Genetic Evidence of SpGH9A3 in Leaf Morphology Variation of Spathiphyllum ‘Mojo’
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Morphological Traits of Leaves Measurement
2.3. Determination of Leaf Cellulose Content and Cellulase Activity
2.4. Analysis of Differential Gene and Quantitative Real-Time PCR Assays
2.5. Correlation Analysis
2.6. Sequence Analysis
2.7. Subcellular Localization
2.8. Phenotypic Analysis of Transgenic Plants
2.9. Statistical Analysis
3. Results
3.1. Comparison of the Leaf Morphologies of Spathiphyllum ‘Mojo’ (Wild) and ‘Mojo’-Ssm-1 (Mutant)
3.2. Comparison of Cellulose Content and Cellulase Activity
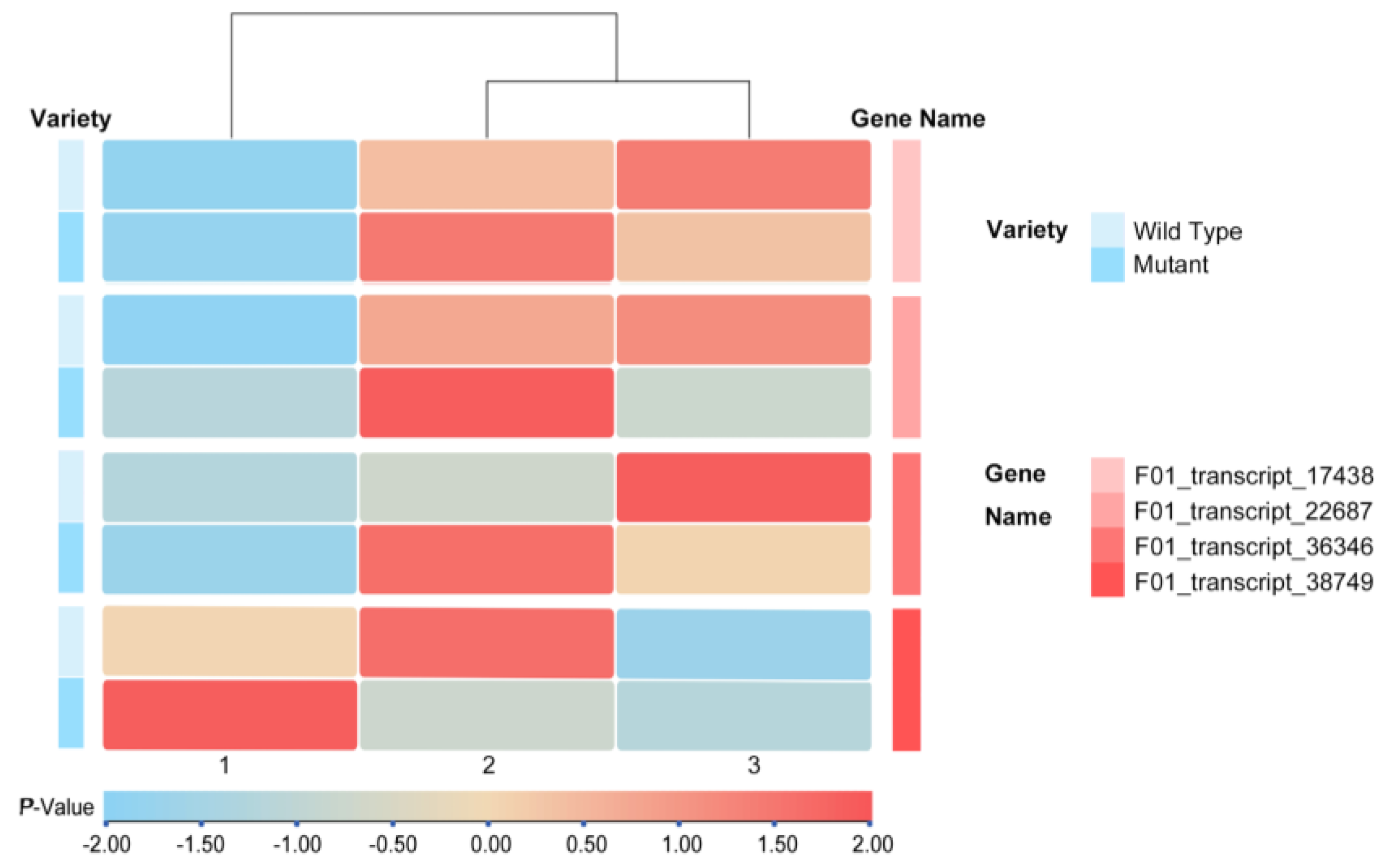
3.3. Screening of Differential Genes for Leaf Morphology
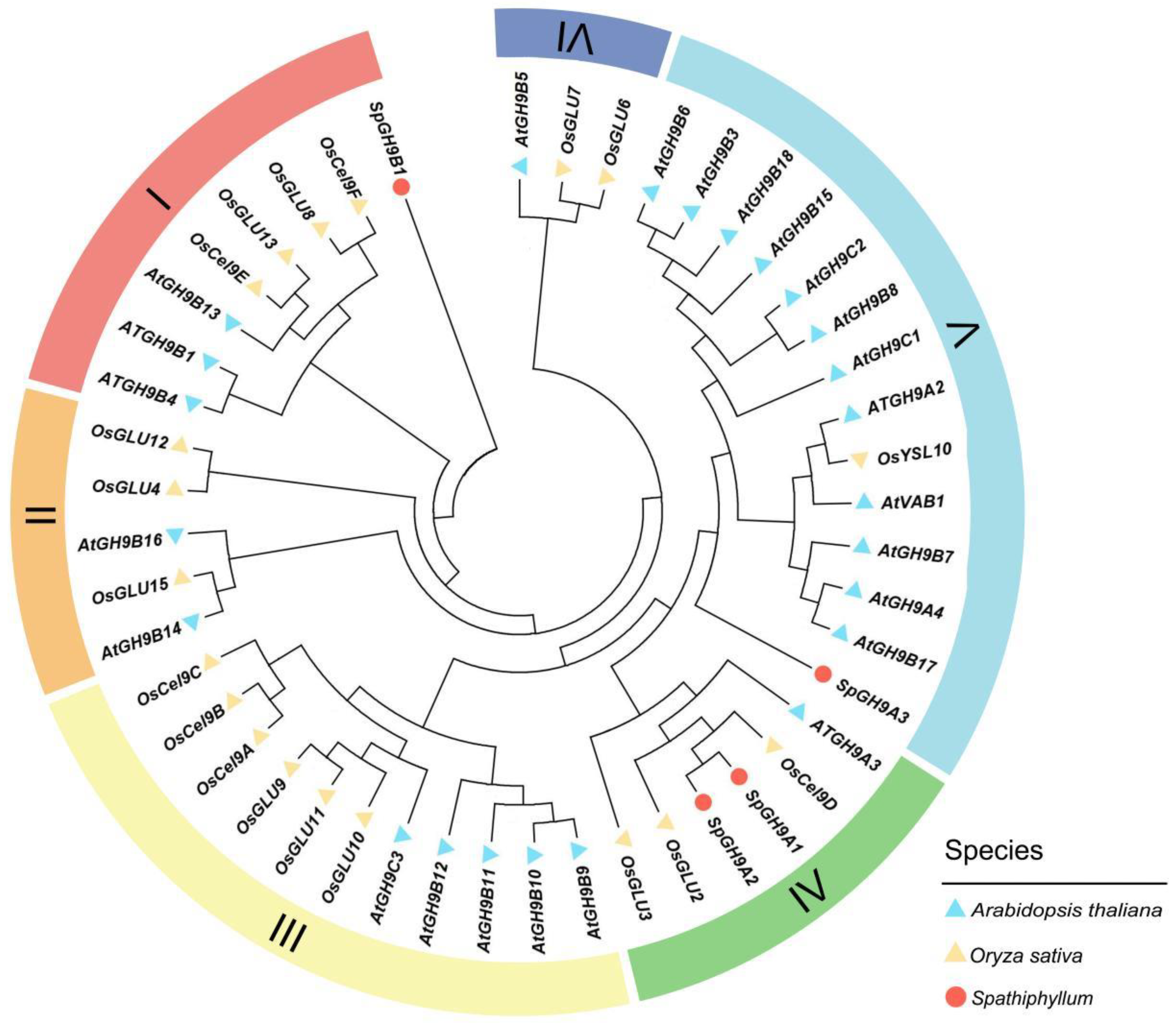
3.4. Bioinformatics Study of SpGH9A3 in Spathiphyllum
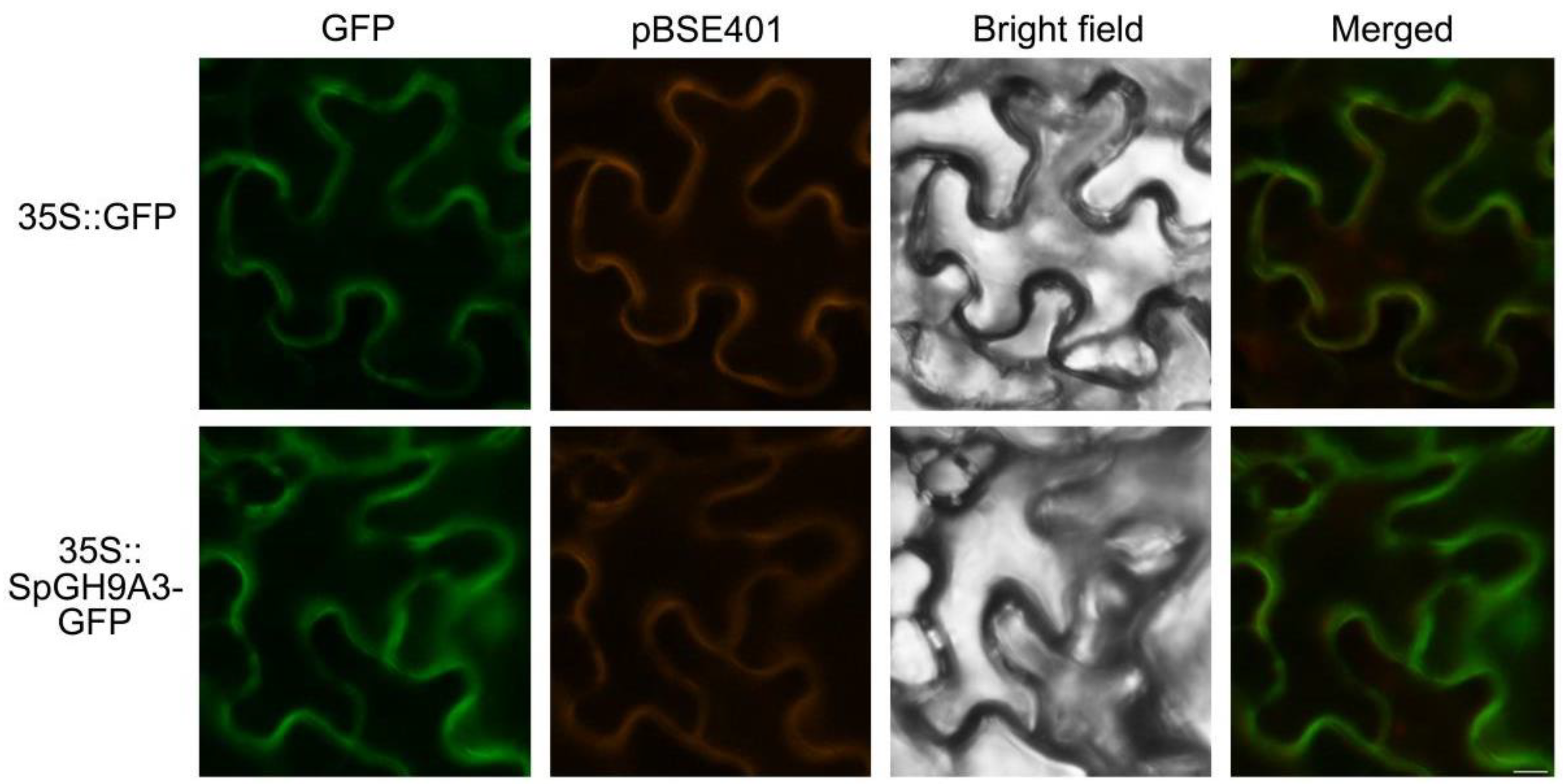
3.5. SpGH9A3 Protein Is Localized to the Cell Membrane
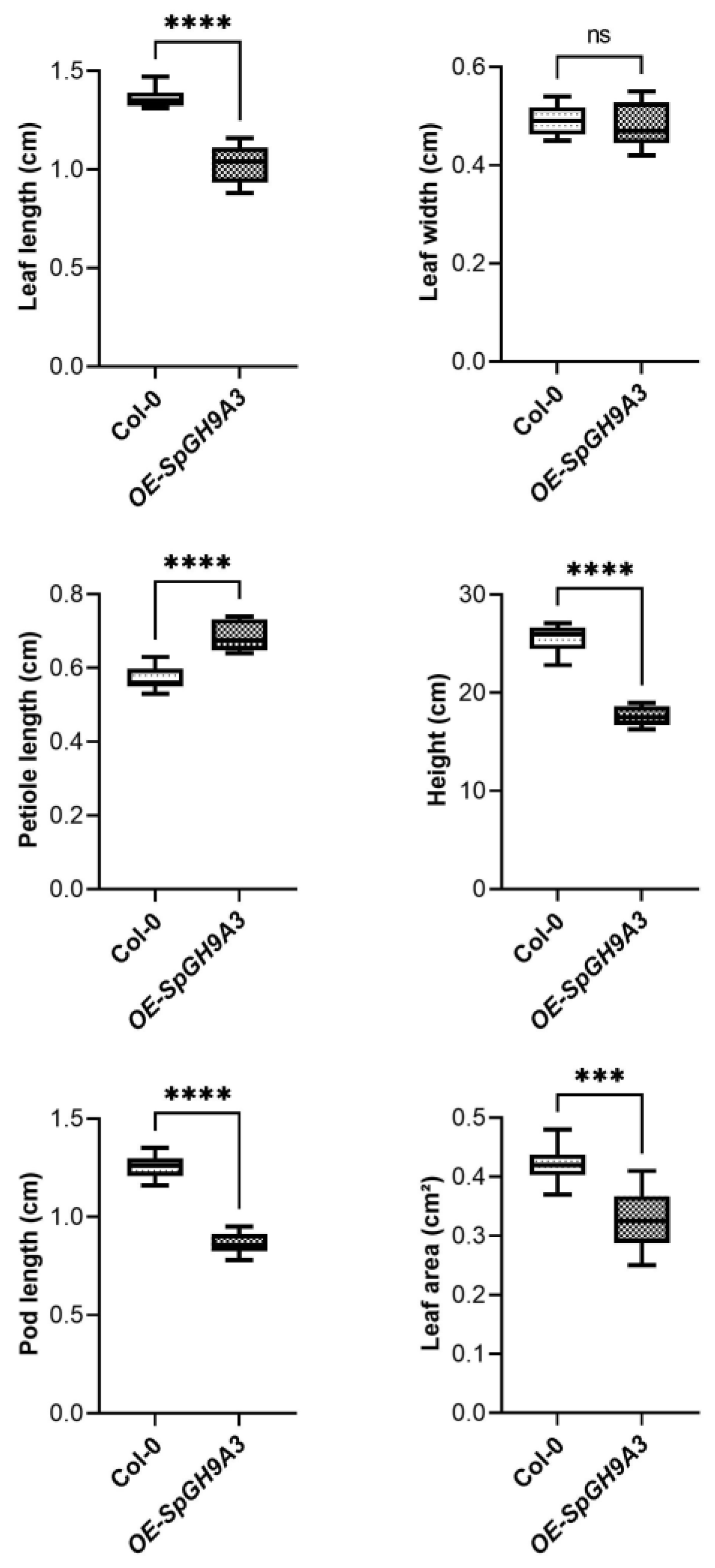
3.6. Overexpressing SpGH9A3 Changed the Leaf Morphology and Decreased Cellulose Content
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, P.; He, Y.L.; Xiao, L.; Quan, M.Y.; Gu, M.Y.; Jin, Z.Y.; Zhou, J.X.; Li, L.Z.; Bo, W.H.; Qi, W.N.; et al. Temporal dynamics of genetic architecture governing leaf development in Populus. New Phytol. 2024, 242, 1113–1130. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.B.; Wang, J.; Wang, L.; Shang, G.D.; Xu, Z.G.; Mai, Y.X.; Liu, Y.T.; Zhang, T.Q.; Wang, J.W. Anisotropic cell growth at the leaf base promotes age-related changes in leaf shape in Arab. thaliana. Plant Cell 2023, 35, 1386–1407. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Yan, C.J.; Gu, M. Molecular mechanism of leaf development. Hereditas 2008, 30, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
- Arun, S.; Ryan, G.; Heather, E.M.; Martin, B.; Jelmer, L.; Anne, M.E.; Lacey, S.; Tijs, K.; David, W.E.; Staffan, P. Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol. 2013, 162, 675–688. [Google Scholar] [CrossRef]
- Anne, E.; Staffan, P. Cellulose synthases and synthesis in Arabidopsis. Mol. Plant 2011, 4, 199–211. [Google Scholar] [CrossRef]
- Michael, M.; Silvere, P.; Herman, H. Towards understanding the role of membrane-bound Endo-β-1,4-glucanases in cellulose biosynthesis. Plant Cell Physiol. 2002, 43, 1399–1406. [Google Scholar] [CrossRef]
- Junko, T.; Rudsander, U.J.; Mattias, H.; Alicja, B.; Jesper, H.; Nicolas, A.; Peter, I.; Peter, R.; Satoshi, E.; Ibatullin, F.M. KORRIGAN1 and its aspen homolog PttCel9A1 decrease cellulose crystallinity in Arabidopsis stems. Plant Cell Physiol. 2009, 50, 1099–1115. [Google Scholar] [CrossRef]
- Jane, G.; Matt, G.; Pedro, M.C.; Bo, S.; Nobuyuki, N.; Junko, T.; Henrik, A.; Soraya, D.; Emma, M.; Sara, A.; et al. Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 2006, 140, 946–962. [Google Scholar] [CrossRef]
- Brummell, D.A.; Catala, C.; Lashbrook, C.C.; Bennett, A.B. A membrane-anchored E-type endo-1,4-β-glucanase is localized on Golgi and plasma membranes of higherplants. Proc. Natl. Acad. Sci. USA 1997, 94, 4794–4799. [Google Scholar] [CrossRef]
- Shani, Z.; Dekel, M.; Tsabary, G.; Goren, R.; Shoseyov, O. Growth enhancement of transgenic poplar plants by overexpression of Arabidopsis thaliana endo-1,4–β-glucanase (cel1). Mol. Breed. 2004, 14, 321–330. [Google Scholar] [CrossRef]
- Suchita, B.; Takeshi, F.; Shiv, T.; Zhang, D.Y.; Xu, F.Y.; Chandrashekhar, P.J. Xylem-specific and tension stress-responsive coexpression of KORRIGAN endoglucanase and three secondary wall-associated cellulose synthase genes in aspen trees. Planta 2006, 224, 828–837. [Google Scholar] [CrossRef]
- Zhou, H.L.; He, S.J.; Cao, Y.R.; Chen, T.; Du, B.X.; Chu, C.C.; Zhang, J.S. OsGLU1, a putative membrane-bound endo-1,4--D-glucanase from Rice, affects plant internode elongation. Plant Mol. Biol. 2006, 60, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Pio, M.J.S.; Iain, M.; Neil, G.T.; John, G.; Mike, C.J.; Simon, R.T. The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J. 2010, 37, 730–740. [Google Scholar] [CrossRef]
- Liao, F.X.; Wang, D.R. Tropical and Subtropical Ornamental Plant Industry in Guangdong Province, China; Acta Horticulturae: Beijing, China, 2017; pp. 31–36. ISBN 0513-353X. [Google Scholar]
- Duquenne, B.; Eeckhaut, T.; Werbrouck, S.; Huylenbroeck, J.V. Effect of enzyme concentrations on protoplast isolation and protoplast culture of Spathiphyllum and Anthurium. Plant Cell Tissue Organ Cult. 2007, 91, 165–173. [Google Scholar] [CrossRef]
- Henny, R.J.; Chen, D.J.N. Progress in ornamental Aroid breeding research. Ann. Mo. Bot. Gard. October 2004, 91, 464–472. [Google Scholar]
- Ogasawara, R.; Ochiai, M.; Nishikawa, K.; Fukui, H. Induction of Polyploidy by In Vitro Colchicine Treatment of Apical Meristem of Spathiphyllum wallisii Regel ‘New Merry’; Horticultural Research: Tokyo, Japan, 2014; pp. 213–219. ISBN 2052-7276. [Google Scholar]
- Liu, X.F.; Sun, Y.B.; Huang, L.L.; Yu, B. Screening of SSR Primer of Spathiphyllum and Analysis of Genetic Relationship of 12 Germplasm Resources; Molecular Plant Breeding: Haikou, China, 2022; pp. 1–16. ISBN 1672-416X. [Google Scholar]
- Hou, Z.W.; Zhang, H.; Li, X.Y.; Liao, F.X. Mining and developing SSR and SNP molecular markers based on transcroptome sequences of Spathiphyllum. In Advances in Ornamental Horticulture of China, 2020, 1st ed.; Zhang, Q.X., Ed.; China Forestry Publishing House: Beijing, China, 1 November 2020; Volume 1, pp. 357–365. ISBN 9787503894077. [Google Scholar]
- Li, X.Y.; Hou, Z.W.; Zhang, H. A Quantitative Analysis of Leaf Growth and Establishment of Leaf Growth Logistics Model for Spathiphyllum, 4th ed.; Journal of Tropical Biology: Haikou, China, 2020; pp. 461–469. ISBN 1000-2561. [Google Scholar]
- Liu, J.; Chang, W.R.; Pan, L.; Liu, X.; Su, L.; Zhang, W.; Li, Q.; Zheng, Y. An improved method of preparing high efficiency transformation Escherichia coli with both plasmids and larger DNA fragments. Indian J. Microbiol. 2018, 58, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, H.; Jeon, H.; Kim, H.R. Agrobacterium-mediated Capsicum annuum gene editing in two cultivars, hot pepper CM334 and bell pepper Dempsey. Int. J. Mol. Sci. 2021, 22, 3921. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Liu, J.X.; Li, J.Y.; Wang, H.N.; Fu, Z.D.; Liu, J.; Yu, Y. Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments. J. Exp. Bot. 2011, 62, 825–840. [Google Scholar] [CrossRef]
- Jo, H.; Michael, L. Regulation of plant lateral-organ growth by modulating cell number and size. Curr. Opin. Plant Biol. 2014, 17, 36–42. [Google Scholar] [CrossRef]
- Jasmien, V.; Alexandra, B.; Nathalie, G.; Dirk, I. Molecular networks regulating the cell division during leaf growth in Arabidopsis. J. Exp. Bot. 2019, 71, 2365–2378. [Google Scholar] [CrossRef]
- Hadrien, O.; Jan, T.; Christophe, G.; Olivier, A. Regulation of plant cell wall stiffness by mechanical stress: A mesoscale physical model. J. Math. Biol. 2018, 78, 625–653. [Google Scholar] [CrossRef]
- Joanna, K.P.; Joseph, J.K. The regulation of cellulose biosynthesis in plants. Plant Cell 2019, 31, 282–296. [Google Scholar] [CrossRef]
- Luo, L.Q.; Bai, J.F.; Yuan, S.H.; Guo, L.P.; Liu, Z.H.; Guo, H.Y.; Zhang, T.B.; Duan, W.J.; Li, Y.M.; Zhao, C.P.; et al. Genome wide identification and characterization of Wheat GH9 genes reveals their roles in Pollen development and anther dehiscence. Int. J. Mol. Sci. 2022, 23, 6324. [Google Scholar] [CrossRef]
- Galit, T.; Ziv, S.; Levava, R.; Ilan, L.; Joseph, R.; Oded, S. Abnormal ‘wrinkled’ cell walls and retarded development of transgenic Arabidopsis thaliana plants expressing endo-1,4-β-glucanase (cel1) antisense. Plant Mol. Biol. 2003, 51, 213–224. [Google Scholar] [CrossRef]
- Mølhøj, M.; Jørgensen, B.; Ulvskov, P.; Borkhardt, B. Two Arabidopsis thaliana genes, KOR2 and KOR3, which encode membrane-anchored endo-1,4-β-D-glucanases, are differentially expressed in developing leaf trichomes and their support cells. Plant Mol. Biol. 2001, 46, 263–275. [Google Scholar] [CrossRef]
- Aggarwal, D.; Kumar, A.; Reddy, M.S. Genetic transformation of endo-1,4-β-glucanase (Korrigan) for cellulose enhancement in Eucalyptus tereticornis. Plant Cell Tissue Organ Cult. 2015, 122, 363–371. [Google Scholar] [CrossRef]
- Lei, L.; Zhang, T.; Richard, S.; Christopher, M.L.; Martine, G.; Lukas, M.; Samantha, V.; Seong, H.K.; Daniel, J.C.; Li, S.D.; et al. The jiaoyao1 mutant is an allele of korrigan1 that abolishes endoglucanase activity and affects the organization of both cellulose microfibrils and microtubules in Arabidopsis. Plant Cell 2014, 26, 2601–2616. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.L.; Sun, J.Y.; Li, L.G. PtrCel9A6, an Endo-1,4-β-Glucanase, is required for cell wall formation during xylem differentiation in Populus. Mol. Plant 2013, 6, 1904–1917. [Google Scholar] [CrossRef]






| Stage | Number | Primary Growth and Morphological Characteristics |
|---|---|---|
| The curled leaf stage | W1/M1 | Approximately ten days after the leaf buds transition to white, the tips of the leaves gradually change from white to yellow/white, while the base of the leaves remains white. During this period, curled leaves develop, with the leaves being enveloped in leaf sheaths. |
| The early spread stage | W2/M2 | Approximately 10 to 13 days after the formation of curled leaves, the leaves emerge from the leaf sheath and begin to gradually unfold. During this process, the leaf color transitions from yellow/white to a delicate green. |
| The late spread stage | W3/M3 | Approximately ten days after the onset of leaf color change, the leaves transition from light green to a deeper green. During this period, the leaves gradually expand, completing their growth. At this stage, the characteristics of the leaves stabilize and exhibit no further changes. |
| WT | OE of SpGH9A3 | SpGH9A3/WT (%) | |
|---|---|---|---|
| Leaf length (cm) | 1.36 ± 0.05 | 1.02 ± 0.10 * | 75.00% |
| Leaf width (cm) | 0.49 ± 0.03 | 0.48 ± 0.05 | 97.96% |
| Length–width ratio | 2.78 ± 0.19 | 2.14 ± 0.28 * | 76.98% |
| Petiole length (cm) | 0.57 ± 0.03 | 0.69 ± 0.04 * | 121.05% |
| Leaf area (cm2) | 0.42 ± 0.03 | 0.33 ± 0.05 * | 78.57% |
| Silique length (cm) | 1.25 ± 0.06 | 0.86 ± 0.06 * | 68.80% |
| Plant height (cm) | 25.49 ± 1.42 | 17.63 ± 1.00 * | 69.16% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Hu, M.; Wu, R.; Hou, Z.; Zhang, H.; He, W.; Gao, L.; Liao, F. Genetic Evidence of SpGH9A3 in Leaf Morphology Variation of Spathiphyllum ‘Mojo’. Genes 2024, 15, 1132. https://doi.org/10.3390/genes15091132
Yang S, Hu M, Wu R, Hou Z, Zhang H, He W, Gao L, Liao F. Genetic Evidence of SpGH9A3 in Leaf Morphology Variation of Spathiphyllum ‘Mojo’. Genes. 2024; 15(9):1132. https://doi.org/10.3390/genes15091132
Chicago/Turabian StyleYang, Songlin, Minghua Hu, Runxin Wu, Zhiwen Hou, Huan Zhang, Wenying He, Lili Gao, and Feixiong Liao. 2024. "Genetic Evidence of SpGH9A3 in Leaf Morphology Variation of Spathiphyllum ‘Mojo’" Genes 15, no. 9: 1132. https://doi.org/10.3390/genes15091132
APA StyleYang, S., Hu, M., Wu, R., Hou, Z., Zhang, H., He, W., Gao, L., & Liao, F. (2024). Genetic Evidence of SpGH9A3 in Leaf Morphology Variation of Spathiphyllum ‘Mojo’. Genes, 15(9), 1132. https://doi.org/10.3390/genes15091132





