Genome-Wide Identification and Functional Analysis of the Genes of the ATL Family in Maize during High-Temperature Stress in Maize
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Plasmid Construction and Plant Transformation
2.3. Identification of ZmATL Genes in Maize
2.4. Analysis of the Physicochemical Properties of Maize ATL Family Proteins
2.5. Analysis of Gene Structure and Promoter Conserved Motifs
2.6. High-Temperature, Drought-Stress, Salt-Stress Treatment
2.7. RNA Extraction and qRT-PCR Analysis
2.8. Abiotic Stress Responses in the ZmATL Family
2.9. Biological Stress Responses in the ZmATL Family
2.10. Tissue Expression Analysis of ZmATL Family Gene
2.11. Quantitation of Hydrogen Peroxide Radical
3. Results
3.1. Genome-Wide Identification of ZmATL Family Genes in Maize
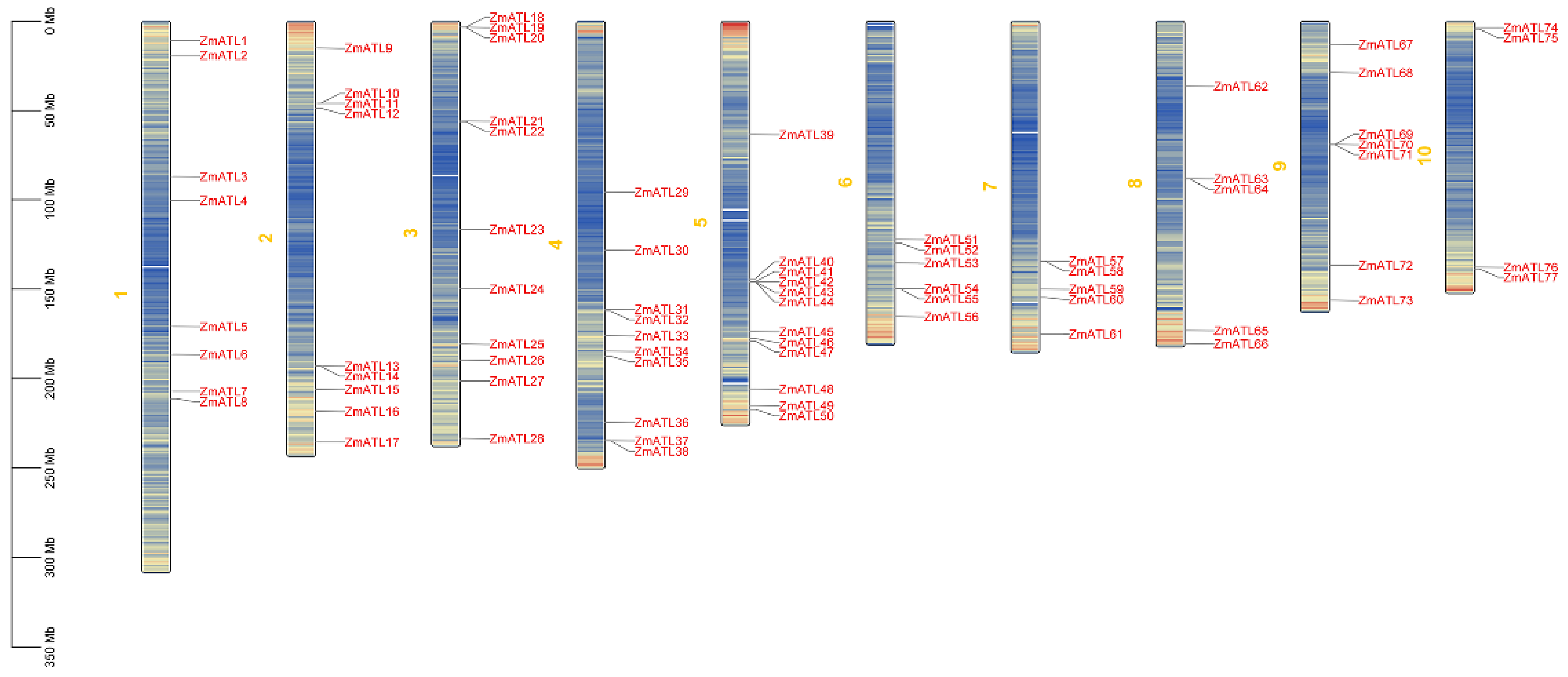
3.2. Positioning Analysis of ZmATL Family Members
3.3. Motif Composition and Gene Structure of ZmATL Genes
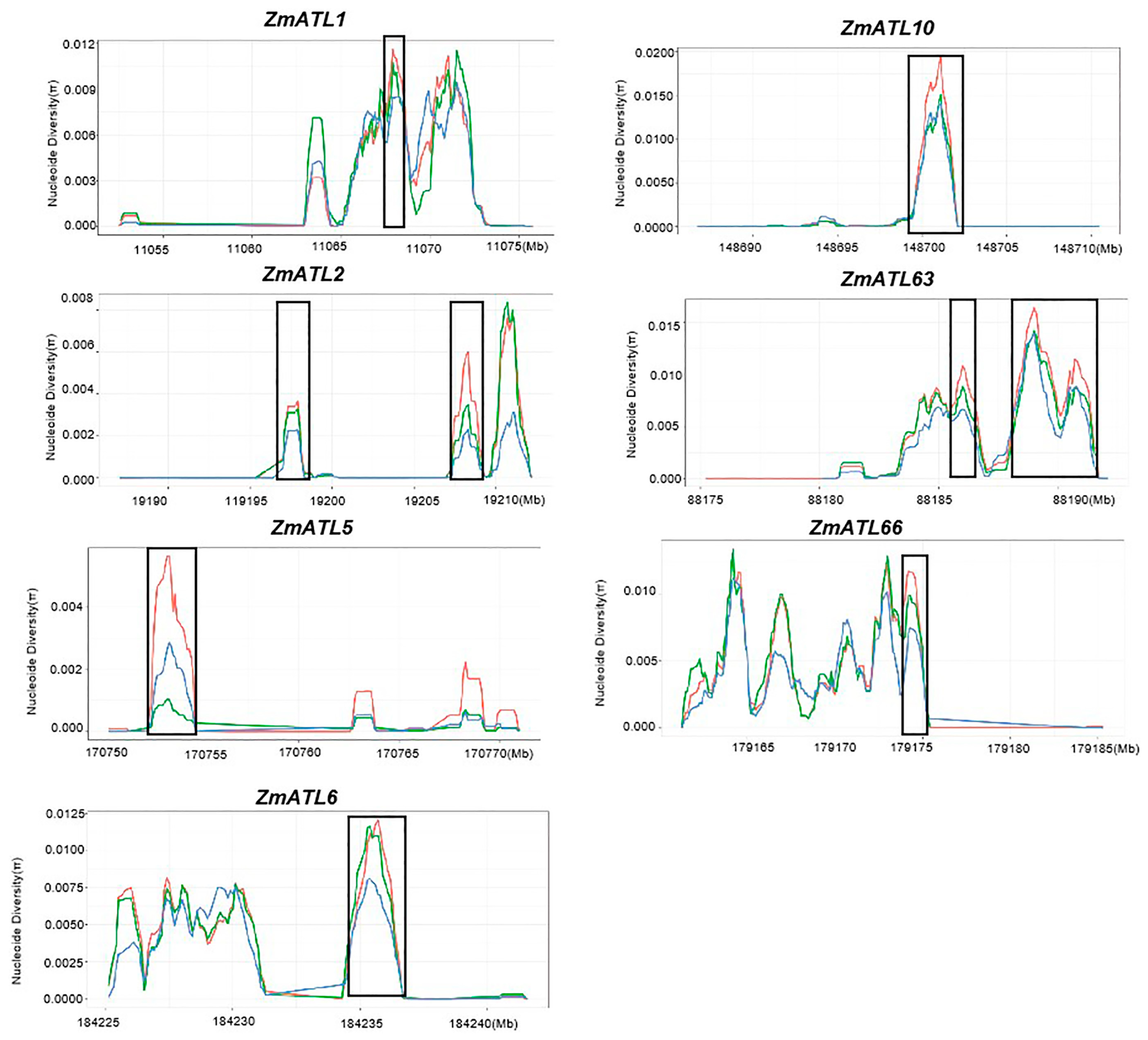
3.4. ZmATLs Underwent Selection during Maize Domestication and Improvement
3.5. Expression Patterns of the ZmATL Gene Family in Various Sugarcane Tissues
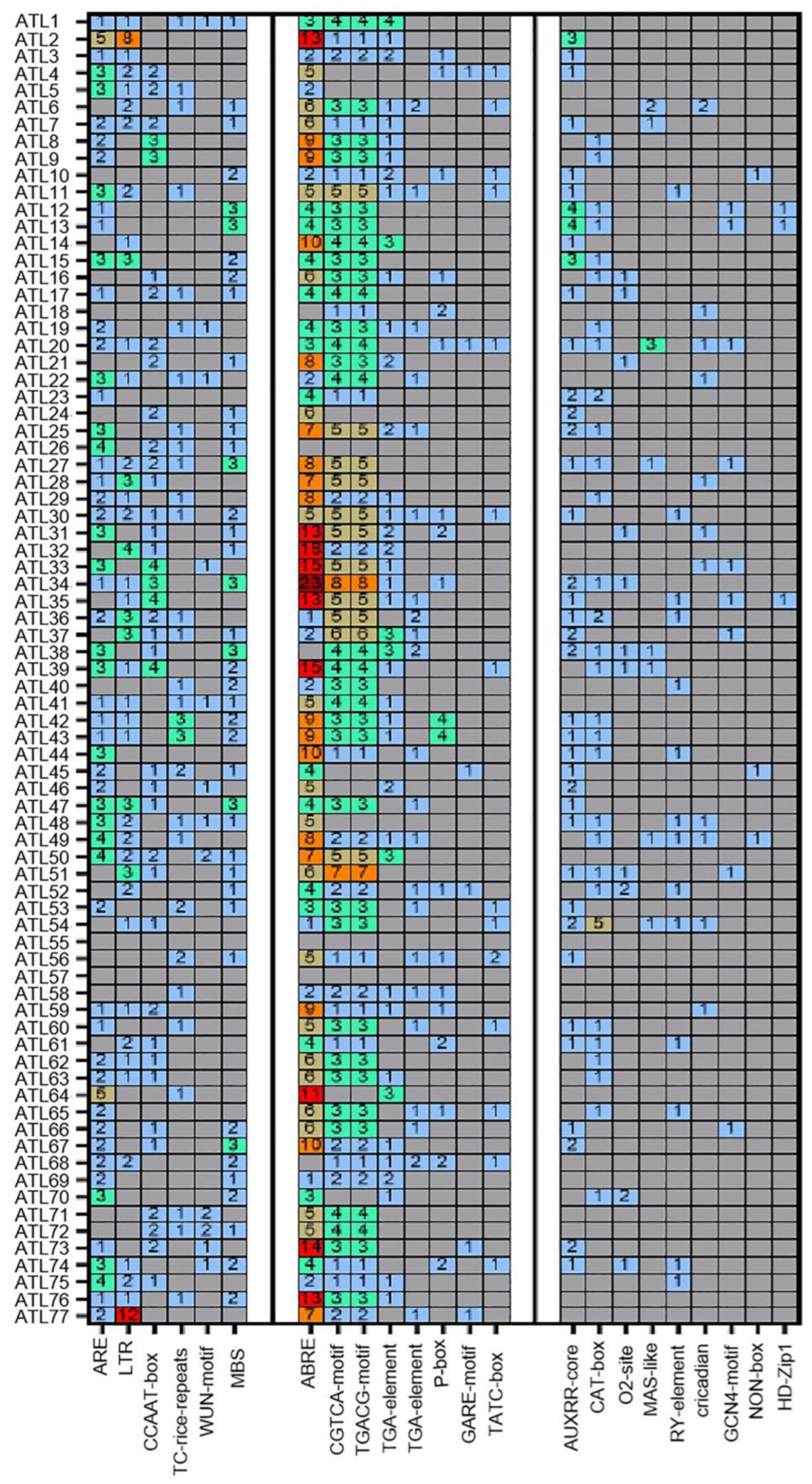
3.6. cis-Element Analysis of the ZmATL Genes in Maize
3.7. Abiotic and Biotic Stress Analysis of ZmATL Genes Family Members
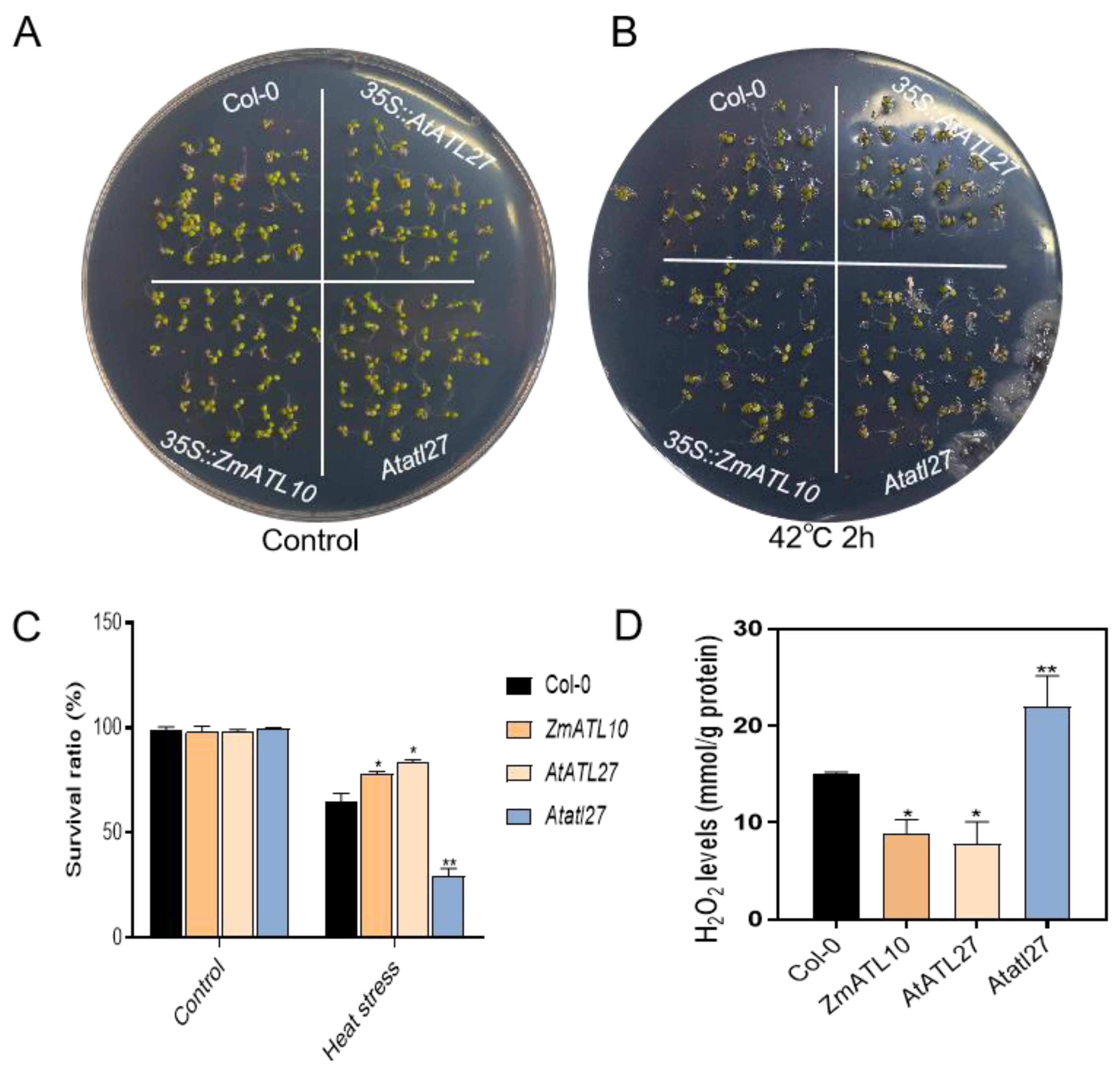
3.8. Functional Study of ZmATL10 and AtATL27 Resistance to High-Temperature Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Y.; Gu, J.-Y.; Wang, C.; Wang, W.-L.; Zhang, W.-Y.; Gu, J.-F.; Liu, L.-J.; Yang, J.-C.; Zhang, H. Carbon footprint of major grain crops in the middle and lower reaches of the Yangtze River during 2011–2020. Ying Yong Sheng Tai Xue Bao 2023, 34, 3364–3372. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hartmann, T.E.; Wang, X.; Cui, Z.; Hou, Y.; Meng, F.; Yu, X.; Wu, J.; Zhang, F. Phosphorus Flow Analysis in the Maize Based Food-Feed-Energy Systems in China. Environ. Res. 2020, 184, 109319. [Google Scholar] [CrossRef]
- Gao, C.; Tang, D.; Wang, W. The Role of Ubiquitination in Plant Immunity: Fine-Tuning Immune Signaling and Beyond. Plant Cell Physiol. 2022, 63, 1405–1413. [Google Scholar] [CrossRef]
- Li, Z.; Howell, S.H. Heat Stress Responses and Thermotolerance in Maize. Int. J. Mol. Sci. 2021, 22, 948. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Shen, Y.; Xing, Y.; Wang, Z.; Han, S.; Li, S.; Yang, C.; Hao, D.; Zhang, Y. QTL Mapping of Fusarium Ear Rot Resistance in Maize. Plant Dis. 2021, 105, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Colombini, F.d.S.; Ceccato-Antonini, S.R.; Rosa-Magri, M.M. Maize Treatment with Yeast Cells Induces Resistance against Fusarium Rot. Lett. Appl. Microbiol. 2023, 76, ovac072. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Nykiel, M.; Gietler, M.; Fidler, J.; Prabucka, B.; Labudda, M. Abiotic Stress Signaling and Responses in Plants. Plants 2023, 12, 3405. [Google Scholar] [CrossRef]
- Mei, F.; Chen, B.; Du, L.; Li, S.; Zhu, D.; Chen, N.; Zhang, Y.; Li, F.; Wang, Z.; Cheng, X.; et al. A Gain-of-Function Allele of a DREB Transcription Factor Gene Ameliorates Drought Tolerance in Wheat. Plant Cell 2022, 34, 4472–4494. [Google Scholar] [CrossRef]
- Hu, W.; Yan, Y.; Shi, H.; Liu, J.; Miao, H.; Tie, W.; Ding, Z.; Ding, X.; Wu, C.; Liu, Y.; et al. The Core Regulatory Network of the Abscisic Acid Pathway in Banana: Genome-Wide Identification and Expression Analyses during Development, Ripening, and Abiotic Stress. BMC Plant Biol. 2017, 17, 145. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB Transcription Factors in Abiotic and Biotic Stress Tolerance in Plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA Co-Receptor ABI1 by PUB12/13 U-Box E3 Ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant Hormone Regulation of Abiotic Stress Responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Qiao, Z.; Li, Y.; Yang, Z.; Wang, C.; Zhang, Y.; Liu, L.; Wang, B. RING Zinc Finger Proteins in Plant Abiotic Stress Tolerance. Front. Plant Sci. 2022, 13, 877011. [Google Scholar] [CrossRef]
- Haakonsen, D.L.; Heider, M.; Ingersoll, A.J.; Vodehnal, K.; Witus, S.R.; Uenaka, T.; Wernig, M.; Rapé, M. Stress Response Silencing by an E3 Ligase Mutated in Neurodegeneration. Nature 2024, 626, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Saxena, H.; Negi, H.; Sharma, B. Role of F-Box E3-Ubiquitin Ligases in Plant Development and Stress Responses. Plant Cell Rep. 2023, 42, 1133–1146. [Google Scholar] [CrossRef]
- Lau, O.S.; Deng, X.W. Effect of Arabidopsis COP10 Ubiquitin E2 Enhancement Activity across E2 Families and Functional Conservation among Its Canonical Homologues. Biochem. J. 2009, 418, 683–690. [Google Scholar] [CrossRef]
- Ban, Z.; Estelle, M. CUL3 E3 Ligases in Plant Development and Environmental Response. Nat. Plants 2021, 7, 6–16. [Google Scholar] [CrossRef]
- Cho, S.K.; Ryu, M.Y.; Kim, J.H.; Hong, J.S.; Oh, T.R.; Kim, W.T.; Yang, S.W. RING E3 Ligases: Key Regulatory Elements Are Involved in Abiotic Stress Responses in Plants. BMB Rep. 2017, 50, 393–400. [Google Scholar] [CrossRef]
- Al-Saharin, R.; Hellmann, H.; Mooney, S. Plant E3 Ligases and Their Role in Abiotic Stress Response. Cells 2022, 11, 890. [Google Scholar] [CrossRef]
- Sun, J.; Sun, Y.; Ahmed, R.I.; Ren, A.; Xie, M. Research Progress on Plant RING-Finger Proteins. Genes 2019, 10, 973. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-López, D.; Muñóz-Belman, F.; González-Prieto, J.M.; Aguilar-Hernández, V.; Guzmán, P. Repertoire of Plant RING E3 Ubiquitin Ligases Revisited: New Groups Counting Gene Families and Single Genes. PLoS ONE 2018, 13, e0203442. [Google Scholar] [CrossRef] [PubMed]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The Ubiquitin-Proteasome System: Central Modifier of Plant Signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Xiong, L.; Gong, Z.; Ishitani, M.; Stevenson, B.; Zhu, J.-K. The Arabidopsis HOS1 Gene Negatively Regulates Cold Signal Transduction and Encodes a RING Finger Protein That Displays Cold-Regulated Nucleo–Cytoplasmic Partitioning. Genes Dev. 2001, 15, 912–924. [Google Scholar] [CrossRef]
- Lan, W.; Miao, Y. New Aspects of HECT-E3 Ligases in Cell Senescence and Cell Death of Plants. Plants 2019, 8, 483. [Google Scholar] [CrossRef]
- Yang, L.; Wu, L.; Chang, W.; Li, Z.; Miao, M.; Li, Y.; Yang, J.; Liu, Z.; Tan, J. Overexpression of the Maize E3 Ubiquitin Ligase Gene ZmAIRP4 Enhances Drought Stress Tolerance in Arabidopsis. Plant Physiol. Biochem. 2018, 123, 34–42. [Google Scholar] [CrossRef]
- Xia, Z.; Liu, Q.; Wu, J.; Ding, J. ZmRFP1, the Putative Ortholog of SDIR1, Encodes a RING-H2 E3 Ubiquitin Ligase and Responds to Drought Stress in an ABA-Dependent Manner in Maize. Gene 2012, 495, 146–153. [Google Scholar] [CrossRef]
- Brugière, N.; Zhang, W.; Xu, Q.; Scolaro, E.J.; Lu, C.; Kahsay, R.Y.; Kise, R.; Trecker, L.; Williams, R.W.; Hakimi, S.; et al. Overexpression of RING Domain E3 Ligase ZmXerico1 Confers Drought Tolerance through Regulation of ABA Homeostasis. Plant Physiol. 2017, 175, 1350–1369. [Google Scholar] [CrossRef]
- Davis, J.G.; Hamuro, J.; Shim, C.Y.; Samanta, A.; Greene, M.I.; Dobashi, K. Isolation and Characterization of a Neu Protein-Specific Activating Factor from Human ATL-2 Cell Conditioned Medium. Biochem. Biophys. Res. Commun. 1991, 179, 1536–1542. [Google Scholar] [CrossRef]
- Serrano, M.; Guzmán, P. Isolation and Gene Expression Analysis of Arabidopsis Thaliana Mutants with Constitutive Expression of ATL2, an Early Elicitor-Response RING-H2 Zinc-Finger Gene. Genetics 2004, 167, 919–929. [Google Scholar] [CrossRef]
- Liu, S.; Wang, K.; Li, J.; Liu, Y.; Zhang, Z.; Meng, D. MiR-30e-5p Deficiency Exerts an Inhibitory Effect on Inflammation in Rheumatoid Arthritis via Regulating Atl2 Expression. Arch. Rheumatol. 2023, 38, 119–128. [Google Scholar] [CrossRef]
- Reynisdottir, I.; Arason, A.; Freysteinsdottir, E.S.; Kristjansdottir, S.B.; Hilmarsdottir, B.; Traustadottir, G.A.; Johannsson, O.T.; Agnarsson, B.A.; Barkardottir, R.B. High Atlastin 2-2 (ATL2-2) Expression Associates with Worse Prognosis in Estrogen-Receptor-Positive Breast Cancer. Genes 2023, 14, 1559. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Jeon, S.J.; Hong, J.K.; Kim, M.G.; Kim, S.H.; Kadam, U.S.; Kim, W.-Y.; Chung, W.S.; Stacey, G.; Hong, J.C. The Auto-Regulation of ATL2 E3 Ubiquitin Ligase Plays an Important Role in the Immune Response against Alternaria Brassicicola in Arabidopsis Thaliana. Int. J. Mol. Sci. 2024, 25, 2388. [Google Scholar] [CrossRef] [PubMed]
- Ariani, P.; Regaiolo, A.; Lovato, A.; Giorgetti, A.; Porceddu, A.; Camiolo, S.; Wong, D.; Castellarin, S.; Vandelle, E.; Polverari, A. Genome-Wide Characterisation and Expression Profile of the Grapevine ATL Ubiquitin Ligase Family Reveal Biotic and Abiotic Stress-Responsive and Development-Related Members. Sci. Rep. 2016, 6, 38260. [Google Scholar] [CrossRef]
- Qanmber, G.; Yu, D.; Li, J.; Wang, L.; Ma, S.; Lu, L.; Yang, Z.; Li, F. Genome-Wide Identification and Expression Analysis of Gossypium RING-H2 Finger E3 Ligase Genes Revealed Their Roles in Fiber Development, and Phytohormone and Abiotic Stress Responses. J. Cotton Res. 2018, 1, 1. [Google Scholar] [CrossRef]
- Cui, L.H.; Min, H.J.; Yu, S.G.; Byun, M.Y.; Oh, T.R.; Lee, A.; Yang, H.W.; Kim, W.T. OsATL38 Mediates Mono-Ubiquitination of the 14-3-3 Protein OsGF14d and Negatively Regulates the Cold Stress Response in Rice. J. Exp. Bot. 2022, 73, 307–323. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, W.T. Suppression of Arabidopsis RING E3 Ubiquitin Ligase AtATL78 Increases Tolerance to Cold Stress and Decreases Tolerance to Drought Stress. FEBS Lett. 2013, 587, 2584–2590. [Google Scholar] [CrossRef]
- Suh, J.Y.; Kim, S.J.; Oh, T.R.; Cho, S.K.; Yang, S.W.; Kim, W.T. Arabidopsis Tóxicos En Levadura 78 (AtATL78) Mediates ABA-Dependent ROS Signaling in Response to Drought Stress. Biochem. Biophys. Res. Commun. 2016, 469, 8–14. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, X.-H.; Guo, Q.-H.; Liu, P.; Li, Y.; Wu, C.-A.; Yang, G.-D.; Huang, J.-G.; Zhang, S.-Z.; Zheng, C.-C.; et al. Salt Responsive Alternative Splicing of a RING Finger E3 Ligase Modulates the Salt Stress Tolerance by Fine-Tuning the Balance of COP9 Signalosome Subunit 5A. PLoS Genet. 2021, 17, e1009898. [Google Scholar] [CrossRef]
- Wu, M.; Musazade, E.; Yang, X.; Yin, L.; Zhao, Z.; Zhang, Y.; Lu, J.; Guo, L. ATL Protein Family: Novel Regulators in Plant Response to Environmental Stresses. J. Agric. Food Chem. 2023, 71, 20419–20440. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Xia, R. A Painless Way to Customize Circos Plot: From Data Preparation to Visualization Using TBtools. Imeta 2022, 1, e35. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Dong, T.; Park, Y.; Hwang, I. Abscisic Acid: Biosynthesis, Inactivation, Homoeostasis and Signalling. Essays Biochem. 2015, 58, 29–48. [Google Scholar] [CrossRef]
- Zhu, M.; Yan, B.; Hu, Y.; Cui, Z.; Wang, X. Genome-Wide Identification and Phylogenetic Analysis of Rice FTIP Gene Family. Genomics 2020, 112, 3803–3814. [Google Scholar] [CrossRef]
- Serrano, M.; Parra, S.; Alcaraz, L.D.; Guzmán, P. The ATL Gene Family from Arabidopsis Thaliana and Oryza Sativa Comprises a Large Number of Putative Ubiquitin Ligases of the RING-H2 Type. J. Mol. Evol. 2006, 62, 434–445. [Google Scholar] [CrossRef]
- Du, M.; Lu, D.; Liu, X. The Arabidopsis Ubiquitin Ligases ATL31 and ATL6 Regulate Plant Response to Salt Stress in an ABA-Independent Manner. Biochem. Biophys. Res. Commun. 2023, 685, 149156. [Google Scholar] [CrossRef]
- Guzmán, P. The Prolific ATL Family of RING-H2 Ubiquitin Ligases. Plant Signal. Behav. 2012, 7, 1014–1021. [Google Scholar] [CrossRef]
- Aguilar-Hernández, V.; Guzmán, P. The Fate of Tandemly Duplicated Genes Assessed by the Expression Analysis of a Group of Arabidopsis Thaliana RING-H2 Ubiquitin Ligase Genes of the ATL Family. Plant Mol. Biol. 2014, 84, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Dyaa, A.; Soliman, H.; Abdelrazak, A.; Samra, B.N.; Khojah, E.; Ahmed, A.F.; El-Esawi, M.A.; Elsayed, A. Optimization of Carotenoids Production from Rhodotorula Sp. Strain ATL72 for Enhancing Its Biotechnological Applications. J. Fungi 2022, 8, 160. [Google Scholar] [CrossRef]
- Shu, K.; Yang, W. E3 Ubiquitin Ligases: Ubiquitous Actors in Plant Development and Abiotic Stress Responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant Hormone-Mediated Regulation of Stress Responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Li, Z.; Shi, Y.; Fu, D.; Yin, P.; Cheng, J.; Jiang, C.; Yang, S. Natural Variation in a Type-A Response Regulator Confers Maize Chilling Tolerance. Nat. Commun. 2021, 12, 4713. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, W.T. The Arabidopsis RING E3 Ubiquitin Ligase AtAIRP3/LOG2 Participates in Positive Regulation of High-Salt and Drought Stress Responses. Plant Physiol. 2013, 162, 1733–1749. [Google Scholar] [CrossRef] [PubMed]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant. Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Wright, S.I.; Bi, I.V.; Schroeder, S.G.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S. The Effects of Artificial Selection on the Maize Genome. Science 2005, 308, 1310–1314. [Google Scholar] [CrossRef]
- Serrano, I.; Gu, Y.; Qi, D.; Dubiella, U.; Innes, R.W. The Arabidopsis EDR1 Protein Kinase Negatively Regulates the ATL1 E3 Ubiquitin Ligase to Suppress Cell Death. Plant Cell 2014, 26, 4532–4546. [Google Scholar] [CrossRef]
- He, W.; Wang, R.; Zhang, Q.; Fan, M.; Lyu, Y.; Chen, S.; Chen, D.; Chen, X. E3 Ligase ATL5 Positively Regulates Seed Longevity by Mediating the Degradation of ABT1 in Arabidopsis. New Phytol. 2023, 239, 1754–1770. [Google Scholar] [CrossRef]
- Maekawa, S.; Sato, T.; Asada, Y.; Yasuda, S.; Yoshida, M.; Chiba, Y.; Yamaguchi, J. The Arabidopsis Ubiquitin Ligases ATL31 and ATL6 Control the Defense Response as Well as the Carbon/Nitrogen Response. Plant Mol. Biol. 2012, 79, 217–227. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Y.; Du, M.; Liang, X.; Fan, F.; Huang, G.; Zou, Y.; Bai, J.; Lu, D. The Calcium-Dependent Protein Kinase CPK28 Is Targeted by the Ubiquitin Ligases ATL31 and ATL6 for Proteasome-Mediated Degradation to Fine-Tune Immune Signaling in Arabidopsis. Plant Cell 2022, 34, 679–697. [Google Scholar] [CrossRef]
- Noda, S.; Takahashi, Y.; Tsurumaki, Y.; Yamamura, M.; Nishikubo, N.; Yamaguchi, M.; Sakurai, N.; Hattori, T.; Suzuki, H.; Demura, T.; et al. ATL54, a RING-H2 Domain Protein Selected by a Gene Co-Expression Network Analysis, Is Associated with Secondary Cell Wall Formation in Arabidopsis. Plant Biotechnol. 2013, 30, 169–177. [Google Scholar] [CrossRef]
- Jiménez-Morales, E.; Aguilar-Hernández, V.; Aguilar-Henonin, L.; Guzmán, P. Molecular Basis for Neofunctionalization of Duplicated E3 Ubiquitin Ligases Underlying Adaptation to Drought Tolerance in Arabidopsis Thaliana. Plant J. 2020, 104, 474–492. [Google Scholar] [CrossRef]





| Gene Name | Gene ID | CDS (bp) | AA | MV (Da) | PI | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| ZmATL1 | Zm00001eb004000 | 1701 | 302 | 31,455.85 | 8.03 | 0.092 | endosome |
| ZmATL2 | Zm00001eb006670 | 1694 | 362 | 38,658.42 | 6.76 | −0.349 | membrane |
| ZmATL3 | Zm00001eb022630 | 1279 | 230 | 23,747.31 | 6.76 | 0.425 | membrane |
| ZmATL4 | Zm00001eb024440 | 1093 | 232 | 24,741.12 | 7.19 | −0.024 | membrane |
| ZmATL5 | Zm00001eb030780 | 2202 | 398 | 42,205.35 | 10.74 | −0.125 | membrane |
| ZmATL6 | Zm00001eb033500 | 1383 | 377 | 40,128.52 | 9.04 | −0.154 | endosome |
| ZmATL7 | Zm00001eb038810 | 960 | 319 | 33,829.16 | 7.19 | −0.118 | membrane |
| ZmATL8 | Zm00001eb039970 | 1226 | 239 | 24,284.78 | 4.81 | 0.458 | membrane |
| ZmATL9 | Zm00001eb072750 | 1222 | 173 | 18,512.32 | 7.99 | −0.07 | membrane |
| ZmATL10 | Zm00001eb081320 | 3022 | 419 | 43,467.62 | 6.73 | −0.388 | membrane |
| ZmATL11 | Zm00001eb081330 | 2038 | 388 | 40,688.78 | 8.9 | −0.301 | membrane |
| ZmATL12 | Zm00001eb081980 | 1442 | 289 | 29,534.54 | 8.71 | −0.025 | membrane |
| ZmATL13 | Zm00001eb101180 | 1570 | 393 | 41,084.32 | 9.52 | −0.197 | membrane |
| ZmATL14 | Zm00001eb101200 | 1739 | 357 | 38,217.32 | 5.9 | −0.154 | membrane |
| ZmATL15 | Zm00001eb104970 | 1280 | 246 | 26,290.32 | 7.98 | 0.283 | endosome |
| ZmATL16 | Zm00001eb109400 | 1182 | 252 | 25,968.3 | 6.06 | −0.129 | endosome |
| ZmATL17 | Zm00001eb115110 | 1791 | 390 | 40,387.1 | 9.26 | −0.303 | membrane |
| ZmATL18 | Zm00001eb120050 | 1395 | 348 | 36,805.09 | 10.36 | −0.052 | membrane |
| ZmATL19 | Zm00001eb120060 | 1212 | 214 | 22,058.23 | 8.32 | 0.176 | membrane |
| ZmATL20 | Zm00001eb120080 | 892 | 176 | 18,341.22 | 8.99 | 0.242 | endosome |
| ZmATL21 | Zm00001eb130410 | 1169 | 254 | 26,296.79 | 6.31 | −0.07 | endosome |
| ZmATL22 | Zm00001eb130430 | 963 | 151 | 15,987.74 | 6.49 | −0.187 | endosome |
| ZmATL23 | Zm00001eb135320 | 2923 | 414 | 43,420.32 | 11 | −0.19 | membrane |
| ZmATL24 | Zm00001eb140640 | 1405 | 283 | 30,593.24 | 5.69 | −0.417 | membrane |
| ZmATL25 | Zm00001eb146800 | 675 | 224 | 23,323.6 | 8.72 | 0.013 | membrane |
| ZmATL26 | Zm00001eb149500 | 1126 | 191 | 19,386.2 | 8.48 | 0.232 | endosome |
| ZmATL27 | Zm00001eb152830 | 1149 | 241 | 24,828.17 | 8.14 | 0.005 | membrane |
| ZmATL28 | Zm00001eb162790 | 1636 | 374 | 42,236 | 6.63 | 0.133 | membrane |
| ZmATL29 | Zm00001eb180470 | 1207 | 279 | 29,126.6 | 4.99 | 0.466 | membrane |
| ZmATL30 | Zm00001eb182770 | 1595 | 331 | 35,780.18 | 5.3 | −0.343 | membrane |
| ZmATL31 | Zm00001eb187290 | 1326 | 220 | 23,625.18 | 5.39 | 0.092 | endosome |
| ZmATL32 | Zm00001eb187620 | 945 | 186 | 19,311.48 | 8.61 | 0.206 | membrane |
| ZmATL33 | Zm00001eb191210 | 1588 | 310 | 32,599.92 | 5.26 | −0.123 | endosome |
| ZmATL34 | Zm00001eb193860 | 1610 | 415 | 45,209.22 | 9.49 | −0.324 | endosome |
| ZmATL35 | Zm00001eb194620 | 1226 | 230 | 24,275.47 | 5.28 | 0.123 | membrane |
| ZmATL36 | Zm00001eb202890 | 1938 | 267 | 27,958.04 | 5.58 | 0.316 | membrane |
| ZmATL37 | Zm00001eb204430 | 441 | 146 | 15,227.8 | 8.78 | 0.589 | membrane |
| ZmATL38 | Zm00001eb204450 | 666 | 221 | 23,063.7 | 6.86 | 0.295 | membrane |
| ZmATL39 | Zm00001eb228180 | 1554 | 262 | 28,113.4 | 9.32 | 0.091 | endosome |
| ZmATL40 | Zm00001eb238220 | 1058 | 210 | 21,367.65 | 6.5 | 0.276 | membrane |
| ZmATL41 | Zm00001eb238400 | 776 | 152 | 15,603 | 8.03 | 0.363 | membrane |
| ZmATL42 | Zm00001eb238420 | 645 | 182 | 19,124.97 | 8.29 | 0.151 | membrane |
| ZmATL43 | Zm00001eb238430 | 579 | 192 | 19,995.74 | 6.87 | 0.114 | membrane |
| ZmATL44 | Zm00001eb238440 | 652 | 201 | 20,704.2 | 6.63 | 0.532 | membrane |
| ZmATL45 | Zm00001eb243170 | 1019 | 226 | 23,412.03 | 8.97 | 0.13 | membrane |
| ZmATL46 | Zm00001eb243960 | 1083 | 297 | 30,661.64 | 6.21 | −0.041 | membrane |
| ZmATL47 | Zm00001eb244640 | 1630 | 347 | 36,916.78 | 5.85 | −0.103 | membrane |
| ZmATL48 | Zm00001eb250960 | 894 | 185 | 19,146.14 | 8.45 | 0.189 | membrane |
| ZmATL49 | Zm00001eb254430 | 1586 | 353 | 38,628.32 | 10.24 | −0.197 | membrane |
| ZmATL50 | Zm00001eb255510 | 2791 | 419 | 44,824.49 | 9.33 | −0.289 | endosome |
| ZmATL51 | Zm00001eb279270 | 2018 | 421 | 45,215.67 | 5.73 | −0.438 | endosome |
| ZmATL52 | Zm00001eb279770 | 1127 | 218 | 23,139.15 | 5.03 | 0.04 | endosome |
| ZmATL53 | Zm00001eb282260 | 869 | 182 | 19,018.83 | 5.07 | 0.324 | endosome |
| ZmATL54 | Zm00001eb285770 | 1322 | 198 | 20,760.37 | 6.29 | −0.045 | membrane |
| ZmATL55 | Zm00001eb285790 | 1402 | 198 | 20,760.37 | 6.29 | −0.073 | membrane |
| ZmATL56 | Zm00001eb290630 | 1701 | 206 | 20,530.3 | 6.12 | 0.211 | membrane |
| ZmATL57 | Zm00001eb315600 | 1565 | 404 | 42,765.13 | 9.01 | −0.298 | endosome |
| ZmATL58 | Zm00001eb315620 | 1665 | 361 | 38,460.51 | 5.91 | −0.174 | membrane |
| ZmATL59 | Zm00001eb319810 | 1206 | 191 | 19,657.09 | 5.09 | −0.122 | membrane |
| ZmATL60 | Zm00001eb321320 | 1274 | 246 | 26,371.41 | 8.26 | 0.274 | endosome |
| ZmATL61 | Zm00001eb327570 | 858 | 271 | 28,116.63 | 4.98 | −0.176 | membrane |
| ZmATL62 | Zm00001eb340160 | 970 | 251 | 25,820.13 | 6.07 | −0.123 | membrane |
| ZmATL63 | Zm00001eb346450 | 1091 | 202 | 21,313.86 | 6.74 | −0.199 | membrane |
| ZmATL64 | Zm00001eb346460 | 603 | 200 | 21,186.75 | 4.88 | −0.009 | endosome |
| ZmATL65 | Zm00001eb366740 | 993 | 219 | 22,852.13 | 7.59 | 0.113 | endosome |
| ZmATL66 | Zm00001eb370730 | 1154 | 237 | 24,601.94 | 8.16 | 0.004 | endosome |
| ZmATL67 | Zm00001eb374170 | 1525 | 331 | 34,576.53 | 8.95 | −0.045 | membrane |
| ZmATL68 | Zm00001eb379190 | 1152 | 154 | 16,678.46 | 8.46 | −0.001 | endosome |
| ZmATL69 | Zm00001eb383450 | 797 | 224 | 23,890.18 | 8.51 | 0.391 | endosome |
| ZmATL70 | Zm00001eb383460 | 1227 | 216 | 22,714.36 | 6.35 | 0.317 | membrane |
| ZmATL71 | Zm00001eb383470 | 725 | 174 | 18,105.23 | 8.14 | 0.591 | endosome |
| ZmATL72 | Zm00001eb395020 | 924 | 166 | 17,299.07 | 7.54 | 0.446 | membrane |
| ZmATL73 | Zm00001eb401490 | 1696 | 380 | 40,216.01 | 6.16 | −0.318 | membrane |
| ZmATL74 | Zm00001eb406080 | 1084 | 207 | 21,246.06 | 4.85 | 0.173 | membrane |
| ZmATL75 | Zm00001eb406570 | 3407 | 279 | 29,411.12 | 7 | −0.314 | endosome |
| ZmATL76 | Zm00001eb428110 | 1070 | 200 | 20,678.66 | 8.49 | −0.003 | membrane |
| ZmATL77 | Zm00001eb428450 | 1034 | 187 | 20,404.67 | 5.57 | 0.119 | endosome |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, H.; Li, X.; Zhuge, S.; Du, J.; Wu, M.; Li, W.; Li, Y.; Ma, H.; Zhang, P.; Wang, X.; et al. Genome-Wide Identification and Functional Analysis of the Genes of the ATL Family in Maize during High-Temperature Stress in Maize. Genes 2024, 15, 1106. https://doi.org/10.3390/genes15081106
Ding H, Li X, Zhuge S, Du J, Wu M, Li W, Li Y, Ma H, Zhang P, Wang X, et al. Genome-Wide Identification and Functional Analysis of the Genes of the ATL Family in Maize during High-Temperature Stress in Maize. Genes. 2024; 15(8):1106. https://doi.org/10.3390/genes15081106
Chicago/Turabian StyleDing, Haiping, Xiaohu Li, Shilin Zhuge, Jiyuan Du, Min Wu, Wenlong Li, Yujing Li, Haoran Ma, Peng Zhang, Xingyu Wang, and et al. 2024. "Genome-Wide Identification and Functional Analysis of the Genes of the ATL Family in Maize during High-Temperature Stress in Maize" Genes 15, no. 8: 1106. https://doi.org/10.3390/genes15081106






