Loss of ZC4H2, an Arthrogryposis Multiplex Congenita Associated Gene, Promotes Osteoclastogenesis in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Experimentation
2.2. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.3. MSCs, BMDM Isolation and In Vitro Differentiation
2.4. RNA-Seq Analysis
2.5. Von Kossa Staining
2.6. Skeletal Staining
2.7. Micro-Computed Tomography (Micro-CT)
2.8. Alkaline Phosphatase (ALP) and Alizarin Red S (ARS) Staining
2.9. Tartrate Resistant Acid Phosphatase (TRAP) Staining Assay
2.10. F-Actin Ring Formation Assay
2.11. Scanning Electron Microscopy
2.12. ELISA Assay of CTX-1
2.13. Cell Counting Kit-8 (CCK-8) Assay
2.14. BrdU Incorporation Assay
2.15. Statistical Analysis
3. Results
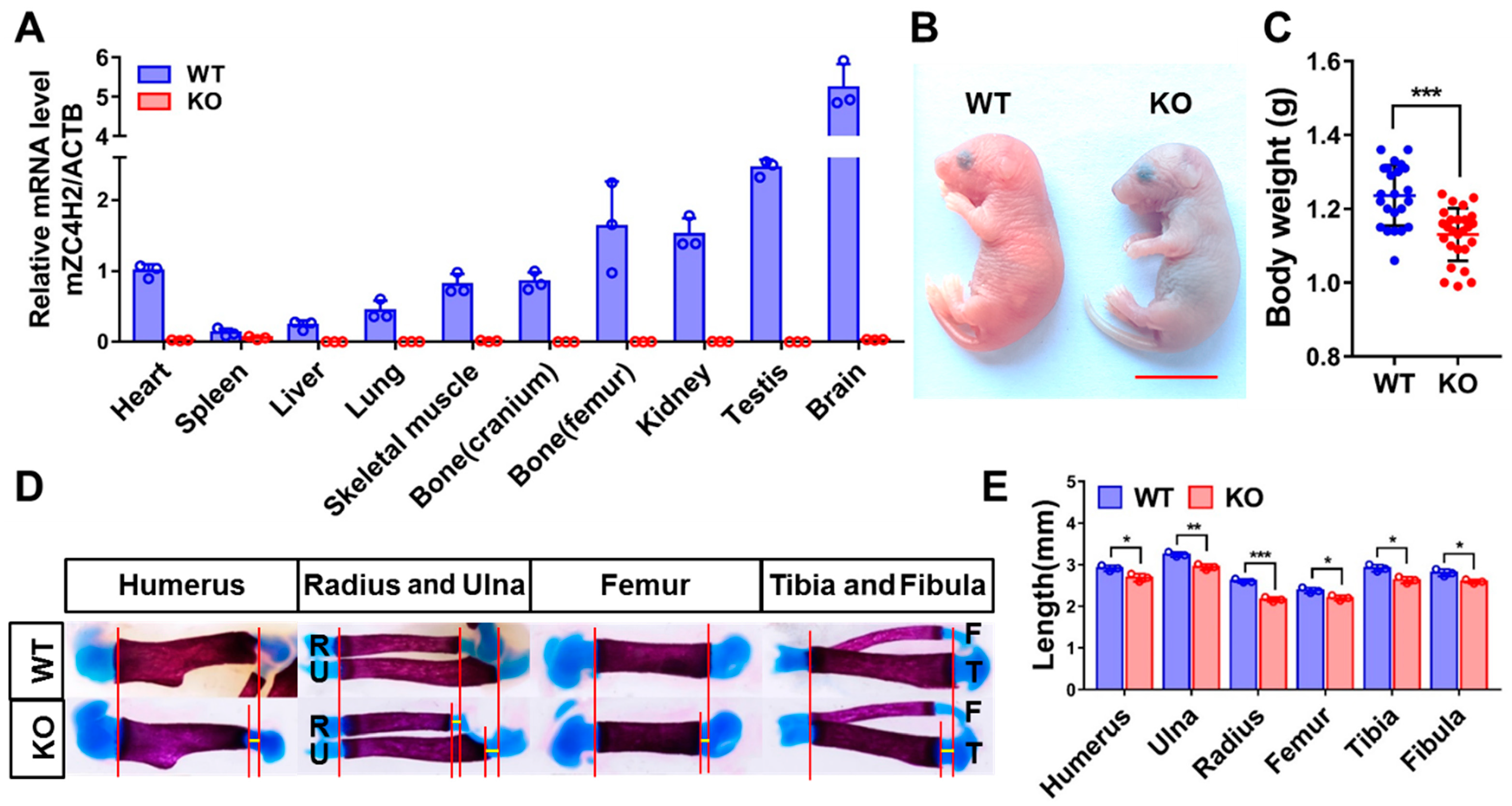
3.1. Loss of ZC4H2 Reduces Long Bone Calcification in Mice
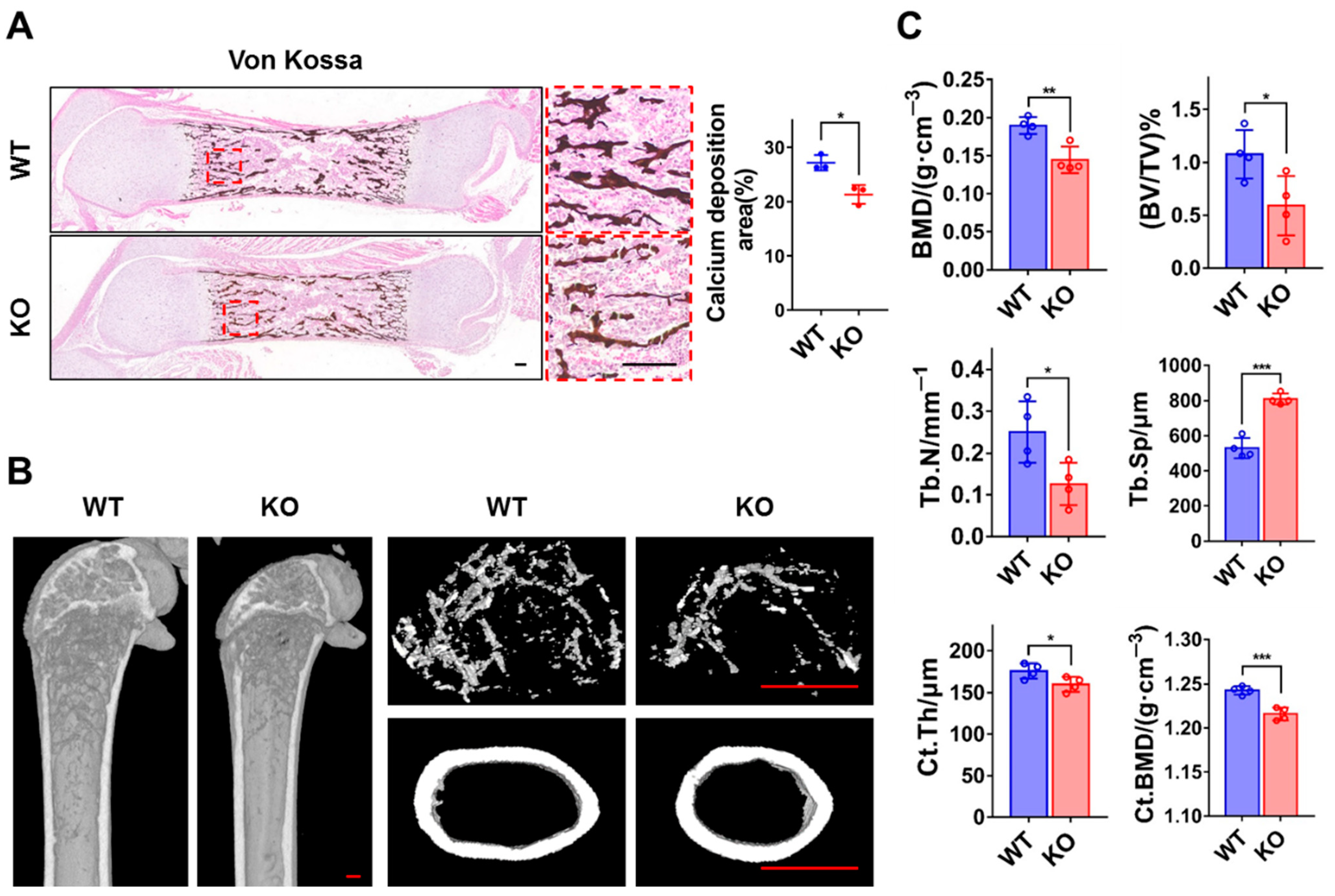
3.2. Loss of ZC4H2 Reduces Femoral Bone Mass
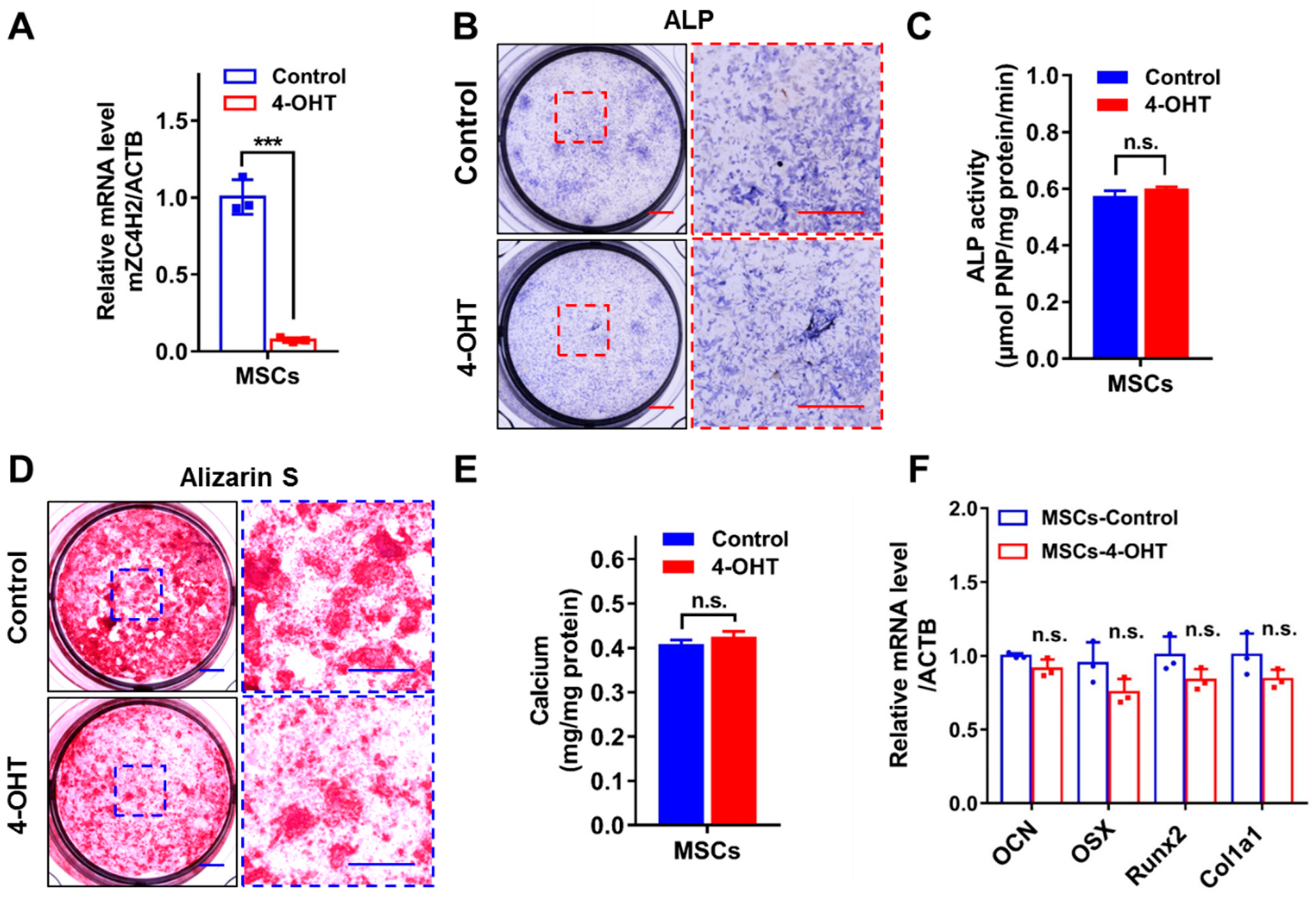
3.3. Loss of ZC4H2 Does Not Affect Osteoblast Differentiation
3.4. Loss of ZC4H2 Promotes Osteoclastogenesis and Bone Resorption
3.5. Loss of ZC4H2 Inhibits Proliferation of MSCs and BMDM
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lombard, Z.; Park, C.; Makova, K.D.; Ramsay, M. A computational approach to candidate gene prioritization for x-linked mental retardation using annotation-based binary filtering and motif-based linear discriminatory analysis. Biol. Direct 2011, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Nanda, I.; van Riesen, A.; McMichael, G.; Hu, H.; Hambrock, M.; Papon, M.A.; Fischer, U.; Marouillat, S.; Ding, C.; et al. Zc4h2 mutations are associated with arthrogryposis multiplex congenita and intellectual disability through impairment of central and peripheral synaptic plasticity. Am. J. Hum. Genet. 2013, 92, 681–695. [Google Scholar] [CrossRef]
- Wieacker, P.; Wolff, G.; Wienker, T.F.; Sauer, M.; Opitz, J.M.; Reynolds, J.F. A new x-linked syndrome with muscle atrophy, congenital contractures, and oculomotor apraxia. Am. J. Med. Genet. 1985, 20, 597–606. [Google Scholar] [CrossRef]
- Wongkittichote, P.; Choi, T.I.; Kim, O.H.; Riley, K.; Koeberl, D.; Narayanan, V.; Ramsey, K.; Balak, C.; Schwartz, C.E.; Cueto-Gonzalez, A.M.; et al. Expanding allelic and phenotypic spectrum of zc4h2-related disorder: A novel hypomorphic variant and high prevalence of tethered cord. Clin. Genet. 2023, 103, 167–178. [Google Scholar] [CrossRef]
- Wakabayashi, T.; Mizukami, M.; Terada, K.; Ishikawa, A.; Hinotsu, S.; Kobayashi, M.; Kato, K.; Ogi, T.; Tsugawa, T.; Sakurai, A. A novel ZC4H2 variant in a female with severe respiratory complications. Brain Dev. 2022, 44, 571–577. [Google Scholar] [CrossRef]
- May, M.; Hwang, K.; Miles, J.; Williams, C.; Niranjan, T.; Kahler, S.; Chiurazzi, P.; Steindl, K.; Van Der Spek, P.; Swagemakers, S.; et al. ZC4H2, an xlid gene, is required for the generation of a specific subset of cns interneurons. Hum. Mol. Genet. 2015, 24, 4848–4861. [Google Scholar] [CrossRef]
- Song, N.; Ma, P.; Zhang, Q.; Zhang, L.; Wang, H.; Zhang, L.; Zhu, L.; He, C.H.; Mao, B.; Ding, Y.Q. Rnf220/zc4h2-mediated monoubiquitylation of phox2 is required for noradrenergic neuron development. Development 2020, 147, dev185199. [Google Scholar] [CrossRef]
- Zhang, L.; Ye, M.; Zhu, L.; Cha, J.; Li, C.; Yao, Y.G.; Mao, B. Loss of zc4h2 and rnf220 inhibits neural stem cell proliferation and promotes neuronal differentiation. Cells 2020, 9, 1600. [Google Scholar] [CrossRef]
- Kim, J.; Choi, T.I.; Park, S.; Kim, M.H.; Kim, C.H.; Lee, S. Rnf220 cooperates with zc4h2 to specify spinal progenitor domains. Development 2018, 145, dev165340. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Ren, B.; Yang, X.; Sun, B.; Liu, X.; Kong, Q.; Li, C.; Mao, B. Zc4h2 stabilizes smads to enhance bmp signalling, which is involved in neural development in. Open Biol. 2017, 7, 170122. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Song, N.N.; Cheng, X.; Zhu, L.; Zhang, Q.; Zhang, L.L.; Yang, X.; Wang, H.; Kong, Q.; Shi, D.; et al. Zc4h2 stabilizes rnf220 to pattern ventral spinal cord through modulating shh/gli signaling. J. Mol. Cell Biol. 2020, 12, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Mao, B. The many faces of the e3 ubiquitin ligase, rnf220, in neural development and beyond. Dev. Growth Differ. 2022, 64, 98–105. [Google Scholar] [CrossRef]
- Li, Y.; Yang, C.; Wang, H.; Zhao, L.; Kong, Q.; Cang, Y.; Zhao, S.; Lv, L.; Li, Y.; Mao, B.; et al. Sequential stabilization of rnf220 by rlim and ZC4H2 during cerebellum development and shh-group medulloblastoma progression. J. Mol. Cell Biol. 2022, 14, mjab082. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, D.; Guo, Z.; Hu, R.; Wang, Q.; Liu, Y.; Liu, M.; Meng, Z.; Yang, H.; Zhang, Y.; et al. A novel de novo nonsense mutation in ZC4H2 causes wieacker-wolff syndrome. Mol. Genet. Genom. Med. 2020, 8, e1100. [Google Scholar] [CrossRef]
- Frints, S.G.M.; Hennig, F.; Colombo, R.; Jacquemont, S.; Terhal, P.; Zimmerman, H.H.; Hunt, D.; Mendelsohn, B.A.; Kordass, U.; Webster, R.; et al. Deleterious de novo variants of x-linked zc4h2 in females cause a variable phenotype with neurogenic arthrogryposis multiplex congenita. Hum. Mutat. 2019, 40, 2270–2285. [Google Scholar] [CrossRef]
- Veis, D.J.; Brien, C.A. Osteoclasts, master sculptors of bone. Annu. Rev. Pathol. Pathol. Mech. Dis. 2023, 18, 257–281. [Google Scholar] [CrossRef]
- Yahara, Y.; Nguyen, T.; Ishikawa, K.; Kamei, K.; Alman, B.A. The origins and roles of osteoclasts in bone development, homeostasis and repair. Development 2022, 149, dev199908. [Google Scholar] [CrossRef]
- Salhotra, A.; Shah, H.N.; Levi, B.; Longaker, M.T. Mechanisms of bone development and repair. Nat. Rev. Mol. Cell Biol. 2020, 21, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Lin, C.; Stavre, Z.; Greenblatt, M.B.; Shim, J.H. Osteoblast-osteoclast communication and bone homeostasis. Cells 2020, 9, 2073. [Google Scholar] [CrossRef]
- Zhen, G.; Dan, Y.; Wang, R.; Dou, C.; Guo, Q.; Zarr, M.; Liu, L.N.; Chen, L.; Deng, R.; Li, Y.; et al. An antibody against siglec-15 promotes bone formation and fracture healing by increasing trap+ mononuclear cells and pdgf-bb secretion. Bone Res. 2021, 9, 47. [Google Scholar] [CrossRef]
- Zhou, S.; Dai, Q.; Huang, X.; Jin, A.; Yang, Y.; Gong, X.; Xu, H.; Gao, X.; Jiang, L. Stat3 is critical for skeletal development and bone homeostasis by regulating osteogenesis. Nat. Commun. 2021, 12, 6891. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Li, P.; Che, M.; Liu, J.; Biswas, S.; Ma, G.; He, L.; Wei, Z.; Zhang, Z.; Yang, Y.; et al. Activation of hedgehog signaling in mesenchymal stem cells induces cartilage and bone tumor formation via wnt/β-catenin. Elife 2019, 8, e50208. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. Soap: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. Rsem: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Abdi, H. The bonferonni and šidák corrections for multiple comparisons. In The Bonferonni and Šidák Corrections for Multiple Comparisons, 1st ed.; Salkind, N.J., Ed.; SAGE Publications, Inc.: Thousand Oaks, CA, USA, 2007; Volume 3, pp. 103–106. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Rauner, M.; Murray, M.; Thiele, S.; Watts, D.; Neumann, D.; Gabet, Y.; Hofbauer, L.; Wielockx, B. Epo/epor signaling in osteoprogenitor cells is essential for bone homeostasis and epo-induced bone loss. Bone Res. 2021, 9, 42. [Google Scholar] [CrossRef]
- Guo, X.; Ma, P.; Li, Y.; Yang, Y.; Wang, C.; Xu, T.; Wang, H.; Li, C.; Mao, B.; Qi, X. Rnf220 mediates k63-linked polyubiquitination of stat1 and promotes host defense. Cell Death Differ. 2020, 28, 640–656. [Google Scholar] [CrossRef]
- Kim, J.; Kim, K.; Kim, I.; Seong, S.; Koh, J.; Kim, N. Nodal negatively regulates osteoclast differentiation by inducing stat1 phosphorylation. J. Cell. Physiol. 2024, 239, e31268. [Google Scholar] [CrossRef] [PubMed]


| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| ACTB | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
| ZC4H2 | AAAGATCAAGGCCCGTTTG | TTGTATTCTTTCAGGTGCCTCTC |
| OSX | ACTCATCCCTATGGCTCGTG | GGTAGGGAGCTGGGTTAAGG |
| OCN | ACTCCGGCGCTACCTTGGAGCC | GCAGGGTTAAGCTCACACTG |
| Runx2 | CCTAGTTAGAGTGGTAGCAGA | ACAGACAACGAAGAAAGTTCC |
| Col1a1 | GCTCCTCTTAGGGGCCACT | CCACGTCTCACCATTGGGG |
| Ctsk | CTCGGCGTTTAATTTGGGAGA | TCGAGAGGGAGGTATTCTGAGT |
| Mmp9 | CTGGACAGCCAGACACTAAAG | CTCGCGGCAAGTCTTCAGAG |
| Acp5 | CACTCCCACCCTGAGATTTGT | CATCGTCTGCACGGTTCTG |
| Dcstamp | GGGGACTTATGTGTTTCCACG | ACAAAGCAACAGACTCCCAAAT |
| Ocstamp | CTGTAACGAACTACTGACCCAG | CCCAGGCTTAGGAAGACGAAG |
| Atp6v0d2 | CAGAGCTGTACTTCAATGTGGA | AGGTCTCACACTGCACTAGGT |
| Oscar | CCTAGCCTCATACCCCCAG | CGTTGATCCCAGGAGTCACAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Zhang, L.; Cha, J.; Li, C.; Mao, B. Loss of ZC4H2, an Arthrogryposis Multiplex Congenita Associated Gene, Promotes Osteoclastogenesis in Mice. Genes 2024, 15, 1134. https://doi.org/10.3390/genes15091134
Zhu L, Zhang L, Cha J, Li C, Mao B. Loss of ZC4H2, an Arthrogryposis Multiplex Congenita Associated Gene, Promotes Osteoclastogenesis in Mice. Genes. 2024; 15(9):1134. https://doi.org/10.3390/genes15091134
Chicago/Turabian StyleZhu, Liang, Longlong Zhang, Jingmei Cha, Chaocui Li, and Bingyu Mao. 2024. "Loss of ZC4H2, an Arthrogryposis Multiplex Congenita Associated Gene, Promotes Osteoclastogenesis in Mice" Genes 15, no. 9: 1134. https://doi.org/10.3390/genes15091134






