role of miRNA in reprogramming plant responses to drought stress. Crit. Rev. Biotechnol. 2022, 43, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Teotia, S.; Lan, T.; Tang, G. MicroRNA Techniques: Valuable Tools for Agronomic Trait Analyses and Breeding in Rice. Front. Plant Sci. 2021, 12, 744357. [Google Scholar] [CrossRef]
- Li, J.; Cao, Y.; Zhang, J.; Zhu, C.; Tang, G.; Yan, J. The miR165/166-PHABULOSA module promotes thermotolerance by transcriptionally and posttranslationally regulating HSFA1. Plant Cell 2023, 35, 2952–2971. [Google Scholar] [CrossRef]
- Lei, X.; Chen, M.; Xu, K.; Sun, R.; Zhao, S.; Wu, N.; Zhang, S.; Yang, X.; Xiao, K.; Zhao, Y. The miR166d/TaCPK7-D Signaling Module Is a Critical Mediator of Wheat (Triticum aestivum L.) Tolerance to K+ Deficiency. Int. J. Mol. Sci. 2023, 24, 7926. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Yang, N.; Tian, C.; Wen, S.; Zhang, C.; Zheng, A.; Hu, X.; Fang, J.; Zhang, Z.; Lai, Z.; et al. The miR166 targets CsHDZ3 genes to negatively regulate drought tolerance in tea plant (Camellia sinensis). Int. J. Biol. Macromol. 2024, 264, 130735. [Google Scholar] [CrossRef] [PubMed]
- Salvador-Guirao, R.; Hsing, Y.I.; San Segundo, B. The Polycistronic miR166k-166h Positively Regulates Rice Immunity via Post-transcriptional Control of EIN2. Front. Plant Sci. 2018, 9, 337. [Google Scholar] [CrossRef]
- Wang, K.; Su, X.; Cui, X.; Du, Y.; Zhang, S.; Gao, J. Identification and Characterization of microRNA during Bemisia tabaci Infestations in Solanum lycopersicum and Solanum habrochaites. Hortic. Plant J. 2018, 4, 62–72. [Google Scholar] [CrossRef]
- Prasad, A.; Sharma, N.; Chirom, O.; Prasad, M. The sly-miR166-SlyHB module acts as a susceptibility factor during ToLCNDV infection. Theor. Appl. Genet. 2022, 135, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Candar-Cakir, B.; Arican, E.; Zhang, B. Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol. J. 2016, 14, 1727–1746. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Sun, S.; Hua, S.; Shen, E.; Ye, C.Y.; Cai, D.; Timko, M.P.; Zhu, Q.H.; Fan, L. Analysis of transcriptional and epigenetic changes in hybrid vigor of allopolyploid Brassica napus uncovers key roles for small RNAs. Plant J. 2017, 91, 874–893. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Ragupathy, R.; Edwards, T.; Domaratzki, M.; Cloutier, S. MicroRNA-guided regulation of heat stress response in wheat. BMC Genom. 2019, 20, 488. [Google Scholar] [CrossRef] [PubMed]
- Mangrauthia, S.K.; Bhogireddy, S.; Agarwal, S.; Prasanth, V.V.; Voleti, S.R.; Neelamraju, S.; Subrahmanyam, D. Genome-wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. J. Exp. Bot. 2017, 68, 2399–2412. [Google Scholar] [CrossRef]
- Peng, Y.; Zhang, X.; Liu, Y.; Chen, X. Exploring Heat-Response Mechanisms of MicroRNAs Based on Microarray Data of Rice Post-meiosis Panicle. Int. J. Genom. 2020, 2020, 7582612. [Google Scholar] [CrossRef]
- He, J.; Jiang, Z.; Gao, L.; You, C.; Ma, X.; Wang, X.; Xu, X.; Mo, B.; Chen, X.; Liu, L. Genome-Wide Transcript and Small RNA Profiling Reveals Transcriptomic Responses to Heat Stress. Plant Physiol. 2019, 181, 609–629. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.; Saxena, S.; Chauhan, A.S.; Mathur, P.; Rani, V.; Chakrabaroty, D. Identification and expression analysis of conserved microRNAs during short and prolonged chromium stress in rice (Oryza sativa). Environ. Sci. Pollut. Res. Int. 2020, 27, 380–390. [Google Scholar] [CrossRef]
- Yin, Z.; Murawska, Z.; Xie, F.; Pawełkowicz, M.; Michalak, K.; Zhang, B.; Lebecka, R. microRNA response in potato virus Y infected tobacco shows strain-specificity depending on host and symptom severity. Virus Res. 2019, 260, 20–32. [Google Scholar] [CrossRef]
- Gorshkov, O.; Chernova, T.; Mokshina, N.; Gogoleva, N.; Suslov, D.; Tkachenko, A.; Gorshkova, T. Intrusive Growth of Phloem Fibers in Flax Stem: Integrated Analysis of miRNA and mRNA Expression Profiles. Plants 2019, 8, 47. [Google Scholar] [CrossRef]
- Bai, B.; Shi, B.; Hou, N.; Cao, Y.; Meng, Y.; Bian, H.; Zhu, M.; Han, N. microRNAs participate in gene expression regulation and phytohormone cross-talk in barley embryo during seed development and germination. BMC Plant Biol. 2017, 17, 150. [Google Scholar] [CrossRef]
- DeBoer, K.; Melser, S.; Sperschneider, J.; Kamphuis, L.G.; Garg, G.; Gao, L.L.; Frick, K.; Singh, K.B. Identification and profiling of narrow-leafed lupin (Lupinus angustifolius) microRNAs during seed development. BMC Genom. 2019, 20, 135. [Google Scholar] [CrossRef]
- Li, D.; Liu, Z.; Gao, L.; Wang, L.; Gao, M.; Jiao, Z.; Qiao, H.; Yang, J.; Chen, M.; Yao, L.; et al. Genome-Wide Identification and Characterization of microRNAs in Developing Grains of Zea mays L. PLoS ONE 2016, 11, e0153168. [Google Scholar] [CrossRef]
- Li, D.; Wang, L.; Liu, X.; Cui, D.; Chen, T.; Zhang, H.; Jiang, C.; Xu, C.; Li, P.; Li, S.; et al. Deep sequencing of maize small RNAs reveals a diverse set of microRNA in dry and imbibed seeds. PLoS ONE 2013, 8, e55107. [Google Scholar] [CrossRef] [PubMed]
- Puchta, M.; Groszyk, J.; Małecka, M.; Koter, M.D.; Niedzielski, M.; Rakoczy-Trojanowska, M.; Boczkowska, M. Barley Seeds miRNome Stability during Long-Term Storage and Aging. Int. J. Mol. Sci. 2021, 22, 4315. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Dai, X.; Wang, R.; Wang, J.; Liu, Z.; Xiang, F. ARGONAUTE10 inhibits in vitro shoot regeneration via repression of miR165/166 in Arabidopsis thaliana. Plant Cell Physiol. 2017, 58, 1789–1800. [Google Scholar] [CrossRef]
- Du, F.; Gong, W.; Boscá, S.; Tucker, M.; Vaucheret, H.; Laux, T. Dose-Dependent AGO1-Mediated Inhibition of the miRNA165/166 Pathway Modulates Stem Cell Maintenance in Arabidopsis Shoot Apical Meristem. Plant Commun. 2020, 1, 100002. [Google Scholar] [CrossRef] [PubMed]
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big impact on plant development. Trends Plant Sci. 2017, 22, 1056–1068. [Google Scholar] [CrossRef] [PubMed]
- Wójcik, A.M.; Nodine, M.D.; Gaj, M.D. miR160 and miR166/165 Contribute to the LEC2-Mediated Auxin Response Involved in the Somatic Embryogenesis Induction in Arabidopsis. Front. Plant Sci. 2017, 8, 2024. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Wang, Y.; Teotia, S.; Wang, Z.; Shi, C.; Sun, H.; Gu, Y.; Zhang, Z.; Tang, G. The interaction between miR160 and miR165/166 in the control of leaf development and drought tolerance in Arabidopsis. Sci. Rep. 2019, 9, 2832. [Google Scholar] [CrossRef] [PubMed]
- Kitazumi, A.; Kawahara, Y.; Onda, T.S.; De Koeyer, D.; de los Reyes, B.G. Implications of miR166 and miR159 induction to the basal response mechanisms of an andigena potato (Solanum tuberosum subsp. andigena) to salinity stress, predicted from network models in Arabidopsis. Genome 2015, 58, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Bian, S.; Tang, M.; Lu, Q.; Li, S.; Liu, X.; Tian, G.; Nguyen, V.; Tsang, E.W.; Wang, A.; et al. MicroRNA-mediated repression of the seed maturation program during vegetative development in Arabidopsis. PLoS Genet. 2012, 8, e1003091. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Liu, X.; Yan, J.; Wang, W.; Yumul, R.E.; Kim, Y.J.; Dinh, T.T.; Liu, J.; Cui, X.; Zheng, B.; et al. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 2011, 7, e1001358. [Google Scholar] [CrossRef] [PubMed]
- Betti, F.; Ladera-Carmona, M.J.; Weits, D.A.; Ferri, G.; Iacopino, S.; Novi, G.; Svezia, B.; Kunkowska, A.B.; Santaniello, A.; Piaggesi, A.; et al. Exogenous miRNAs induce post-transcriptional gene silencing in plants. Nat. Plants 2021, 7, 1379–1388. [Google Scholar] [CrossRef]
- Lauressergues, D.; Couzigou, J.-M.; Clemente, H.S.; Martinez, Y.; Dunand, C.; Bécard, G.; Combier, J.-P. Primary transcripts of microRNAs encode regulatory peptides. Nature 2015, 520, 90–93. [Google Scholar] [CrossRef]
- Ormancey, M.; Guillotin, B.; San Clemente, H.; Thuleau, P.; Plaza, S.; Combier, J.P. Use of microRNA-encoded peptides to improve agronomic traits. Plant Biotechnol. J. 2021, 19, 1687–1689. [Google Scholar] [CrossRef]
- Chen, Q.-J.; Deng, B.-H.; Gao, J.; Zhao, Z.-Y.; Chen, Z.-L.; Song, S.-R.; Wang, L.; Zhao, L.-P.; Xu, W.-P.; Zhang, C.-X.; et al. A miRNA-Encoded Small Peptide, vvi-miPEP171d1, Regulates Adventitious Root Formation. Plant Physiol. 2020, 183, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Couzigou, J.M.; André, O.; Guillotin, B.; Alexandre, M.; Combier, J.P. Use of microRNA-encoded peptide miPEP172c to stimulate nodulation in soybean. New Phytol. 2016, 211, 379–381. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, R.; Jia, X.; Tang, X.; Guo, Y.; Yang, H.; Zheng, X.; Qian, Q.; Qi, Y.; Zhang, Y. CRISPR-Cas9 mediated OsMIR168a knockout reveals its pleiotropy in rice. Plant Biotechnol. J. 2021, 20, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Teotia, S.; Singh, D.; Tang, X.; Tang, G. Essential RNA-Based Technologies and Their Applications in Plant Functional Genomics. Trends Biotechnol. 2016, 34, 106–123. [Google Scholar] [CrossRef]
- Wang, X.-W.; Hu, L.-F.; Hao, J.; Liao, L.-Q.; Chiu, Y.-T.; Shi, M.; Wang, Y. A microRNA-inducible CRISPR-Cas9 platform serves as a microRNA sensor and cell-type-specific genome regulation tool. Nat. Cell Biol. 2019, 21, 522–530. [Google Scholar] [CrossRef]


| Species/Members | Sequence Alignment | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arabidopsis thaliana | ath-miR165a,b | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | C | C | C | C | C | 21 | ||
| 9 | ath-miR166a-g | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | ||
| Brassica napus | bna-miR166a-e | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | ||
| 6 | bna-miR166f | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | C | C | C | C | C | 21 | ||
| Glycine max | gma-miR166h,k | U | C | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | 21 | ||
| 21 | gma-miR166u | U | C | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | 20 | |||
| gma-miR166a-g,i | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | |||
| gma-miR166m | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 20 | ||||
| gma-miR166n,o | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | |||
| gma-miR166j | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | G | 21 | |||
| gma-miR166p-t | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | 20 | ||||
| Gossypium hirsutum 2 | ghr-miR166a,b | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | ||
| Medicago truncatula | mtr-miR166a,b,d,e,g | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | ||
| 7 | mtr-miR166c,f | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | U | C | 21 | ||
| Solanum lycopersicum | sly-miR166a,b | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | ||
| 3 | sly-miR166c | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | U | C | 21 | ||
| Brachypodium distachyon | bdi-miR166g | U | G | U | G | G | U | G | A | ||||||||||||||||
| U | C | U | C | G | G | A | C | C | A | G | G | C | 21 | ||||||||||||
| 10 | bdi-miR166h | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | A | U | C | C | C | U | 21 | ||
| bdi-miR166f | U | C | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | 21 | |||
| bdi-miR166a-d,i | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | |||
| bdi-miR166e | C | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | 21 | |||
| bdi-miR166j | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | U | U | 21 | |||
| Oryza sativa | osa-miR166g-i | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | U | C | 21 | ||
| 13 | osa-miR166a-d,f,j | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | ||
| osa-miR166e | U | C | G | A | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | |||
| osa-miR166k-m | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | A | U | C | C | C | U | 21 | |||
| Sorghum bicolor | sbi-miR166f | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | U | C | 21 | ||
| 11 | sbi-miR166k | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | U | 20 | |||
| sbi-miR166a-d,h-j | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | 20 | ||||
| sbi-miR166e,g | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | A | U | C | C | C | U | 21 | |||
| Zea mays | zma-miR166l,m | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | U | C | 21 | ||
| 14 | zma-miR166j,k,n | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | A | U | C | C | C | U | 21 | ||
| zma-miR166a | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | |||
| zma-miR166b-i | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | * | 20 | |||
| Class | Species | Polycistronic MIR166 | Location |
|---|---|---|---|
| Dicots | Brassica napus | bna-MIR166b,c | Scaffold2676:6222~6333 Scaffold2676:6215~6341 |
| Glycine max | gma-MIR166e,q | 4:46797931~46798040 4:46798188~46798339 | |
| Gossypium hirsutum | ghr-MIR166a,b | D12:41573882~41574028 D12:41573879~41574032 | |
| Medicago truncatula | mtr-MIR166c,d | 3:47901757~47901861 3:47901931~47902021 | |
| Monocots | Brachypodium distachyon | bdi-MIR166h,j | 3:57184726~57184865 3:57184616~57184767 |
| Oryza sativa | osa-MIR166i,j | 3:25294953~25295097 3:25294953~25295092 | |
| osa-MIR166d,h,k | 2:32435174~32435292 2:32435003~32435129 | ||
| Sorghum bicolor | sbi-MIR166d,f,g | 4:64857783~64857921 4:64857514~64857647 | |
| Zea mays | zma-MIR166k,m | 5:219021288~219021455 5:219021559~219021714 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Yang, T.; Li, N.; Tang, G.; Tang, J. MicroRNA166: Old Players and New Insights into Crop Agronomic Traits Improvement. Genes 2024, 15, 944. https://doi.org/10.3390/genes15070944
Zhang Z, Yang T, Li N, Tang G, Tang J. MicroRNA166: Old Players and New Insights into Crop Agronomic Traits Improvement. Genes. 2024; 15(7):944. https://doi.org/10.3390/genes15070944
Chicago/Turabian StyleZhang, Zhanhui, Tianxiao Yang, Na Li, Guiliang Tang, and Jihua Tang. 2024. "MicroRNA166: Old Players and New Insights into Crop Agronomic Traits Improvement" Genes 15, no. 7: 944. https://doi.org/10.3390/genes15070944
APA StyleZhang, Z., Yang, T., Li, N., Tang, G., & Tang, J. (2024). MicroRNA166: Old Players and New Insights into Crop Agronomic Traits Improvement. Genes, 15(7), 944. https://doi.org/10.3390/genes15070944






